Hypomyelinating Leukodystrophy 7 (HLD7)-Associated Mutation of POLR3A Is Related to Defective Oligodendroglial Cell Differentiation, Which Is Ameliorated by Ibuprofen
Abstract
:1. Introduction
2. Material and Methods
2.1. Primary and Secondary Antibodies, Inhibitors, and Plasmid Constructions
2.2. Cell Culture and Differentiation
2.3. Transfection into Cells
2.4. Confocal Microscopic Images
2.5. Polyacrylamide Gel Electrophoresis and Immunoblotting
2.6. Statistical Analysis
2.7. Ethics Statement
3. Results
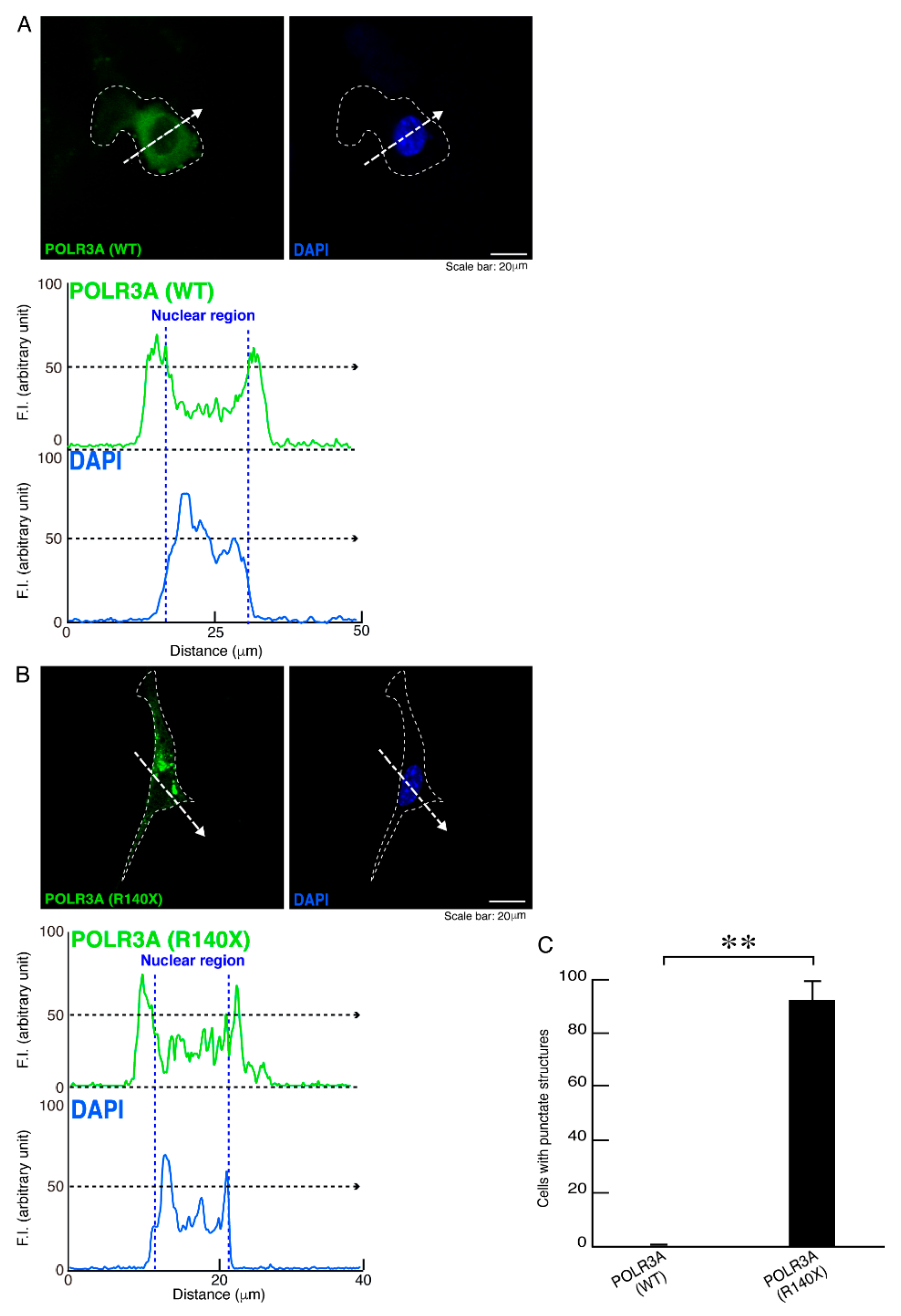
3.1. The R140X Mutant Proteins of POLR3A Are Aggregated and Present in Lysosomes in FBD−102b Cells
3.2. Morphological Differentiation Is Inhibited in Cells Harboring the R140X Mutant Constructs of POLR3A
3.3. Ibuprofen Reverses Phenotypes in Cells Harboring the R140X Mutant Constructs of POLR3A
4. Discussion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Garbern, J.; Cambi, F.; Shy, M.; Kamholz, J. The molecular pathogenesis of Pelizaeus-Merzbacher disease. Arch. Neurol. 1999, 56, 1210–1214. [Google Scholar] [CrossRef]
- Inoue, K. Pelizaeus-Merzbacher disease: Molecular and cellular pathologies and associated phenotypes. Adv. Exp. Med. Biol. 2019, 1190, 201–216. [Google Scholar]
- Wolf, N.I.; Ffrench-Constant, C.; Van der Knaap, M.S. Hypomyelinating leukodystrophies-unravelling myelin biology. Nat. Rev. Neurol. 2021, 17, 88–103. [Google Scholar] [CrossRef]
- Dhaunchak, A.S.; Colman, D.R.; Nave, K.A. Misalignment of PLP/DM20 transmembrane domains determines protein misfolding in Pelizaeus-Merzbacher disease. J. Neurosci. 2011, 31, 14961–14971. [Google Scholar] [CrossRef] [Green Version]
- Simons, M.; Lyons, D.A. Axonal selection and myelin sheath generation in the central nervous system. Curr. Opin. Cell Biol. 2013, 25, 512–519. [Google Scholar] [CrossRef]
- Morton, P.D.; Ishibashi, N.; Jonas, R.A.; Gallo, V. Congenital cardiac anomalies and white matter injury. Trends Neurosci. 2015, 38, 3535–3563. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saab, A.S.; Nave, K.A. Myelin dynamics: Protecting and shaping neuronal functions. Curr. Opin. Neurobiol. 2017, 47, 104–112. [Google Scholar] [CrossRef]
- Abu-Rub, M.; Miller, R.H. Emerging cellular and molecular strategies for enhancing central nervous system (CNS) remyelination. Brain Sci. 2018, 8, E111. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bernard, G.; Chouery, E.; Putorti, M.L.; Tétreault, M.; Takanohashi, A.; Carosso, G.; Clément, I.; Boespflug-Tanguy, O.; Rodriguez, D.; Delague, V.; et al. Mutations of POLR3A encoding a catalytic subunit of RNA polymerase Pol III cause a recessive hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011, 89, 415–423. [Google Scholar] [CrossRef] [Green Version]
- Saitsu, H.; Osaka, H.; Sasaki, M.; Takanashi, J.; Hamada, K.; Yamashita, A.; Shibayama, H.; Shiina, M.; Kondo, Y.; Nishiyama, K.; et al. Mutations in POLR3A and POLR3B encoding RNA Polymerase III subunits cause an autosomal-recessive hypomyelinating leukoencephalopathy. Am. J. Hum. Genet. 2011, 89, 644–651. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tétreault, M.; Choquet, K.; Orcesi, S.; Tonduti, D.; Balottin, U.; Teichmann, M.; Fribourg, S.; Schiffmann, R.; Brais, B.; Vanderver, A.; et al. Recessive mutations in POLR3B, encoding the second largest subunit of Pol III, cause a rare hypomyelinating leukodystrophy. Am. J. Hum. Genet. 2011, 89, 652–655. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Daoud, H.; Tétreault, M.; Gibson, W.; Guerrero, K.; Cohen, A.; Gburek-Augustat, J.; Synofzik, M.; Brais, B.; Stevens, C.A.; Sanchez-Carpintero, R.; et al. Mutations in POLR3A and POLR3B are a major cause of hypomyelinating leukodystrophies with or without dental abnormalities and/or hypogonadotropic hypogonadism. J. Med. Genet. 2013, 50, 194–197. [Google Scholar] [CrossRef] [PubMed]
- Merheb, E.; Cui, M.H.; DuBois, J.C.; Branch, C.A.; Gulinello, M.; Shafit-Zagardo, B.; Moir, R.D.; Willis, I.M. Defective myelination in an RNA polymerase III mutant leukodystrophic mouse. Proc. Natl. Acad. Sci. USA 2021, 118, e2024378118. [Google Scholar] [CrossRef] [PubMed]
- Graham, G.G.; Scott, K.F. Limitations of drug concentrations used in cell culture studies for understanding clinical responses of NSAIDs. Inflammopharmacology 2021, 29, 1261–1278. [Google Scholar] [CrossRef]
- Singh, A.; Tripathi, P.; Singh, S. Neuroinflammatory responses in Parkinson’s disease: Relevance of Ibuprofen in therapeutics. Inflammopharmacology 2021, 29, 51–54. [Google Scholar] [CrossRef]
- Markworth, J.F.; Vella, L.D.; Figueiredo, V.C.; Cameron-Smith, D. Ibuprofen treatment blunts early translational signaling responses in human skeletal muscle following resistance exercise. J. Appl. Physiol. 2014, 117, 20–28. [Google Scholar] [CrossRef] [Green Version]
- Chi, Y.; Ma, Q.; Ding, X.-Q.; Qin, X.; Wang, C.; Zhang, J. Research on protective mechanism of ibuprofen in myocardial ischemia-reperfusion injury in rats through the PI3K/Akt/mTOR signaling pathway. Eur. Rev. Med. Pharmacol. Sci. 2019, 23, 4465–4473. [Google Scholar]
- Matsumoto, N.; Watanabe, N.; Iibe, N.; Tatsumi, Y.; Hattori, K.; Takeuchi, Y.; Oizumi, H.; Ohbuchi, K.; Torii, T.; Miyamoto, Y.; et al. Hypomyelinating leukodystrophy-associated mutation of RARS leads it to the lysosome, inhibiting oligodendroglial morphological differentiation. Biochem. Biophys. Rep. 2019, 20, 100705. [Google Scholar] [CrossRef]
- Matsumoto, N.; Miyamoto, Y.; Hattori, K.; Ito, A.; Harada, H.; Oizumi, H.; Ohbuchi, K.; Mizoguchi, K.; Yamauchi, J. PP1C and PP2A are p70S6K phosphatases whose inhibition ameliorates HLD12-associated inhibition of oligodendroglial cell morphological differentiation. Biomedicines 2020, 8, 89. [Google Scholar] [CrossRef]
- Hattori, K.; Tago, K.; Memezawa, S.; Ochiai, A.; Sawaguchi, S.; Kato, Y.; Sato, T.; Tomizuka, K.; Ooizumi, H.; Ohbuchi, K.; et al. The infantile leukoencephalopathy-associated mutation of C11ORF73/HIKESHI proteins generates de novo interactive activity with Filamin A, Inhibiting oligodendroglial cell morphological differentiation. Medicines 2021, 8, 9. [Google Scholar] [CrossRef]
- Sawaguchi, S.; Goto, M.; Kato, Y.; Tanaka, M.; Tago, K.; Oizumi, H.; Ohbuchi, K.; Mizoguchi, K.; Miyamoto, Y.; Yamauchi, J. Hypomyelinating leukodystrophy 15 (HLD15)-associated mutation of EPRS1 leads to its polymeric aggregation in Rab7-positive vesicle structures, inhibiting oligodendroglial cell morphological differentiation. Polymers 2021, 13, 1074. [Google Scholar] [CrossRef] [PubMed]
- Ochiai, A.; Sawaguchi, S.; Memezawa, S.; Seki, Y.; Morimoto, T.; Oizumi, H.; Ohbuchi, K.; Yamamoto, M.; Mizoguchi, K.; Miyamoto, Y.; et al. Knockdown of golgi stress-responsive caspase-2 ameliorates HLD17-associated AIMP2 mutant-mediated inhibition of oligodendroglial cell morphological differentiation. Neurochem. Res. 2021, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Miyamoto, Y.; Torii, T.; Tanoue, A.; Yamauchi, J. VCAM1 acts in parallel with CD69 and is required for the initiation of oligodendrocyte myelination. Nat. Commun. 2016, 7, 13478. [Google Scholar] [CrossRef] [Green Version]
- Miyamoto, Y.; Torii, T.; Tago, K.; Tanoue, A.; Takashima, S.; Yamauchi, J. BIG1/Arfgef1 and Arf1 regulate the initiation of myelination by Schwann cells in mice. Sci. Adv. 2018, 4, eaar4471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wyant, G.A.; Abu-Remaileh, M.; Wolfson, R.L.; Chen, W.W.; Freinkman, E.; Danai, L.V.; Vander Heiden, M.G.; Sabatini, D.M. mTORC1 activator SLC38A9 is required to efflux essential amino acids from lysosomes and use protein as a nutrient. Cell 2017, 171, 642–654. [Google Scholar] [CrossRef]
- Condon, K.J.; Sabatini, D.M. Nutrient regulation of mTORC1 at a glance. J. Cell Sci. 2019, 132, jcs222. [Google Scholar] [CrossRef]
- Tyler, W.A.; Gangoli, N.; Gokina, P.; Kim, H.A.; Covey, M.; Levison, S.W.; Wood, T.L. Activation of the mammalian target of rapamycin (mTOR) is essential for oligodendrocyte differentiation. J. Neurosci. 2009, 29, 6367–6378. [Google Scholar] [CrossRef] [Green Version]
- Narayanan, S.P.; Flores, A.I.; Wang, F.; Macklin, W.B. Akt signals through the mammalian target of rapamycin pathway to regulate CNS myelination. J. Neurosci. 2009, 29, 6860–6870. [Google Scholar] [CrossRef]
- Bibollet-Bahena, O.; Almazan, G. IGF-1-stimulated protein synthesis in oligodendrocyte progenitors requires PI3K/mTOR/Akt and MEK/ERK pathways. J. Neurochem. 2009, 109, 1440–1451. [Google Scholar] [CrossRef]
- Tsai, V.; Parker, W.E.; Orlova, K.A.; Baybis, M.; Chi, A.W.; Berg, B.D.; Birnbaum, J.F.; Estevez, J.; Okochi, K.; Sarnat, H.B.; et al. Fetal brain mTOR signaling activation in tuberous sclerosis complex. Cereb. Cortex 2014, 24, 315–327. [Google Scholar] [CrossRef] [Green Version]
- Prukop, T.; Epplen, D.B.; Nientiedt, T.; Wichert, S.P.; Fledrich, R.; Stassart, R.M.; Rossner, M.J.; Edgar, J.M.; Werner, H.B.; Nave, K.A.; et al. Progesterone antagonist therapy in a Pelizaeus-Merzbacher mouse model. Am. J. Hum. Genet. 2014, 94, 533–546. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Epplen, D.B.; Prukop, T.; Nientiedt, T.; Albrecht, P.; Arlt, F.A.; Stassart, R.M.; Kassmann, C.M.; Methner, A.; Nave, K.A.; Werner, H.B.; et al. Curcumin therapy in a Plp1 transgenic mouse model of Pelizaeus-Merzbacher disease. Ann. Clin. Transl. Neurol. 2015, 2, 787–796. [Google Scholar] [CrossRef] [PubMed]
- Stumpf, S.K.; Berghoff, S.A.; Trevisiol, A.; Spieth, L.; Düking, T.; Schneider, L.V.; Schlaphoff, L.; Dreha-Kulaczewski, S.; Bley, A.; Burfeind, D.; et al. Ketogenic diet ameliorates axonal defects and promotes myelination in Pelizaeus-Merzbacher disease. Acta Neuropathol. 2019, 138, 147–161. [Google Scholar] [CrossRef] [Green Version]
- Gupta, N.; Henry, R.G.; Kang, S.M.; Strober, J.; Lim, D.A.; Ryan, T.; Perry, R.; Farrell, J.; Ulman, M.; Rajalingam, R.; et al. Long-term safety, immunologic response, and imaging outcomes following neural stem cell transplantation for Pelizaeus-Merzbacher disease. Stem Cell Rep. 2019, 13, 254–261. [Google Scholar] [CrossRef] [Green Version]
- Gruenenfelder, F.I.; McLaughlin, M.; Griffiths, I.R.; Garbern, J.; Thomson, G.; Kuzman, P.; Barrie, J.A.; McCulloch, M.L.; Penderis, J.; Stassart, R.; et al. Neural stem cells restore myelin in a demyelinating model of Pelizaeus-Merzbacher disease. Brain 2020, 143, 1383–1399. [Google Scholar] [CrossRef] [PubMed]
- Nobuta, H.; Yang, N.; Ng, Y.H.; Marro, S.G.; Sabeur, K.; Chavali, M.; Stockley, J.H.; Killilea, D.W.; Walter, P.B.; Zhao, C.; et al. Oligodendrocyte death in Pelizaeus-Merzbacher disease is rescued by iron chelation. Cell Stem Cell 2019, 25, 531–541. [Google Scholar] [CrossRef] [PubMed]
- Galloway, D.A.; Moore, C.S. miRNAs as emerging regulators of oligodendrocyte development and differentiation. Front. Cell Dev. Biol. 2016, 4, 59. [Google Scholar] [CrossRef] [Green Version]
- Gleason, A.M.; Woo, E.G.; McKinney, C.; Sidransky, E. The role of exosomes in lysosomal storage disorders. Biomolecules 2021, 11, 576. [Google Scholar] [CrossRef]














| Product Name | Target or Description | Company | Cat. No. | Lot. No. | Final Conc. for Antibody |
|---|---|---|---|---|---|
| Anti-eIF4EBP1 (phospho T37) antibody | A phospho-peptide corresponding to residues surrounding theronine 37 of eIF4EBP1 | abcam | ab75767 | GR88680-14 | IF, 1/100 |
| Anti-F-actin antibody | Filamentous actin (F-actin) | abcam | ab205 | GR319251-7 | IB, 1/80,000 |
| Anti-GFP mAb | Green fluorescent protein (GFP) | MBL | 598 | “078” | IB, 1/80,000 |
| Anti-Halo Tag pAb | Halo Tag | Promega | G9281 | “0000028445” | IB, 1/20,000 |
| HaloTag Oregon Green Ligand | Halo-tag ligand | Promega | G2802 | “0000391849” | IF, 1/1000 |
| Anti-KDEL mAb | KDEL-containing peptide of the endoplasmic reticulum (ER)-resident glucose-regulated protein (GRP78) | MBL | M181-3 | “004” | IF, 1/500 |
| Anti-PLP1 antibody | Myelin proteolipid protein 1 (PLP1) | Atlas Antibodies | HPA004128 | B115828 | IB, 1/500 |
| Anti-RPS6 (phospho S240 + S244) antibody | Synthetic peptide within human S6 protein to the C-terminus (phospho S240 + S244) | abcam | ab215214 | GR3205097-3 | IF, 1/100 |
| CNPase (D83E10) XP Rabbit mAb | 2’, 3’-cyclic nucleotide 3’-phospho-diesterase (CNPase) | Cell Signaling Technology | #5664 | 1 | IB, 1/500 |
| Anti-LAMP-1 antibody (H4A3) | Lysosomal-associated membrane protein1 (LAMP1) | Santa Cruz Biotechnology | sc-20011 | J0919 | IF, 1/200 |
| Purified Mouse Anti-GM130 antibody | Golgi matrix protein of 130 kDa (GM130) | BD Biosciences | 610823 | 8352796 | IF, 1/500 |
| Anti-Sox-10 antibody (A-2) | SRY-related HMG-box 10 (Sox10) | Santa Cruz Biotechnology | sc-365692 | J0720 | IB, 1/500 |
| Anti-LC3 protein | Microtubule-associated protein 1 light chain 3 (LC3) | MBL | M152-3 | “057” | IF, 1/500 |
| Anti-mTOR antibody (A30) | Mechanistic target of rapamycin (mTOR [kinase ]) | Santa Cruz Biotechnology | sc-517464 | H2621 | IB, 1/100 |
| Alexa Fluor 488 goat anti-mouse IgG (H+L) | Mouse IgG (H+L) conjugated with Alexa Fluor 488 | Thermo Fisher Scientific | A-11001 | 774904 | IF, 1/500 |
| Alexa Fluor 488 goat anti-rabbit IgG (H+L) | Rabbit IgG (H+L) conjugated with Alexa Fluor 488 | Thermo Fisher Scientific | A-11008 | 751094 | IF, 1/500 |
| Alexa Fluor 594 goat anti-mouse IgG (H+L) | Mouse IgG (H+L) conjugated with Alexa Fluor 594 | Thermo Fisher Scientific | A-11005 | 2043369 | IF, 1/500 |
| Alexa Fluor 594 goat anti-rabbit IgG (H+L) | Rabbit IgG (H+L) conjugated with Alexa Fluor 594 | Thermo Fisher Scientific | A-11012 | 2018240 | IF, 1/500 |
| Anti-IgG (H+L chain) (Mouse) pAb-HRP | Mouse IgG F(ab’) conjugated with horseradish peroxidase | MBL | 330 | 366 | IB, 1/5000 |
| Anti-IgG (H+L chain) (Mouse) pAb-HRP | Mouse IgG F(ab’) conjugated with horseradish peroxidase | MBL | 330 | 366 | IB, 1/5000 |
| AH6809 | Prostaglandin receptor antagonist | Med Chem Express | HY-10418 | 27839 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sawaguchi, S.; Tago, K.; Oizumi, H.; Ohbuchi, K.; Yamamoto, M.; Mizoguchi, K.; Miyamoto, Y.; Yamauchi, J. Hypomyelinating Leukodystrophy 7 (HLD7)-Associated Mutation of POLR3A Is Related to Defective Oligodendroglial Cell Differentiation, Which Is Ameliorated by Ibuprofen. Neurol. Int. 2022, 14, 11-33. https://doi.org/10.3390/neurolint14010002
Sawaguchi S, Tago K, Oizumi H, Ohbuchi K, Yamamoto M, Mizoguchi K, Miyamoto Y, Yamauchi J. Hypomyelinating Leukodystrophy 7 (HLD7)-Associated Mutation of POLR3A Is Related to Defective Oligodendroglial Cell Differentiation, Which Is Ameliorated by Ibuprofen. Neurology International. 2022; 14(1):11-33. https://doi.org/10.3390/neurolint14010002
Chicago/Turabian StyleSawaguchi, Sui, Kenji Tago, Hiroaki Oizumi, Katsuya Ohbuchi, Masahiro Yamamoto, Kazushige Mizoguchi, Yuki Miyamoto, and Junji Yamauchi. 2022. "Hypomyelinating Leukodystrophy 7 (HLD7)-Associated Mutation of POLR3A Is Related to Defective Oligodendroglial Cell Differentiation, Which Is Ameliorated by Ibuprofen" Neurology International 14, no. 1: 11-33. https://doi.org/10.3390/neurolint14010002
APA StyleSawaguchi, S., Tago, K., Oizumi, H., Ohbuchi, K., Yamamoto, M., Mizoguchi, K., Miyamoto, Y., & Yamauchi, J. (2022). Hypomyelinating Leukodystrophy 7 (HLD7)-Associated Mutation of POLR3A Is Related to Defective Oligodendroglial Cell Differentiation, Which Is Ameliorated by Ibuprofen. Neurology International, 14(1), 11-33. https://doi.org/10.3390/neurolint14010002








