Pedobacter ghigonii sp. nov., Isolated from the Microbiota of the Planarian Schmidtea mediterranea
Abstract
:1. Introduction
2. Materials and Methods
2.1. Culture of Schmidtea mediterranea
2.2. Isolation and Identification of the Bacteria from Schmidtea mediterranea
2.3. DNA Extraction, Sequencing, Assembly, and Annotation
2.4. Phylogenetic Analysis
2.5. Genomic Comparison
2.6. Phenotypic Characteristics of Strain Marseille-Q2390
2.7. Antibiotic Susceptibility
2.8. Analysis of Cellular Fatty Acids of the Strain Marseille-Q2390
3. Results and Discussions
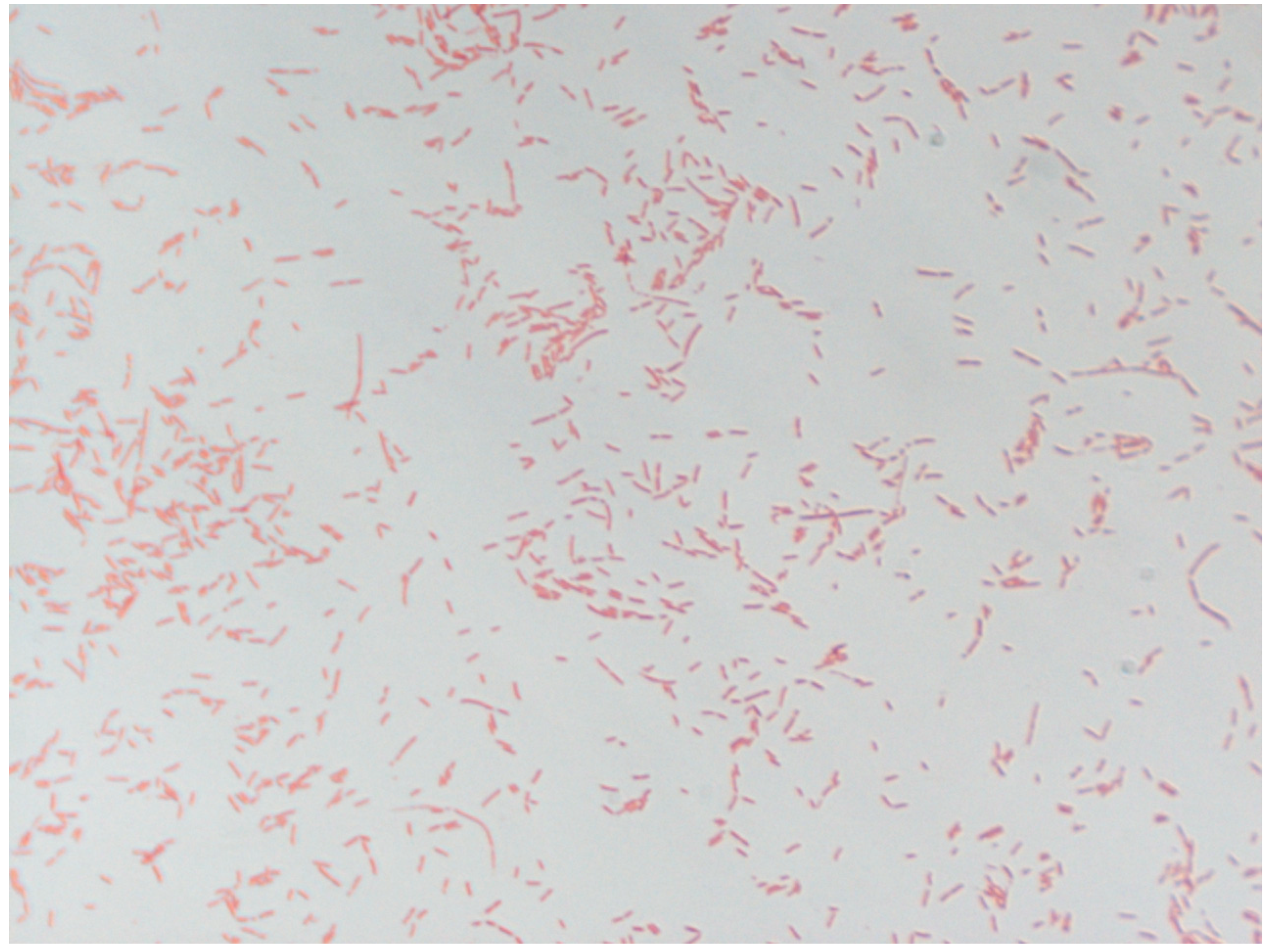
3.1. MALDI-TOF-MS
3.2. Phylogenetic Analysis
3.3. Genomic Comparison
3.4. Phenotypic Characteristics of Strain Marseille-Q2390
3.5. Antibiotic Susceptibility
3.6. Cellular Fatty Acids Analysis
4. Conclusions
4.1. Protologue
4.2. Nucleotide Sequence Accession Numbers
4.3. Deposit in Culture Collections
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Elliott, S.A.; Alvarado, A.S. The history and enduring contributions of planarians to the study of animal regeneration. Wiley Interdiscip. Rev. Dev. Biol. 2012, 2, 301–326. [Google Scholar] [CrossRef]
- Abnave, P.; Mottola, G.; Gimenez, G.; Boucherit, N.; Trouplin, V.; Torre, C.; Conti, F.; Ben Amara, A.; Lepolard, C.; Djian, B.; et al. Screening in Planarians Identifies MORN2 as a Key Component in LC3-Associated Phagocytosis and Resistance to Bacterial Infection. Cell Host Microbe 2014, 16, 338–350. [Google Scholar] [CrossRef] [Green Version]
- Maciel, E.I.; Jiang, C.; Barghouth, P.G.; Nobile, C.J.; Oviedo, N.J. The planarian Schmidtea mediterranea is a new model to study host-pathogen interactions during fungal infections. Dev. Comp. Immunol. 2019, 93, 18–27. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Torre, C.; Ghigo, E. La planaire: Un ver immortel pour élucider la réponse immunitaire de l’homme. Méd. Sci. 2015, 31, 20–22. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arnold, C.P.; Merryman, M.S.; Harris-Arnold, A.; A McKinney, S.; Seidel, C.W.; Loethen, S.; Proctor, K.N.; Guo, L.; Alvarado, A.S. Pathogenic shifts in endogenous microbiota impede tissue regeneration via distinct activation of TAK1/MKK/p38. eLife 2016, 5, e16793. [Google Scholar] [CrossRef] [PubMed]
- Lee, F.J.; Williams, K.B.; Levin, M.; Wolfe, B.E. The Bacterial Metabolite Indole Inhibits Regeneration of the Planarian Flatworm Dugesia japonica. iScience 2018, 10, 135–148. [Google Scholar] [CrossRef] [Green Version]
- Seng, P.; Abat, C.; Rolain, J.M.; Colson, P.; Lagier, J.-C.; Gouriet, F.; Fournier, P.E.; Drancourt, M.; La Scola, B.; Raoult, D. Identification of Rare Pathogenic Bacteria in a Clinical Microbiology Laboratory: Impact of Matrix-Assisted Laser Desorption Ionization-Time of Flight Mass Spectrometry. J. Clin. Microbiol. 2013, 51, 2182–2194. [Google Scholar] [CrossRef] [Green Version]
- Lagier, J.-C.; Armougom, F.; Million, M.; Hugon, P.; Pagnier, I.; Robert, C.; Bittar, F.; Fournous, G.; Gimenez, G.; Maraninchi, M.; et al. Microbial culturomics: Paradigm shift in the human gut microbiome study. Clin. Microbiol. Infect. 2012, 18, 1185–1193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lagier, J.-C.; Hugon, P.; Khelaifia, S.; Fournier, P.-E.; La Scola, B.; Raoult, D. The Rebirth of Culture in Microbiology through the Example of Culturomics To Study Human Gut Microbiota. Clin. Microbiol. Rev. 2015, 28, 237–264. [Google Scholar] [CrossRef] [Green Version]
- Woese, C.R.; Kandler, O.; Wheelis, M.L. Towards a natural system of organisms: Proposal for the domains Archaea, Bacteria, and Eucarya. Proc. Natl. Acad. Sci. USA 1990, 87, 4576–4579. [Google Scholar] [CrossRef] [Green Version]
- La Scola, B.; Raoult, D. Direct Identification of Bacteria in Positive Blood Culture Bottles by Matrix-Assisted Laser Desorption Ionisation Time-of-Flight Mass Spectrometry. PLoS ONE 2009, 4, e8041. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Seng, P.; Drancourt, M.; Gouriet, F.; La Scola, B.; Fournier, P.; Rolain, J.M.; Raoult, D. Ongoing Revolution in Bacteriology: Routine Identification of Bacteria by Matrix-Assisted Laser Desorption Ionization Time-of-Flight Mass Spectrometry. Clin. Infect. Dis. 2009, 49, 543–551. [Google Scholar] [CrossRef]
- Ramasamy, D.; Mishra, A.K.; Lagier, J.-C.; Padhmanabhan, R.; Rossi, M.; Sentausa, E.; Raoult, D.; Fournier, P.-E. A polyphasic strategy incorporating genomic data for the taxonomic description of novel bacterial species. Int. J. Syst. Evol. Microbiol. 2014, 64, 384–391. [Google Scholar] [CrossRef]
- Fournier, P.-E.; Lagier, J.-C.; Dubourg, G.; Raoult, D. From culturomics to taxonomogenomics: A need to change the taxonomy of prokaryotes in clinical microbiology. Anaerobe 2015, 36, 73–78. [Google Scholar] [CrossRef] [PubMed]
- Drancourt, M.; Berger, P.; Raoult, D. Systematic 16S rRNA Gene Sequencing of Atypical Clinical Isolates Identified 27 New Bacterial Species Associated with Humans. J. Clin. Microbiol. 2004, 42, 2197–2202. [Google Scholar] [CrossRef] [Green Version]
- Pei, A.Y.; Oberdorf, W.E.; Nossa, C.W.; Agarwal, A.; Chokshi, P.; Gerz, E.A.; Jin, Z.; Lee, P.; Yang, L.; Poles, M.; et al. Diversity of 16S rRNA Genes within Individual Prokaryotic Genomes. Appl. Environ. Microbiol. 2010, 76, 3886–3897. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ochman, H.; Elwyn, S.; Moran, N.A. Calibrating bacterial evolution. Proc. Natl. Acad. Sci. USA 1999, 96, 12638–12643. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Auch, A.F.; Von Jan, M.; Klenk, H.-P.; Göker, M. Digital DNA-DNA hybridization for microbial species delineation by means of genome-to-genome sequence comparison. Stand. Genom. Sci. 2010, 2, 117–134. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Lee, I.; Kim, Y.O.; Park, S.-C.; Chun, J. OrthoANI: An improved algorithm and software for calculating average nucleotide identity. Int. J. Syst. Evol. Microbiol. 2016, 66, 1100–1103. [Google Scholar] [CrossRef] [PubMed]
- Margesin, R.; Shivaji, S. Pedobacter. In Bergey’s Manual of Systematics of Archaea and Bacteria; American Cancer Society: Atlanta, GA, USA, 2015; pp. 1–17. [Google Scholar]
- Steyn, P.L.; Segers, P.; Vancanneyt, M.; Sandra, P.; Kersters, K.; Joubert, J.J. Classification of heparinolytic bacteria into a new genus, Pedobacter, comprising four species: Pedobacter heparinus comb. nov., Pedobacter piscium comb. nov., Pedobacter africanus sp. nov. and Pedobacter saltans sp. nov. Proposal of the family Sphingobacteriaceae fam. nov. Int. J. Syst. Bacteriol. 1998, 48, 165–177. [Google Scholar] [CrossRef] [PubMed]
- Viana, A.T.; Caetano, T.; Covas, C.; Santos, T.; Mendo, S. Environmental superbugs: The case study of Pedobacter spp. Environ. Pollut. 2018, 241, 1048–1055. [Google Scholar] [CrossRef]
- Kangale, L.J.; Raoult, D.; Ghigo, E.; Fournier, P.-E. Pedobacter schmidteae sp. nov., a new bacterium isolated from the microbiota of the planarian Schmidtea mediterranea. Sci. Rep. 2020, 10, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Ravi, R.K.; Walton, K.; Khosroheidari, M. MiSeq: A Next Generation Sequencing Platform for Genomic Analysis. Methods Mol. Biol. 2018, 1706, 223–232. [Google Scholar] [CrossRef] [PubMed]
- Wingett, S.W.; Andrews, S. FastQ Screen: A tool for multi-genome mapping and quality control. F1000Research 2018, 7, 1338. [Google Scholar] [CrossRef] [PubMed]
- Bolger, A.M.; Lohse, M.; Usadel, B. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bankevich, A.; Nurk, S.; Antipov, D.; Gurevich, A.A.; Dvorkin, M.; Kulikov, A.S.; Lesin, V.M.; Nikolenko, S.I.; Pham, S.; Prjibelski, A.D.; et al. SPAdes: A New Genome Assembly Algorithm and Its Applications to Single-Cell Sequencing. J. Comput. Biol. 2012, 19, 455–477. [Google Scholar] [CrossRef] [Green Version]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef]
- Meier-Kolthoff, J.P.; Göker, M.; Spröer, C.; Klenk, H.-P. When should a DDH experiment be mandatory in microbial taxonomy? Arch. Microbiol. 2013, 195, 413–418. [Google Scholar] [CrossRef]
- Kumar, S.; Stecher, G.; Li, M.; Knyaz, C.; Tamura, K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018, 35, 1547–1549. [Google Scholar] [CrossRef] [PubMed]
- Tamura, K.; Nei, M. Estimation of the number of nucleotide substitutions in the control region of mitochondrial DNA in humans and chimpanzees. Mol. Biol. Evol. 1993, 10, 512–526. [Google Scholar] [CrossRef] [PubMed]
- Tatusov, R.L.; Galperin, M.Y.; Natale, D.A.; Koonin, E.V. The COG database: A tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2000, 28, 33–36. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Unaogu, I.C.; Gugnani, H.C.; Boiron, P. The enzymatic profile of some pathogenic aerobic actinomycetes as determined by api-zym method. J. Med. Mycol. 1999, 9, 235. [Google Scholar]
- Gruner, E.; Von Graevenitz, A.; Altwegg, M. The API ZYM system: A tabulated review from 1977 to date. J. Microbiol. Methods 1992, 16, 101–118. [Google Scholar] [CrossRef]
- Humble, M.W.; King, A.; Phillips, I. API ZYM: A simple rapid system for the detection of bacterial enzymes. J. Clin. Pathol. 1977, 30, 275–277. [Google Scholar] [CrossRef] [Green Version]
- Søgaard, P.; Gahrn-Hansen, B.; Zhou, H.P.; Frederiksen, W.; Hui-Ping, Z. An Investigation of Three Commercial Methods for Rapid Identification of Non-Enteric Gram-Negative Rods. Acta Pathol. Microbiol. Scand. Ser. B Microbiol. 2009, 94, 357–363. [Google Scholar] [CrossRef]
- Bilkey, M.K.; Bremner, D.A.; Cameron, G.L.; Garner, J.G. Comparison of five commercial methods for the identification of non- fermentative and oxidase positive fermentative gram negative Bacilli. N. Z. J. Med. Lab. Technol. 1988, 42, 8–12. [Google Scholar]
- Swanson, E.C.; Collins, M.T. Use of the API 20E system to identify veterinary Enterobacteriaceae. J. Clin. Microbiol. 1980, 12, 10–14. [Google Scholar] [CrossRef] [Green Version]
- Smith, P.B.; Tomfohrde, K.M.; Rhoden, D.L.; Balows, A. API System: A multitube micromethod for identification of Enterobacteriaceae. Appl. Microbiol. 1972, 24, 449–452. [Google Scholar] [CrossRef]
- Véron, M.; Le Minor, L. Nutrition and taxonomy of “enterobacteriaceae” and related bacteria. III. Nutritional characters and differentiation of the taxonomic groups (author’s transl). Ann. Microbiol. 1975, 126, 125–147. [Google Scholar]
- Bergey, D.H.; Krieg, N.R.; Holt, J.G. Bergey’s Manual of Systematic Bacteriology; Williams and Wilkins: Baltimore, MD, USA, 1984. [Google Scholar]
- Rogosa, M.; Sharpe, M.E. An approach to the classification of the Lactobacilli. J. Appl. Bacteriol. 1960, 22, 329–340. [Google Scholar]
- Sharpe, M.E.; Hill, L.R.; LaPage, S.P. Pathogenic Lactobacilli. J. Med. Microbiol. 1973, 6, 281–286. [Google Scholar] [CrossRef] [Green Version]
- Jorgensen, J.H.; Turnidge, J.D. Susceptibility Test Methods: Dilution and Disk Diffusion Methods. 15.6 Pack. Shipp. Infect. Subst. 2015, 1253–1273. [Google Scholar] [CrossRef]
- Sasser, M. Identification of Bacteria by Gas Chromatography of Cellular Fatty Acids; MIDI Technical note #101: Newark, DE, USA, 1990; revised 2001. [Google Scholar]
- Dione, N.; Sankar, S.; Lagier, J.-C.; Khelaifia, S.; Michele, C.; Armstrong, N.; Richez, M.; Abrahão, J.; Raoult, D.; Fournier, P.-E. Genome sequence and description of Anaerosalibacter massiliensis sp. nov. New Microb. New Infect. 2016, 10, 66–76. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yang, J.-E.; Shin, J.-Y.; Park, S.-Y.; Mavlonov, G.T.; Yi, E.-J.; Lee, E.-H.; Lee, J.M.; Yi, T.-H. Pedobacter kyungheensis sp. nov., with ginsenoside converting activity. J. Gen. Appl. Microbiol. 2012, 58, 309–316. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hwang, C.Y.; Choi, D.H.; Cho, B.C. Pedobacter roseus sp. nov., isolated from a hypertrophic pond, and emended description of the genus Pedobacter. Int. J. Syst. Evol. Microbiol. 2006, 56, 1831–1836. [Google Scholar] [CrossRef]
- Kwon, S.-W.; Son, J.-A.; Kim, S.-J.; Kim, Y.-S.; Park, I.-C.; Bok, J.-I.; Weon, H.-Y. Pedobacter rhizosphaerae sp. nov. and Pedobacter soli sp. nov., isolated from rhizosphere soil of Chinese cabbage (Brassica campestris). Int. J. Syst. Evol. Microbiol. 2011, 61, 2874–2879. [Google Scholar] [CrossRef]
- Gordon, N.S.; Valenzuela, A.; Adams, S.M.; Ramsey, P.W.; Pollock, J.L.; Holben, W.E.; Gannon, J.E. Pedobacter nyackensis sp. nov., Pedobacter alluvionis sp. nov. and Pedobacter borealis sp. nov., isolated from Montana flood-plain sediment and forest soil. Int. J. Syst. Evol. Microbiol. 2009, 59, 1720–1726. [Google Scholar] [CrossRef]
- Chen, Z.; Tian, W.; Sun, F.; Chen, Y.; Han, H.; Yao, L.; Zhang, Z. Pedobacter miscanthi sp. nov., isolated from Miscanthus sinensis. Int. J. Syst. Evol. Microbiol. 2019, 69, 3344–3349. [Google Scholar] [CrossRef] [PubMed]
- Yang, J.-E.; Son, H.-M.; Lee, J.M.; Shin, H.-S.; Park, S.-Y.; Lee, D.-G.; Kook, M.; Yi, T.-H. Pedobacter ginsenosidimutans sp. nov., with ginsenoside-converting activity. Int. J. Syst. Evol. Microbiol. 2013, 63, 4396–4401. [Google Scholar] [CrossRef] [Green Version]
- Kwon, S.-W.; Kim, B.-Y.; Lee, K.-H.; Jang, K.-Y.; Seok, S.-J.; Kwon, J.-S.; Kim, W.-G.; Weon, H.-Y. Pedobacter suwonensis sp. nov., isolated from the rhizosphere of Chinese cabbage (Brassica campestris). Int. J. Syst. Evol. Microbiol. 2007, 57, 480–484. [Google Scholar] [CrossRef]
- Kook, M.; Park, Y.; Yi, T.-H. Pedobacter jejuensis sp. nov., isolated from soil of a pine grove, and emended description of the genus Pedobacter. Int. J. Syst. Evol. Microbiol. 2014, 64, 1789–1794. [Google Scholar] [CrossRef] [PubMed]
- Chaudhary, D.K.; Lee, S.D.; Kim, J. Pedobacter kyonggii sp. nov., a psychrotolerant bacterium isolated from forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 5120–5127. [Google Scholar] [CrossRef]
- Kämpfer, P.; Irgang, R.; Fernández-Negrete, G.; Busse, H.-J.; Poblete-Morales, M.; Fuentes-Messina, D.; Glaeser, S.P.; Avendaño-Herrera, R. Proposal of Pedobacter nototheniae sp. nov., isolated from the spleen of a black rock cod (Notothenia coriiceps, Richardson 1844) from the Chilean Antarctica. Antonie Leeuwenhoek 2019, 112, 1465–1475. [Google Scholar] [CrossRef] [PubMed]
- Gao, J.-L.; Sun, P.; Mao, X.-J.; Du, Y.-L.; Liu, B.-Y.; Sun, J.-G. Pedobacter zeae sp. nov., an endophytic bacterium isolated from maize root. Int. J. Syst. Evol. Microbiol. 2017, 67, 231–236. [Google Scholar] [CrossRef]
- Roh, S.W.; Quan, Z.-X.; Nam, Y.-D.; Chang, H.-W.; Kim, K.-H.; Kim, M.-K.; Im, W.-T.; Jin, L.; Kim, S.-H.; Lee, S.-T.; et al. Pedobacter agri sp. nov., from soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 1640–1643. [Google Scholar] [CrossRef] [Green Version]
- Yoon, J.-H.; Kang, S.-J.; Oh, T.-K. Pedobacter terrae sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2007, 57, 2462–2466. [Google Scholar] [CrossRef]
- Jung, J.; Park, W. Pedobacter jeongneungensis sp. nov., isolated from forest soil. J. Microbiol. 2012, 50, 660–664. [Google Scholar] [CrossRef]
- Wang, Z.; Tan, Y.; Xu, D.; Wang, G.; Yuan, J.; Zheng, S. Pedobacter vanadiisoli sp. nov., isolated from soil of a vanadium mine. Int. J. Syst. Evol. Microbiol. 2016, 66, 5112–5117. [Google Scholar] [CrossRef]
- Dahal, R.H.; Kim, J. Pedobacter humicola sp. nov., a member of the genus Pedobacter isolated from soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2205–2211. [Google Scholar] [CrossRef] [PubMed]
- Švec, P.; Králová, S.; Busse, H.-J.; Kleinhagauer, T.; Pantůček, R.; Mašlaňová, I.; Cnockaert, M.; Vandamme, P.; Staňková, E.; Gelbíčová, T.; et al. Pedobacter jamesrossensis sp. nov., Pedobacter lithocola sp. nov., Pedobacter mendelii sp. nov. and Pedobacter petrophilus sp. nov., isolated from the Antarctic environment. Int. J. Syst. Evol. Microbiol. 2017, 67, 1499–1507. [Google Scholar] [CrossRef]
- Yoon, J.-H.; Lee, M.-H.; Kang, S.-J.; Park, S.-Y.; Oh, T.-K. Pedobacter sandarakinus sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2006, 56, 1273–1277. [Google Scholar] [CrossRef] [Green Version]
- Hoang, V.-A.; Kim, Y.-J.; Nguyen, N.-L.; Min, J.-W.; Yang, D.-C. Pedobacter ginsengiterrae sp. nov., isolated from soil of a ginseng field. Int. J. Syst. Evol. Microbiol. 2013, 63, 1273–1279. [Google Scholar] [CrossRef]
- He, R.-H.; Liu, Z.-W.; Yu, Y.; Li, H.-R.; Du, Z.-J. Pedobacter changchengzhani sp. nov., isolated from soil of Antarctica. Antonie Leeuwenhoek 2019, 112, 1747–1754. [Google Scholar] [CrossRef]
- Ngo, H.T.T.; Son, H.-M.; Park, S.-Y.; Kim, K.-Y.; Yi, T.-H. Pedobacter seoulensis sp. nov., isolated from soil of a bamboo field. Antonie Leeuwenhoek 2014, 105, 961–970. [Google Scholar] [CrossRef]
- Corsaro, D.; Wylezich, C.; Walochnik, J.; Venditti, D.; Michel, R. Molecular identification of bacterial endosymbionts of Sappinia strains. Parasitol. Res. 2016, 116, 549–558. [Google Scholar] [CrossRef]
- Krieg, N.R.; Ludwig, W.; Euzéby, J.P.; Whitman, W.B. Bacteroidetes phyl. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–2. [Google Scholar]
- Kämpfer, P. Sphingobacteriia class. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; p. 1. [Google Scholar]
- Kämpfer, P. Sphingobacteriales ord. nov. In Bergey’s Manual of Systematics of Archaea and Bacteria; Wiley: Hoboken, NJ, USA, 2015; pp. 1–11. [Google Scholar] [CrossRef]




| Marseille-Q2390 | |||
|---|---|---|---|
| Names | Cover | Identification Percentage | Accession |
| Pedobacter kyungheensis strain THG-T17T | 82% | 99.36% | NR_132668.1 |
| Pedobacter roseus strain CL-GP80T | 89% | 98.68% | NR_043555.1 |
| Pedobacter soli strain 15-51T | 97% | 98.59% | NR_115008.1 |
| Pedobacter borealis strain G-1T | 92% | 98.23% | NR_044381.1 |
| Pedobacter alluvionis strain NWER-II11T | 92% | 98.21% | NR_044382.1 |
| Pedobacter miscanthi strain RS10T | 95% | 97.99% | NR_164958.1 |
| Pedobacter ginsenosidimutans strain THG-45T | 95% | 97.93% | NR_108685.1 |
| Pedobacter suwonensis strain 15-52T | 94% | 97.78% | NR_043543.1 |
| Pedobacter jejuensis strain THG-DR3T | 93% | 97.68% | NR_133810.1 |
| Pedobacter kyonggii strain K-4-11-1T | 95% | 97.65% | NR_159165.1 |
| Pedobacter nototheniae strain 36B243T | 91% | 97.62% | NR_164976.1 |
| Pedobacter psychrotolerant strain V5RDT | 89% | 97.57% | NR_152669.1 |
| Pedobacter zeae strain 22T | 97% | 97.57% | NR_156064.1 |
| Pedobacter agri PB92T | 95% | 97.45% | NR_044339.1 |
| Pedobacter terrae strain DS-57T | 97% | 97.43% | NR_044005.1 |
| Pedobacter rhizosphaerae strain 01-96T | 97% | 97.31% | NR_122096.1 |
| Pedobacter jeongneungensis strain BH45T | 95% | 97.30% | NR_132685.1 |
| Pedobacter vanadiisoli strain XNV015T | 95% | 97.30% | NR_153693.1 |
| Pedobacter humicola strain R135T | 95% | 97.03% | NR_149278.1 |
| Pedobacter lithocola strain CCM 8691T | 97% | 96.90% | NR_156883.1 |
| Pedobacter sandarakinus strain DS-27T | 97% | 96.82% | NR_043665.1 |
| Pedobacter jamesrossensis strain CCM 8689T | 97% | 96.68% | NR_156882.1 |
| Pedobacter petrophilus strain CCM 8687T | 97% | 96.61% | NR_156885.1 |
| Pedobacter ginsengiterrae strain DCY49T | 91% | 96.55% | NR_109023.1 |
| Pedobacter heparinus strain DSM 2366T | 99% | 96.49% | NR_074519.1 |
| Pedobacter changchengzhani strain E01020T | 99% | 96.09% | NR_164993.1 |
| Pedobacter seoulensis strain THG-G12T | 92% | 96.08% | NR_145561.1 |
| Pedobacter schmidteae EGT | 100% | 96.12% | LS453293.1 |
| Property | Term |
|---|---|
| Current classification | Domain: Bacteria [10] |
| Phylum: Bacteroidetes [69] | |
| Class: Sphingobacteriia [70] | |
| Order: Sphingobacteriales [71] | |
| Family: Sphingobacteriaceae [21] | |
| Genus name: Pedobacter [20] | |
| Species name: ghigonii | |
| Specific epithet: Pedobacter ghigonii | |
| Type strain: Marseille-Q2390 | |
| Species status | sp. nov. |
| Gram stain | Negative |
| Cell shape | Rod-shaped |
| Motility | Motile |
| Sporulation | Non-spore-forming |
| Temperature range for growth | 4–30 |
| Temperature optimum | 28 |
| pH range for growth | 5.5–10 |
| pH optimum | 7.5 |
| pH category | Neutro-alkalophilic |
| Lowest NaCl concentration for growth | 0 |
| Highest NaCl concentration for growth | 20 g/L |
| Salinity optimum | 9 g/L |
| O2 conditions for strain testing | Aerobiosis |
| Catalase | Positive |
| Oxydase | Positive |
| Habitat | Gut microbiota of Schmidtea mediterranea |
| Biotic relationship | Symbiotic |
| Name | Contigs | Size (bp) | CDSs | GC% | tRNAs | rRNAs | Refseq |
|---|---|---|---|---|---|---|---|
| P. soli | 38 | 6,006,420 | 4,923 | 40.5 | 49 | 6 | NZ_FMZH00000000.1 |
| P. ghigonii | 41 | 5,921,534 | 4,870 | 40.3 | 49 | 7 | CAESCM000000000.1 |
| P. kyungheensis | 67 | 6,358,642 | 5,270 | 40.5 | 52 | 6 | NZ_JSYN00000000.1 |
| P. borealis | 216 | 5,544,917 | 4,610 | 38.4 | 50 | 3 | NZ_JAUG00000000.1 |
| P. zeae | 15 | 5,444,802 | 4,567 | 40.3 | 49 | 3 | NZ_JACIEF000000000.1 |
| P. alluvionis | 20 | 6,037,645 | 5,006 | 38.4 | 46 | 4 | NZ_RCCK00000000.1 |
| P. kyonggii | 73 | 6,186,183 | 5,107 | 38.8 | 50 | 8 | NZ_SIXF00000000.1 |
| P. ginsenosidimutans | 86 | 6,517,553 | 5,301 | 38.7 | 52 | 5 | NZ_LMZQ00000000.1 |
| P. suwonensis | 40 | 5,803.831 | 4,738 | 39.5 | 47 | 3 | NZ_FOJM00000000.1 |
| P. terrae | 63 | 5,755,101 | 4,.783 | 38.8 | 46 | 3 | NZ_FNCH00000000.1 |
| Digital DNA-DNA Hybridization | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 |
|---|---|---|---|---|---|---|---|---|---|
| P. borealis | 23.90 | ||||||||
| P. ginsenosidimutans | 23.30 | 45.70 | |||||||
| P. zeae | 24.50 | 26.10 | 25.70 | ||||||
| P. kyonggii | 23.60 | 45.80 | 44.40 | 25.50 | |||||
| P. ghigonii | 30.50 | 24.50 | 23.80 | 24.00 | 23.80 | ||||
| P. alluvionis | 23.50 | 31.70 | 31.10 | 26.30 | 31.10 | 23.90 | |||
| P. soli | 56.40 | 23.70 | 23.50 | 24.40 | 23.50 | 30.50 | 23.40 | ||
| P. suwonensis | 24.20 | 26.70 | 26.30 | 25.20 | 26.00 | 23.60 | 27.50 | 23.90 | |
| P. terrae | 23.60 | 28.40 | 27.60 | 25.30 | 27.50 | 23.60 | 30.30 | 23.70 | 31.70 |
| Characteristics | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|
| Gram-staining | - | - | - | - | - | - | - | - | - | - |
| Sporulation | - | - | - | - | - | - | - | - | - | - |
| Growth temperature range (°C) | 4–30 | 4–35 | 4–30 | 4–40 | 4–35 | 4–30 | 1–37 | 0–32 | 15–30 | 4–30 |
| Aerobic growth | + | + | + | + | + | + | + | + | + | + |
| Source | Planarian | Rhizosphere | Soil | Soil | Soil | Soil | Rhizosphere | Soil | Maize root | Floodplain |
| Colony color | White | Pinkish yellow | Reddish pink | Pink | Orange | Pink | Pinkish yellow | Light salmon | Pinkish yellow | Reddish pink |
| Catalase | + | + | + | + | + | + | + | + | + | + |
| Oxidase | + | + | + | + | + | + | + | + | + | + |
| Enzyme activity (API ZYM): | ||||||||||
| Alkaline phosphatase | + | + | + | + | + | + | + | + | + | + |
| Esterase (C4) | + | + | + | - | + | + | + | + | + | + |
| Esterase lipase (C8) | + | + | + | + | + | + | + | + | + | + |
| Lipase (C14) | - | - | - | - | - | - | - | - | - | - |
| Leucine arylamidase | + | + | + | + | + | + | + | + | + | + |
| Valine arylamidase | + | + | + | + | + | + | + | + | + | + |
| Cystine arylamidase | + | + | + | - | - | - | - | + | + | + |
| Trypsin | + | + | + | - | - | + | + | + | + | + |
| α-chymotrypsin | - | - | - | - | - | + | - | NA | - | - |
| Acid phosphatase | + | + | + | + | + | + | + | + | + | + |
| Naphtol-AS-BI-P.hydrolase | + | + | + | + | + | + | + | - | + | + |
| α-galactosidase | + | + | + | + | - | + | - | + | + | + |
| β-galactosidase | + | + | + | + | + | + | + | - | + | + |
| β-glucuronidase | - | - | + | - | - | - | - | - | + | - |
| α-glucosidase | + | + | + | - | + | + | + | + | + | + |
| β-glucosidase | + | + | v | + | + | + | + | + | + | + |
| N-acetyl-β-glucosaminidase | + | + | + | + | + | + | + | + | + | + |
| α-mannosidase | - | + | - | + | - | + | - | + | + | - |
| α-fucosidase | + | + | + | + | - | + | + | + | + | + |
| Assimilation of (API 50 CH/B): | ||||||||||
| Glycerol | - | - | - | NA | - | - | - | - | - | - |
| Erythritol | - | - | - | NA | - | - | - | - | - | - |
| D-Arabinose | - | - | - | NA | - | - | NA | + | + | |
| L-Arabinose | - | + | + | + | + | + | + | + | + | + |
| D-Ribose | - | - | - | - | - | - | - | - | + | + |
| D-Xylose | - | + | + | NA | + | + | NA | + | - | |
| L-Xylose | - | - | - | NA | - | - | - | - | - | - |
| D-Adonitol | - | - | - | NA | - | - | - | - | - | + |
| Methyl-βD-Xylopyranoside | - | - | - | NA | + | NA | NA | NA | - | + |
| D-Galactose | - | + | + | + | NA | + | - | + | + | |
| D-Glucose | + | + | + | + | + | + | + | + | + | + |
| D-Fructose | - | + | + | NA | + | NA | + | NA | + | + |
| D-Mannose | + | + | + | + | + | + | + | + | + | + |
| L-Sorbose | - | - | - | NA | - | - | - | NA | - | + |
| L-Rhamnose | - | + | + | + | - | + | + | + | - | + |
| Dulcitol | - | - | - | - | - | - | - | NA | - | - |
| Inositol | - | - | - | - | - | - | - | - | - | - |
| D-Mannitol | - | - | - | - | - | - | - | - | - | - |
| D-Sorbitol | - | - | - | - | - | - | - | - | - | - |
| Methyl-αD-Mannopyranoside | - | + | + | NA | + | NA | + | - | + | + |
| Methyl-αD-Glucopyranoside | - | + | + | NA | + | NA | + | NA | + | + |
| N-Acetylglucosamine | - | + | + | + | + | + | + | + | + | + |
| Amygdalin | - | + | + | NA | + | NA | + | - | + | + |
| Arbutin | - | + | + | NA | + | NA | + | NA | + | + |
| Esculin ferric citrate | + | + | + | NA | + | + | + | + | + | + |
| Salicin | - | + | + | + | + | + | + | + | + | + |
| D-Cellobiose | - | + | + | + | + | NA | + | NA | + | + |
| D-Maltose | + | + | + | + | + | + | + | + | + | + |
| D-Lactose | - | + | + | + | + | + | + | + | + | + |
| D-Melibiose | - | + | + | + | + | + | + | + | + | + |
| D-Saccharose | - | + | + | + | + | + | + | + | + | + |
| D-Trehalose | - | + | + | NA | + | NA | + | NA | + | + |
| Inulin | - | - | - | NA | + | NA | - | NA | - | + |
| D-Melezitose | - | + | - | NA | - | NA | + | NA | - | - |
| D-Raffinose | - | + | + | NA | + | NA | + | NA | + | + |
| Starch | - | + | + | - | + | + | + | NA | - | + |
| Glycogen | - | + | - | + | - | - | + | - | + | - |
| Xylitol | - | - | - | NA | - | NA | - | NA | - | - |
| Gentiobiose | - | + | + | NA | + | NA | + | NA | - | + |
| D-Turanose | - | + | + | NA | + | NA | + | NA | + | + |
| D-lyxose | - | - | - | NA | - | NA | - | - | + | - |
| D-Tagatose | - | - | - | NA | - | NA | - | - | + | - |
| D-Fucose | - | - | - | - | - | NA | - | NA | - | - |
| L-Fucose | - | - | - | - | - | - | - | - | + | - |
| D-Arabitol | - | - | - | NA | - | NA | - | NA | - | - |
| L-Arabitol | - | - | - | - | - | - | - | - | ||
| Potassium Gluconate | - | - | - | - | - | - | - | - | - | - |
| Potassium 2-ketoGluconate | - | - | - | - | - | - | - | - | - | - |
| Potassium 5-ketogluconate | - | - | + | - | - | - | - | - | - | - |
| API 20E | ||||||||||
| Natriumthiosulfat | - | NA | NA | NA | - | NA | NA | - | NA | NA |
| L-tryptophan | + | NA | NA | - | - | NA | NA | - | NA | NA |
| Indole production | - | NA | - | - | - | - | - | - | - | - |
| API 20NE | ||||||||||
| Potassium nitrate | - | NA | - | - | - | - | - | - | - | - |
| L-arginine | - | NA | + | - | - | - | - | - | - | + |
| Urea | - | NA | - | - | - | - | - | - | - | - |
| Gelatin | - | + | + | NA | + | + | + | + | + | + |
| Capric acid | + | NA | NA | + | - | - | - | - | - | NA |
| Adipic acid | - | NA | NA | - | - | - | - | - | - | NA |
| Malic acid | + | NA | NA | - | - | + | - | - | - | - |
| Trisodium citrate | + | NA | NA | - | - | - | - | - | + | - |
| Phenylacetic acid | - | NA | NA | - | - | - | - | - | - | NA |
| Drug (Antibiotics) | CC µg/mL | P. ghigonii MIC µg/mL |
|---|---|---|
| Benzylpenicelin | 0.016–256 | >256 |
| Amoxicillin | 0.016–256 | >256 |
| Ampicillin | 0.016–256 | >256 |
| Ceftriaxone | 0.016–256 | 128 |
| Imipenem | 0.002–32 | 0.047 |
| Ciprofloxacin | 0.002–32 | 0.25 |
| Amikacin | 0.016–256 | 1 |
| Gentamicin | 0.64–1024 | 0.5 |
| Streptomicin | 0.064–1024 | 6 |
| Daptomycin | 0.016–256 | >256 |
| Doxycyclin | 0.016–256 | 1.5 |
| Metronidazole | 0.016–256 | >256 |
| Rifampicin | 0.002–32 | 16 |
| Fosfomycin | 0.064–1024 | 192 |
| Vancomycin | 0.016–256 | >256 |
| Tigecyclin | 0.016–256 | 4 |
| Fatty Acids | Name | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| Straight-Chain Saturated | |||||||||||
| 10:0 | Decanoic acid | tr | - | - | 1.92 | - | - | - | - | - | - |
| 14:0 | Tetradecanoic acid | tr | tr | 1.0 | - | - | - | tr | 1.2 | 2.6 | 0.7 |
| 15:0 | Pentadecanoic acid | tr | - | 2.0 | - | - | - | - | - | - | 1.1 |
| 16:0 | Hexadecanoic acid | 1.1 | 4.1 | - | 2.54 | 1.9 | 4.7 | 1.6 | 2.2 | tr | 0.7 |
| 14:0 2-OH | 2-hydroxy-tetradecanoic acid | - | tr | tr | - | - | - | - | tr | tr | - |
| 16:0 3-OH | 3-hydroxy-hexadecanoic acid | tr | 2.1 | 2.2 | - | 1.6 | 1.4 | tr | 1.4 | tr | - |
| 17:0 2-OH | 2-hydroxy-hexadecenoic acid | - | - | - | - | - | 1.7 | - | tr | - | - |
| Branched Saturated | |||||||||||
| 11:0 anteiso | 8-methyl-decanoic acid | 1.1 | - | - | - | - | - | - | - | - | - |
| 13:0 anteiso | 10-methyl-dodecanoic acid | tr | - | - | - | - | - | - | - | - | - |
| 17:1 anteiso | 14-methyl-hexadecenoic acid | 1.1 | 1.0 | 2.3 | - | 1.2 | - | - | 1.5 | tr | - |
| 15:0 anteiso | - | tr | 2.1 | 1.76 | 2.1 | 3.3 | tr | tr | tr | 1.7 | |
| 5:0 iso | 3-methyl-butanoic acid | 3.0 | - | - | - | - | - | - | - | - | - |
| 8:0 iso | 6-methyl-octanoic acid | tr | - | - | - | - | - | - | - | - | - |
| 11:0 iso | 9-methyl-decanoic acid | 1.8 | tr | - | - | - | - | - | - | tr | - |
| 14:0 iso | 12-methyl-tridecanoic acid | tr | - | - | - | - | - | - | - | - | - |
| 15:0 iso | 13-methyl-tetradecanoic acid | 54.5 | 29.6 | 25.4 | 24.8 | 28.3 | 30.1 | 35.4 | 27.0 | 37.0 | 36.6 |
| 17:1 iso | 15-methyl-hexadecenoic acid | 3.5 | tr | tr | - | 1.2 | - | - | tr | tr | - |
| 15:0 3-OH iso | 3-hydroxy-13-methyl-tetradecanoic acid | 3.5 | 2.0 | 3.3 | 4.41 | 3.5 | - | 2.4 | 2.6 | 3.0 | - |
| 16:0 3-OH iso | 3-hydroxy-13-methyl-hexadecanoic acid | - | tr | tr | - | tr | - | - | tr | tr | - |
| 17:0 3-OH iso | 3-hydroxy-15-methyl-hexadecenoic acid | 5.5 | 12.3 | 14.6 | 20.1 | 20.4 | 18.2 | - | 12.8 | 7.9 | 13.7 |
| Mono-Unsaturated | |||||||||||
| 14:1ω5 | 9-tetradecenoic acid | tr | - | - | 1.33 | - | - | - | - | tr | - |
| 15:1ω7 | 8-pentadecenoic acid | 1.7 | - | - | - | - | - | - | - | - | |
| 16:1ω7 | 9-hexadecenoic acid | 11.1 | 33.1 | 30.6 | 30.0 | 27.7 | 24.5 | 27.2 | 29.3 | 31.4 | 20.2 |
| 16:1ω5 | 11-hexadecenoic acid | 8.6 | 1.4 | 2.7 | - | 1.7 | tr | 1.4 | 1.4 | 1.4 | 21.1 |
| 17:1ω9 | 9-hexadecanoic acid | - | 6.9 | 6.6 | 6.51 | 6.1 | 3.4 | 7.4 | 4.7 | 4.5 | 3.4 |
| 18:1ω9 | 9-octadecenoic acid | tr | - | - | - | - | - | - | - | - | - |
| 18:2ω6 | 9,12-octadecadienoic acid | tr | - | - | - | - | - | - | - | - | - |
| 18:1ω5 | 11-octadecenoic acid | - | tr | 1.3 | - | tr | - | - | tr | tr | - |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kangale, L.J.; Raoult, D.; Pierre-Edouard, F. Pedobacter ghigonii sp. nov., Isolated from the Microbiota of the Planarian Schmidtea mediterranea. Microbiol. Res. 2021, 12, 268-287. https://doi.org/10.3390/microbiolres12020019
Kangale LJ, Raoult D, Pierre-Edouard F. Pedobacter ghigonii sp. nov., Isolated from the Microbiota of the Planarian Schmidtea mediterranea. Microbiology Research. 2021; 12(2):268-287. https://doi.org/10.3390/microbiolres12020019
Chicago/Turabian StyleKangale, Luis Johnson, Didier Raoult, and Fournier Pierre-Edouard. 2021. "Pedobacter ghigonii sp. nov., Isolated from the Microbiota of the Planarian Schmidtea mediterranea" Microbiology Research 12, no. 2: 268-287. https://doi.org/10.3390/microbiolres12020019






