Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective
Abstract
:1. Introduction
2. Hidden Potential of Streptomyces: Metagenomic Insights and Evidence
3. Novel Streptomyces Species Isolated from Terrestrial Environments
3.1. Isolation Methods
3.2. Extreme Environments
| Strain | Nature of the Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces boncukensis sp. nov. | Saltern soil | Starch Casein agar, pH 7.0–7.2, supplemented with filter-sterilized cycloheximide (50 μg mL−1) and 3% NaCl | Turkey | [38] |
| Streptomyces taklimakanensis sp. nov. | Desert | Gauze’s No. 1 medium 1 supplemented with Nystatin (100 mg mL−1) and nalidixic acid (50 mg mL−1) | North-West China | [40] |
| Streptomyces alkaliterrae sp. nov. | Alkaline soil close to Soda lake | Starch casein agar adjusted to pH 8.5 with 1N NaOH and supplemented with 5% (w/v) sodium chloride and cycloheximide and nystatin (each at 50 μg mL−1) | India | [37] |
| Streptomyces cahuitamycinicus sp. nov | Desert soil | Minimal medium supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (10 μg mL−1) | Turkmenistan | [53] |
| Streptomyces acidicola sp. nov. | Soil from peat swamp forest | Humic acid vitamin (HV) agar supplemented with nalidixic acid (25 μg mL−1) and nystatin (50 μg mL−1) | Thailand | [51] |
| Streptomyces harenosi sp. nov. | Sand dunes | Actinomycete isolation agar (HiMedia), pH 7.3 | Indonesia | [74] |
| Streptomyces tibetensis sp. nov. | Acid sandy soil sample | ISP medium 7 adjusted to pH 7.3 at 25 °C supplemented with an inhibitor solution containing K2Cr2O7 (25 mg mL−1), calcium propionate (30 mg mL−1) and cycloheximide (50 mg mL−1) | China | [66] |
| Streptomyces abyssomicinicus sp. nov. | Rock soil sample | Humic acid vitamin agar | Mexico | [50] |
| Streptomyces altiplanensis sp. nov. | Arid soil samples | Starch Casein Agar within the pH range of 7.0–7.2, supplemented with 50 μg mL−1 nyastatin and 50 μg mL−1 cycloheximide | Chile | [65] |
| Streptomyces cyaneochromogenes sp. nov. | Soil sampled at a manganese contaminated area | Gause’s synthetic medium 1, supplemented with 0.04 g K2Cr2O7 | China | [64] |
| Streptomyces huasconensis sp. nov. | Arid soil samples | Starch Casein agar within the pH range of 7.0–7.2 | Chile | [48] |
| Streptomyces cadmiisoli sp. nov. | Cadmium-contaminated soil | Modified proline agar medium, supplemented with 2.0–3.0 mL solution (1.775 g L−1) in a 100 mL medium + Gause’s synthetic agar medium no.1 | China | [61] |
| Streptomyces fodineus sp. nov. | Acidic mine area soil | Acidified (pH 5) starch-Casein Agar supplemented with cycloheximide and nystatin, each at 50 μg mL−1 | Korea | [49] |
| Streptomyces dengpaensis sp. nov | Desert soil | ISP 7 medium (HiMedia) supplemented with inhibitor solution containing (25 mg mL−1), calcium propionate (30 mg mL−1) and cycloheximide (50 mg mL−1) | China | [67] |
| Streptomyces durbertensis sp. nov. | Saline–alkali soil | CMKA medium 1 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North-East China | [69] |
| Streptomyces polaris sp. nov. | Frozen soil | Humic acid vitamin (HV) agar supplemented with (50 mg L−1) | High Arctic | [60] |
| Streptomyces septentrionalis sp. nov. | ||||
| Streptomyces desertarenae sp. nov. | Desert Soil | Reasoner’s 2A (R2A; BD) agar adjusted to pH 7.0. | China | [57] |
| Streptomyces manganisoli sp. nov. | Manganese-polluted soil | Modified proline agar medium, supplemented with 2.0–3.0 mL solution (1.775 g L−1) in a 100 mL medium | China | [63] |
| Streptomyces salilacus sp. nov. | Salt lake sediment | ISP (International Streptomyces Project) medium 4 supplemented with 1.5% (w/v) NaCl | China | [52] |
| Streptomyces sediminis sp. nov. | Crater lake sediments | ISP 2 medium supplemented with 10 mg L−1 tetracycline with (50 μg mL−1) of nystatin and (5 μg mL−1) of rifampicin | Turkey | [58] |
| Streptomyces asenjonii sp. nov. | Hyper-arid Atacama desert soils | Humic acid vitamin (HV) agar | Chile, Peru, South America | [73] |
| Streptomyces aridus sp. nov. | Subsurface soil of Atacama desert | Glucose-yeast extract agar (HiMedia) supplemented with cycloheximide and nystatin (each at 25 μg mL−1) | Chile, Peru, South America | [59] |
| Streptomyces jeddahensis sp. nov. | Desert soil | Mineral salt medium (MSM) | Saudi Arabia | [71] |
| Streptomyces caldifontis sp. nov. | Hot water spring sediment | Starch casein agar medium supplemented with 25 μg mL−1 nystatin | Pakistan | [55] |
| Streptomyces daqingensis sp. nov. | Saline–alkaline soil | CMKA medium 2 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North-East China | [56] |
| Streptomyces actinomycinicus sp. nov. | Soil of a peat swamp forest | Humic acid vitamin (HV) agar supplemented with nalidixic acid (25 mg mL−1) and cycloheximide (50 mg mL−1) | Thailand | [68] |
| Streptomyces luozhongensis sp. nov. | Desert soil | Gauze’s No. 1 medium 2 pH 7.2, supplemented with 2.0–3.0 mL of solution (1.775 g L−1) in a 100 mL medium at pH 7.2 | Lop Nur, Xinjiang, North-West China | [54] |
| Streptomyces xiangtanensis sp. nov. | Soil near Xiangtan Manganese mine | Gauze’s synthetic medium 1 adjusted to pH 7.2, supplemented with 2.0–3.0 mL of K2Cr2O7 solution (1.775 g/L) in a 100 mL medium | Central-South China | [62] |
| Streptomyces arcticus sp. nov. | Frozen soil | Mineral agar 1 Gause medium supplemented with (50 mg L−1) | Arctic | [75] |
| Streptomyces canalis sp. nov. | Hypersaline soilsample | B7 medium supplemented with 1.5% (w/v) NaCl | China | [72] |
| Streptomyces alkaliphilus sp. nov. | Saline lake sediment | Solid basal medium, Horikoshi 1 supplemented with 100 mL of sterilized 10% Na2CO3 | Kenya | [76] |
| Streptomyces lonarensis sp. nov. | Lake sediments (alkaline salt water meteorite lake) | Medium for the isolation of alkalophilic actinomycetes at pH 10.0 or 11.0 (after autoclaving) , or NaOH were separately sterilized and used for adjusting the pH | India | [8] |
3.3. Symbionts
| Strain | Nature of Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces bauhiniae sp. nov. | Tree bark of Bauhinia variegata Linn | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1) | Thailand | [79] |
| Streptomyces fuscigenes sp. nov. | Bamboo (Sasa borealis) litter | Bennett’s Agar adjusted to pH 7.3 with NaOH and supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (20 μg mL−1) at pH 5.5 | Republic of Korea | [80] |
| Streptomyces dioscori sp. nov. | Bulbil of Dioscorea bulbifera L. | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (25 mg L−1) | South-West China | [81] |
| Streptomyces carminius sp. nov. | Roots of Sophora alopecuroides | Gauze’s No. 1 medium 3 at pH 7.5 | North-West China | [84] |
| Streptomyces geranii sp. nov. | Root of Geranium carolinianum Linn | Humic acid vitamin (HV) agar supplemented with nystatin (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [83] |
| Streptomyces populi sp. nov. | Stem of Populus adenopoda | Humic acid vitamin (HV) agar supplemented with nalidixic acid (25 mg L−1) and cycloheximide (50 mg L−1) | China | [87] |
| Streptomyces lichenis sp. nov. | Lichen sample | Arginine-vitamin (AV) agar | Thailand | [97] |
| Streptomyces roietensis sp. nov. | Surface-sterilized stem of jasmine rice, Oryza sativa KDML 105 | Humic acid vitamin (HV) agar | Thailand | [85] |
| Streptomyces capparidis sp. nov. | Fruits of Capparis spinosa | Tap water-yeast extract (TWYE) witin the pH range of 7.0–7.2 supplemented with 3% (w/v) NaCl | China | [88] |
| Streptomyces ginkgonis sp. nov. | Aril of a seed of Ginkgo biloba | Gause’s Synthetic agar medium 2 supplemented with streptomycin sulphate (10 μg mL−1) and actidione (50 μg mL−1) | Yangling, China | [89] |
| Streptomyces tremellae sp. nov. | Culture of mushroom Tremella fuciformis | Potato dextrose agar (PDA) medium (200 gpotato tissue, 20 g glucose, 20 g agar and 1000 mL deionized water, pH 5.6); cycloheximide (100 μg mL−1) | China | [98] |
| Streptomyces polygonati sp. nov. | Root of Polygonatum odoratum (Mill.) | Humic acid-vitamin agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [82] |
| Streptomyces pini sp. nov. | Phylloplane of pine (Pinus sylvestris L.) needle-like leaves | Ammonium mineral salts medium amended with 0.5% (v/v) methanol as carbon source and cycloheximide (10 μg mL−1) | India | [90] |
| Streptomyces phyllanthi sp. nov. | Stem of Phyllanthus amarus | Yeast extract-malt extract medium (ISP2 medium) supplemented with 10 μg L−1 tetracycline | Thailand | [86] |
| Streptomyces bryophytorum sp. nov. | Moss (Bryophyta) | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North China | [91] |
| Strain | Nature of Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces smaragdinus sp. nov. | Gut of the fungus-farming termite Macrotermes natalensis | Chitin agar supplemented with 0.05 g L−1 cycloheximide | South Africa | [101] |
| Streptomyces buecherae sp. nov. | Femaloe cave myotis bat (Myotis velifer) | ISP 2 Medium | New Mexico | [108] |
| Streptomyces corynorhini sp. nov. | Male Townsend’s big-eared bat | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1), nalidixic acid (50 mg L−1), trimethoprim (50 mg L−1) | New Mexico | [109] |
| Streptomyces capitiformicae sp. nov. | Head of an ant (Camponotus japonicus Mayr) | Sodium succinate-asparagine agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid 20 mg L−1 | China | [104] |
| Streptomyces lasiicapitis sp. nov. | Head of an ant(Lasius fuliginosus L.) | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [106] |
| Streptomyces camponoti sp. nov. | Cuticle of Camponotus japonicus Mayr | Gause’s synthetic agar no. 1 1 adjusted to pH 7.2 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | Harbin, Heilongjiang, China | [102] |
| Streptomyces cuticulae sp. nov. | ||||
| Streptomyces amphotericinicus sp. nov. | Head of an ant | Sodium succinate-asparagine agar pH 7.2, supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | Harbin, Heilongjiang, China | [103] |
| Streptomyces kronopolitis sp. nov. | Millipede (Kronopolites svenhedind Verhoeff) | Gause’s Synthetic Agar No. 1 1 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [110] |
| Streptomyces camponoticapitis sp. nov. | Head of an ant (Camponotus japonicus Mayr) | Tap Water Yeast Extract Agar (TWYE)2 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [105] |
| Streptomyces formicae sp. nov. | Head of Camponotus japonicus Mayr ant | Gause’s synthetic agar no. 1 1 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [93] |
| Streptomyces fractus sp. nov. | Gut of a South African termite | Medium II at pH 7, supplemented with μg mL−1 cycloheximide and 10 μg mL−1 nalidixic acid | South Africa | [100] |
3.4. Soil and Sediments
| Strain | Nature of Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces triticiradicis sp. nov. | Rhizosphere soil of wheat (Triticum aestivum L.) | cellulose-proline agar (CPA) supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | Central China | [128] |
| Streptomyces coryli sp. nov | Soil from a commercial hazelnutorchard | Stevenson’s medium no. 3 adjusted to pH 7.0 and supplemented with cycloheximide (50 μg mL−1), nalidixic acid (10 μg mL−1), nystatin (50 μg mL−1) and novobiocin (10 μg mL−1) | Turkey | [129] |
| Streptomyces paludis sp. nov. | Alpine wetland soil | Gause’s synthetic agar medium 2 adjusted pH 7.2 | China | [130] |
| Streptomyces boluensis sp. nov. | Lake sediment | M1 agar supplemented with filter-sterilized cycloheximide (50 mg mL−1) and rifampicin (5 mg mL−1) | Turkey | [131] |
| Streptomyces roseicoloratus sp. nov. | Soil in cotton fields | GJ medium adjusted to pH 7.0–7.5 | North-WestChina | [132] |
| Streptomyces soli sp. nov. | Birch forest soil | Streptomyces Project 2 (ISP2) medium (yeast extract–malt extract agar) adjusted to pH 7.2 supplemented with 10 mg L−1 tetracycline | China | [133] |
| Streptomyces albicerus sp. nov. | River sediment | Glycerol-arginine medium adjusted to pH 7.5 and supplemented with 100 μL of 50 mg mL−1 K2Cr2O7 in a 100 mL medium to reduce fungal contamination | China | [134] |
| Streptomyces inhibens sp. nov. | Rhizosphere soil of wheat (Triticum aestivum L.) | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North-East China. | [118] |
| Streptomyces dangxiongensis sp. nov. | Grass soil | Gause’s synthetic agar medium 2 adjusted to pH 7.2 and supplemented with nalidixic acid (25 μg mL−1) | China | [135] |
| Streptomyces rhizosphaericola sp. nov. | Brazilian Cerrado biome (wheat rhizosphere) | Glucose Yeast Extract Agar (GYEA) –HiMedia | Brazil | [119] |
| Streptomyces sporangiiformans sp. nov. | Soil collected from Mount Song | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [136] |
| Streptomyces monticola sp. nov. | Soil from Mount Song | Sodium succinate-asparagine agar adjusted to pH 7.2 and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [137] |
| Streptomyces tritici sp. nov. | Rhizosphere soil of wheat (Triticum aestivum L.) | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg L−1) and nalidixic acid (20 μg L−1) | Central China | [120] |
| Streptomyces venetus sp. nov. | Rhizosphere soil of an oil palm (Elaeis guineensis) | Starch casein agar (SCA) adjusted to pH 7.0–7.2 supplemented with nalidixic acid (25 μg mL−1) and cycloheximide (50 μg mL−1) | Thailand | [138] |
| Streptomyces xiangluensis sp. nov. | Soil from Xianglu Mountain | Sodium succinate-asparagine agar adjusted to pH 7.2 and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [139] |
| Streptomyces urticae sp. nov. | Rhizosphere soilof Urtica urens L. | Cellulose proline agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | northeast China | [140] |
| Streptomyces tunisialbus sp. nov. | Tunisian rhizosphere soil of Lavandula officinalis | Glucose yeast-malt extract agar (DSMZ medium 65) | Tunisia (North America) | [141] |
| Streptomyces flavalbus sp. nov. | Rhizosphere of maize (Zea mays L.) | Humic acid vitamin (HV) agar supplemented with nystatin (50 mg L−1) and nalidixic acid (20 mg L−1) | North-East China | [142] |
| Streptomyces lutosisoli sp. nov. | Muddy soil from stream | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North-East China | [143] |
| Streptomyces boninensis sp. nov. | Soil | Humic acid vitamin (HV) agar supplemented benlate (final conc. 25 μg mL−1 (w/v)) and nalidixic acid (final conc. 25 μg mL−1 (w/v)) | Japan | [123] |
| Streptomyces triticisoli sp. nov. | Rhizosphere soil of wheat | Gause’s Synthetic Agar No. 1 2 adjusted to pH 7.2 supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [144] |
| Streptomyces cerasinus sp. nov. | Soil | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1) | Thailand | [121] |
| Streptomyces solisilvae sp. nov. | Tropical forest soil | Starch–casein–nitrate agar within the pH range of 7.0–7.2 and supplemented with cycloheximide (50 μg mL−1), nystatin (50 μg mL−1) and nalidixic acid (20 μg mL−1) | China | [126] |
| Streptomyces thermoalkaliphilus sp. nov. | Soil of a tropical rainforest | Humic acid vitamin (HV) agar | China | [145] |
| Streptomyces swartbergensis sp. nov. | Soil collected from the banks of the Gamka river | MC agar pH 7.4 | South Africa | [146] |
| Streptomyces luteus sp. nov. | Soil | Mannitol-casein acid hydrolysis (GW1) medium prepared with 5% (w/v) NaCl | SouthernChina | [122] |
| Streptomyces xylanilyticus sp. nov. | Soil | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1) | Thailand | [147] |
| Streptomyces odonnellii sp. nov. | Soil savanna | Malt extract–yeast extract–glucose-agar medium pH 7.0 | Brazil | [148] |
| Streptomyces fuscichromogenes sp. nov. | Soil from a tropical rain forest | Yeast extract-malt extract agar (ISP 2) supplemented with 10 mg L−1 tetracycline | China | [149] |
| Streptomyces krungchingensis sp. nov. | Soil collected from Krung Ching Waterfall National Park | Starch casein nitrate agar within the pH range of 7.0–7.2 and supplemented with nystatin (25 mg L−1) and tetracycline (10 mg L−1) | Thailand | [150] |
| Streptomyces rhizosphaerihabitans sp. nov. | Rhizosphere soil and humus layer from bamboo forest | Starch casein agar at pH 5.5 adjusted with HCl | Korea | [151] |
| Streptomyces adustus sp. nov. | ||||
| Streptomyces indoligenes sp. nov. | Rhizosphere soil of Populus euphratica | Gause’s synthetic agar medium 2 adjusted to pH 7.2 | China | [152] |
| Streptomyces yangpuensis sp. nov. | Soil | Gause’s synthetic agar medium 2 adjusted to pH 7.2 | China | [116] |
| Streptomyces xinjiangensis sp. nov. | Soil | Reasoner’s 2A (R2A) agar medium at pH 7.2; adjust with crystalline before adding agar | China | [153] |
| Streptomyces alfalfae sp. nov. | Rhizosphere soil in an alfalfa field | International Streptomyces Project 2 (ISP2) supplemented with 10 mg L−1 tetracycline | China | [154] |
| Streptomyces palmae sp. nov. | Oil palm (Elaeis guineensis) rhizosphere soil | Starch casein agar (SCA) within the pH range of 7.0–7.2 supplemented with nalidixic acid (25 μg mL−1) and cycloheximide (50 μg mL−1) | Thailand | [155] |
| Streptomyces gamaensis sp. nov. | Tropical soil | Gause’s synthetic agar No. 1 adjusted to pH 7.2 and supplemented with nystatin (50 mg L−1) and nalidixic acid (20 mg L−1) | Gama, Chad | [156] |
| Streptomyces andamanensis sp. nov. | Soil | Starch casein nitrate agar plates (HiMedia) supplemented with 25 mg mL−1 nystatin | Thailand | [157] |
| Streptomyces lacrimifluminis sp. nov. | Soil from river bank | Gause’s synthetic agar medium 3 adjusted to pH 7.2 supplemented with nalidixic acid (25 μg mL−1) | China | [158] |
| Streptomyces olivicoloratus sp. nov. | Forest soil | HV agar adjusted to pH 7.2 and supplemented with 50 mg mL−1 filter-sterilized cycloheximide, 50 mg mL−1 nystatin and 0.5 mg mL−1 rifampicin | Korea | [159] |
| Streptomonospora halotolerans sp. nov. | Muddy soil | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | China | [160] |
| Streptomyces tyrosinilyticus sp. nov. | River sediment | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | North China | [161] |
| Streptomyces albiflavescens sp. nov. | Rainforest soil | ISP 2 medium with 10 mg L−1 tetracycline | South-West China | [124] |
| Streptomyces polymachus sp. nov. | Forest soil | Humic acid vitamin (HV) agar | South Korea | [125] |
| Streptomyces maoxianensis sp. nov. | Pine forest soil | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | South-West China | [162] |
| Streptomyces rubrisoli sp. nov. | Red soil | Modified mineral-medium agar containing 0.5% sorbitol supplemented with cycloheximide, nystatin, nalidixic acid (each at 50 μg mL−1), and novobiocin (at 25 μg mL−1) | China | [163] |
| Streptomyces gilvifuscus sp. nov. | Forest soil | Humic acid vitamin (HV) agar | Republic of Korea | [164] |
| Streptomyces lushanensis sp. nov. | Soil from mount Lushan | ISP media | China | [165] |
| Streptomyces bambusae sp. nov. | Bamboo rhizosphere soil | Humic acid vitamin agar (HV agar) adjusted to pH 7.2 and supplemented with filter-sterilized cycloheximide (50 μg mL−1), nystatin (50 μg mL−1), and rifampicin (0.5 μg mL−1) | Republic of Korea | [166] |
| Streptomyces sasae sp. nov. | Rhizosphere soil of bamboo (Sasa borealis) | Starch casein agar adjusted to pH 8.5 | Republic of Korea | [167] |
4. Novel Streptomycetes Species Isolated from Marine Environments
4.1. Isolation Methods
4.2. Invertebrates
4.3. Sediments
| Strain | Nature of Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces marianii sp. | Subtidal marine sediment | Gause’s inorganic agar media (pH 7.2–7.4) supplemented with 75 mg mL−1 of cycloheximide and 25 mg mL−1 of nystatin | India | [196] |
| Streptomyces otsuchiensis sp. nov. | Marine sediment | Bushnell–Haas medium for 5 hrs and 3.0% (w/v) NaCl | Japan | [205] |
| Streptomyces nigra sp. nov. | Rhizosphere soil Avicennia marina | Modified ZoBell 2216E agar plates (HiMedia) | China | [211] |
| Streptomyces caeni sp. nov. | Mangrove mud | that had been made with 70% aged seawater in distilled water (instead of pure distilled water), and supplemented with cycloheximide (25 mg mL−1), potassium dichromate (50 mg mL−1) and nystatin (50 mg mL−1) | China | [199] |
| Streptomyces qaidamensis sp. nov. | Sand | Gause’s synthetic agar medium 2 at pH 7.2 supplemented with nalidixic acid (25 μg mL−1) | China | [197] |
| Streptomyces monashensis sp. nov. | Mangrove soil | ISP2 agar | Malaysia | [34] |
| Streptomyces euryhalinus sp. nov. | Sediment in a mangrove forest | Enrichment medium at pH 7.5 | India | [198] |
| Streptomyces colonosanans sp. nov. | Sediment in mangrove soil | g mL−1) and nalidixic (20 μg mL−1) | Malaysia | [201] |
| Streptomyces kalpinensis sp. nov. | Salt water beach | GW1 medium | China | [195] |
| Streptomyces humi sp. nov. | Mangrove soil | g mL−1) and nystatin (10 μg mL−1) | Malaysia | [206] |
| Streptomyces litoralis sp. nov. | Salt water beach | GW1 medium prepared with 5% (w/v) NaCl | China | [212] |
| Streptomyces ovatisporus sp. nov. | Marine sediments collected at a depth of 42 m | Non-sporulating medium within the pH range of 7.2–7.4 and supplemented with filter-sterilized rifampicin (5 μg mL−1g mL−1) | Turkey | [204] |
| Streptomyces chitinivorans sp. nov. | Brackish sediment of a fish dumping yard in Chilika lake | Colloidal Chitin agar (CCA) medium supplemented with nystatin (50 mg L−1) | India | [208] |
| Streptomyces verrucosisporus sp. nov. | Marine sediments | Seawater– proline −1−1) | Thailand | [207] |
| Streptomyces antioxidans sp. nov. | Mangrove forest soil | g mL−1) | Malaysia | [213] |
| Streptomyces malaysiense sp. nov. | Mangrove soil | g mL−1) | Malaysia | [202] |
| Streptomyces lonarensis sp. nov. | Lake sediment | Beef extract-yeast extract-glucose agar medium adjusted to a pH between 8 and 10 with addition of an appropriate amount of 10% sterile Na2CO3 solution | India | [8] |
| Streptomyces gilvigriseus sp. nov. | Mangrove sediments | mL−1) | Malaysia | [203] |
| Streptomyces mangrovisoli sp. nov. | Mangrove sediments | g mL−1) | Malaysia | [209] |
| Streptomyces mangrovi sp. nov. | Mangrove sediments | SM3 agar (Gauze’s medium) 2 g mL−1).] supplemented with sterile seawater (3.3%, w/v) | Egypt | [210] |
5. Summary
6. Streptomyces as Source of Antibiotics
6.1. Terrestial Streptomyces as a Source of Antibiotics
6.2. Marine Streptomyces as a Source of Antibiotics
6.3. New Compounds from Streptomyces spp. with Bioactivity
6.4. Antibacterial Activity
6.5. Anticancer Activity
6.6. Enzyme Inhibitor/Inducer Activity
6.7. Antifungal
6.8. Other Biological Activity
7. Conclusions and Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Berdy, J. Thoughts and facts about antibiotics: Where we are now and where we are heading. J. Antibiot. 2012, 65, 385–395. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Demain, A.L.; Fang, A. The natural functions of secondary metabolites. In History of Modern Biotechnology; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2000; Volume 69, pp. 1–39. [Google Scholar]
- Locey, K.J.; Lennon, J.T. Scaling laws predict global microbial diversity. Proc. Natl. Acad. Sci. USA 2016, 113, 5970–5975. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bartlett, J.G.; Gilbert, D.N.; Spellberg, B. Seven ways to preserve the miracle of antibiotics. Clin. Infect. Dis. 2013, 56, 1445–1450. [Google Scholar] [CrossRef] [PubMed]
- Genilloud, O. Actinomycetes: Still a source of novel antibiotics. Nat. Prod. Rep. 2017, 34, 1203–1232. [Google Scholar] [CrossRef]
- Manteca, A.; Yague, P. Streptomyces as a Source of Antimicrobials: Novel Approaches to Activate Cryptic Secondary Metabolite Pathways. In Antimicrobials, Antibiotic Resistance, Antibiofilm Strategies and Activity Methods; IntechOpen: London, UK, 2019; pp. 1–22. [Google Scholar]
- Parte, A.C.; Sardà Carbasse, J.; Meier-Kolthoff, J.P.; Reimer, L.C.; Göker, M. List of Prokaryotic names with Standing in Nomenclature (LPSN) moves to the DSMZ. Int. J. Syst. Evol. Microbiol. 2020, 70, 5607–5612. [Google Scholar] [CrossRef]
- Sharma, T.K.; Mawlankar, R.; Sonalkar, V.V.; Shinde, V.K.; Zhan, J.; Li, W.J.; Rele, M.V.; Dastager, S.G.; Kumar, L.S. Streptomyces lonarensis sp. nov., isolated from Lonar Lake, a meteorite salt water lake in India. Antonie Van Leeuwenhoek 2016, 109, 225–235. [Google Scholar] [CrossRef]
- Li, F.; Liu, S.; Lu, Q.; Zheng, H.; Osterman, I.A.; Lukyanov, D.A.; Sergiev, P.V.; Dontsova, O.A.; Liu, S.; Ye, J.; et al. Studies on Antibacterial Activity and Diversity of Cultivable Actinobacteria Isolated from Mangrove Soil in Futian and Maoweihai of China. Evid.-Based Complement. Altern. Med. 2019, 2019, 3476567. [Google Scholar] [CrossRef] [Green Version]
- Gopalakrishnan, S.; Srinivas, V.; Prasanna, S.L. Streptomyces. In Beneficial Microbes in Agro-Ecology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 55–71. [Google Scholar]
- Lo Grasso, L.; Chillura-Martino, D.; Alduina, R. Production of Antibacterial Compounds from Actinomycetes. In Actinobacteria—Basics and Biotechnological Applications; IntechOpen: London, UK, 2016. [Google Scholar]
- Subramani, R.; Sipkema, D. Marine Rare Actinomycetes: A Promising Source of Structurally Diverse and Unique Novel Natural Products. Mar. Drugs 2019, 17, 249. [Google Scholar] [CrossRef] [Green Version]
- Woodruff, H.B. Selman A. Waksman, winner of the 1952 Nobel Prize for Physiology or Medicine. Appl. Environ. Microbiol. 2014, 80, 2–8. [Google Scholar] [CrossRef] [Green Version]
- Przybylska-Balcerek, A.; Frankowski, J.; Stuper-Szablewska, K. The influence of weather conditions on bioactive compound content in sorghum grain. Eur. Food Res. Technol. 2019, 246, 13–22. [Google Scholar] [CrossRef] [Green Version]
- Quinn, G.A.; Banat, A.M.; Abdelhameed, A.M.; Banat, I.M. Streptomyces from traditional medicine: Sources of new innovations in antibiotic discovery. J. Med. Microbiol. 2020, 69, 1040–1048. [Google Scholar] [CrossRef] [PubMed]
- Procopio, R.E.; Silva, I.R.; Martins, M.K.; Azevedo, J.L.; Araujo, J.M. Antibiotics produced by Streptomyces. Braz. J. Infect. Dis. 2012, 16, 466–471. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Giordano, D. Bioactive Molecules from Extreme Environments. Mar. Drugs 2020, 18, 640. [Google Scholar] [CrossRef]
- Laskaris, P.; Karagouni, A.D. Streptomyces, Greek Habitats and Novel Pharmaceuticals: A Promising Challenge. Microbiol. Res. 2021, 12, 840–846. [Google Scholar] [CrossRef]
- Lewis, K.; Epstein, S.; D’onofrio, A.; Ling, L.L. Uncultured microorganisms as a source of secondary metabolites. J. Antibiot. 2010, 63, 468–476. [Google Scholar] [CrossRef]
- Islam, M.R.; Jeong, Y.T.; Ryu, Y.J.; Song, C.H.; Lee, Y.S. Isolation, identification and optimal culture conditions of Streptomyces albidoflavus C247 producing antifungal agents against Rhizoctonia solani AG2-2. Mycobiology 2009, 37, 114–120. [Google Scholar] [CrossRef] [Green Version]
- Palazzotto, E.; Weber, T. Omics and multi-omics approaches to study the biosynthesis of secondary metabolites in microorganisms. Curr. Opin. Microbiol. 2018, 45, 109–116. [Google Scholar] [CrossRef]
- Doroghazi, J.R.; Metcalf, W.W. Comparative genomics of actinomycetes with a focus on natural product biosynthetic genes. BMC Genom. 2013, 14, 611. [Google Scholar] [CrossRef] [Green Version]
- Van der Meij, A.; Worsley, S.F.; Hutchings, M.I.; van Wezel, G.P. Chemical ecology of antibiotic production by actinomycetes. FEMS Microbiol. Rev. 2017, 41, 392–416. [Google Scholar] [CrossRef]
- Guerrero-Garzón, J.F.; Zehl, M.; Schneider, O.; Rückert, C.; Busche, T.; Kalinowski, J.; Bredholt, H.; Zotchev, S.B. Streptomyces spp. from the marine sponge Antho dichotoma: Analyses of secondary metabolite biosynthesis gene clusters and some of their products. Front. Microbiol. 2020, 11, 437. [Google Scholar] [CrossRef] [Green Version]
- Lee, N.; Hwang, S.; Kim, J.; Cho, S.; Palsson, B.; Cho, B.-K. Mini review: Genome mining approaches for the identification of secondary metabolite biosynthetic gene clusters in Streptomyces. Comput. Struct. Biotechnol. J. 2020, 18, 1548–1556. [Google Scholar] [CrossRef] [PubMed]
- Lee, N.; Hwang, S.; Kim, W.; Lee, Y.; Kim, J.H.; Cho, S.; Kim, H.U.; Yoon, Y.J.; Oh, M.-K.; Palsson, B.O.; et al. Systems and synthetic biology to elucidate secondary metabolite biosynthetic gene clusters encoded in Streptomyces genomes. Nat. Prod. Rep. 2021, 38, 1330–1361. [Google Scholar] [CrossRef] [PubMed]
- Belknap, K.C.; Park, C.J.; Barth, B.M.; Andam, C.P. Genome mining of biosynthetic and chemotherapeutic gene clusters in Streptomyces bacteria. Sci. Rep. 2020, 10, 2003. [Google Scholar] [CrossRef] [PubMed]
- Liu, T.; Ren, Z.; Chunyu, W.-X.; Li, G.-D.; Chen, X.; Zhang, Z.-T.-L.; Sun, H.-B.; Wang, M.; Xie, T.-P.; Wang, M.; et al. Exploration of Diverse Secondary Metabolites From Streptomyces sp. YINM00001, Using Genome Mining and One Strain Many Compounds Approach. Front. Microbiol. 2022, 13, 831174. [Google Scholar] [PubMed]
- Thomy, D.; Culp, E.; Adamek, M.; Cheng, E.Y.; Ziemert, N.; Wright, G.D.; Sass, P.; Brötz-Oesterhelt, H. The ADEP biosynthetic gene cluster in Streptomyces hawaiiensis NRRL 15010 reveals an accessory clpP gene as a novel antibiotic resistance factor. Appl. Environ. Microbiol. 2019, 85, e01292-19. [Google Scholar] [CrossRef]
- Lee, N.; Kim, W.; Hwang, S.; Lee, Y.; Cho, S.; Palsson, B.; Cho, B.-K. Thirty complete Streptomyces genome sequences for mining novel secondary metabolite biosynthetic gene clusters. Sci. Data 2020, 7, 55. [Google Scholar] [CrossRef]
- Choudoir, M.J.; Pepe-Ranney, C.; Buckley, D.H. Diversification of secondary metabolite biosynthetic gene clusters coincides with lineage divergence in Streptomyces. Antibiotics 2018, 7, 12. [Google Scholar] [CrossRef] [Green Version]
- Peng, Q.; Gao, G.; Lü, J.; Long, Q.; Chen, X.; Zhang, F.; Xu, M.; Liu, K.; Wang, Y.; Deng, Z.; et al. Engineered Streptomyces lividans strains for optimal identification and expression of cryptic biosynthetic gene clusters. Front. Microbiol. 2018, 9, 3042. [Google Scholar] [CrossRef] [Green Version]
- Singh, T.A.; Passari, A.K.; Jajoo, A.; Bhasin, S.; Gupta, V.K.; Hashem, A.; Alqarawi, A.A.; Abd Allah, E.F. Tapping into Actinobacterial genomes for natural product discovery. Front. Microbiol. 2021, 12, 1662. [Google Scholar] [CrossRef]
- Law, J.W.; Ser, H.L.; Ab Mutalib, N.S.; Saokaew, S.; Duangjai, A.; Khan, T.M.; Chan, K.G.; Goh, B.H.; Lee, L.H. Streptomyces monashensis sp. nov., a novel mangrove soil actinobacterium from East Malaysia with antioxidative potential. Sci. Rep. 2019, 9, 3056. [Google Scholar] [CrossRef]
- Qin, S.; Li, W.-J.; Dastager, S.G.; Hozzein, W.N. Actinobacteria in special and extreme habitats: Diversity, function roles, and environmental adaptations. Front. Microbiol. 2016, 7, 1415. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “uncultivable” microorganisms in pure culture in a simulated natural environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Swiecimska, M.; Golinska, P.; Nouioui, I.; Wypij, M.; Rai, M.; Sangal, V.; Goodfellow, M. Streptomyces alkaliterrae sp. nov., isolated from an alkaline soil, and emended descriptions of Streptomyces alkaliphilus, Streptomyces calidiresistens and Streptomyces durbertensis. Syst. Appl. Microbiol. 2020, 43, 126153. [Google Scholar] [CrossRef] [PubMed]
- Tatar, D.; Veyisoglu, A.; Saygin, H.; Sahin, N. Streptomyces boncukensis sp. nov., isolated from saltern soil. Arch. Microbiol. 2021, 203, 279–285. [Google Scholar] [CrossRef] [PubMed]
- Shivlata, L.; Satyanarayana, T. Thermophilic and alkaliphilic Actinobacteria: Biology and potential applications. Front. Microbiol. 2015, 6, 1014. [Google Scholar] [CrossRef] [PubMed]
- Yuan, L.L.; Zhang, L.L.; Luo, X.X.; Xia, Z.F.; Sun, B.B.; Zeng, H. Streptomyces taklimakanensis sp. nov., an actinomycete isolated from the Taklimakan desert. Antonie Van Leeuwenhoek 2020, 113, 1023–1031. [Google Scholar] [CrossRef]
- Li, S.H.; Jin, Y.; Cheng, J.; Park, D.J.; Kim, C.J.; Hozzein, W.N.; Wadaan, M.A.; Shu, W.S.; Ding, L.X.; Li, W.J. Gordonia jinhuaensis sp. nov., a novel actinobacterium, isolated from a VBNC (viable but non-culturable) state in pharmaceutical wastewater. Antonie Van Leeuwenhoek 2014, 106, 347–356. [Google Scholar] [CrossRef]
- McIver, L.J.; Abu-Ali, G.; Franzosa, E.A.; Schwager, R.; Morgan, X.C.; Waldron, L.; Segata, N.; Huttenhower, C. bioBakery: A meta’omic analysis environment. Bioinformatics 2018, 34, 1235–1237. [Google Scholar] [CrossRef] [Green Version]
- Zenova, G.M.; Manucharova, N.A.; Zvyagintsev, D.G. Extremophilic and extremotolerant actinomycetes in different soil types. Eurasian Soil Sci. 2011, 44, 417–436. [Google Scholar] [CrossRef]
- Sivalingam, P.; Hong, K.; Pote, J.; Prabakar, K. Extreme Environment Streptomyces: Potential Sources for New Antibacterial and Anticancer Drug Leads? Int. J. Microbiol. 2019, 2019, 5283948. [Google Scholar] [CrossRef] [Green Version]
- Pettit, R.K. Culturability and secondary metabolite diversity of extreme microbes: Expanding contribution of deep sea and deep-sea vent microbes to natural product discovery. Mar. Biotechnol. 2011, 13, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chen, X.; Jiang, Y.; Jiang, C. Morphological Identification of Actinobacteria. In Actinobacteria—Basics and Biotechnological Applications; IntechOpen: London, UK, 2016. [Google Scholar]
- Nikitushkin, V.D.; Demina, G.R.; Kaprelyants, A.S. Rpf Proteins Are the Factors of Reactivation of the Dormant Forms of Actinobacteria. Biochemistry 2016, 81, 1719–1734. [Google Scholar] [CrossRef] [PubMed]
- Cortes-Albayay, C.; Dorador, C.; Schumann, P.; Andrews, B.; Asenjo, J.; Nouioui, I. Streptomyces huasconensis sp. nov., an haloalkalitolerant actinobacterium isolated from a high altitude saline wetland at the Chilean Altiplano. Int. J. Syst. Evol. Microbiol. 2019, 69, 2315–2322. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.K.; Kang, H.J.; Roh, S.G.; Park, J.S.; Kim, S.B. Streptomyces fodineus sp. nov., an actinobacterium with antifungal activity isolated from mine area soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1350–1354. [Google Scholar] [CrossRef] [PubMed]
- Komaki, H.; Sakurai, K.; Hosoyama, A.; Kimura, A.; Trujilo, M.E.; Igarashi, Y.; Tamura, T. Diversity of PKS and NRPS gene clusters between Streptomyces abyssomicinicus sp. nov. and its taxonomic neighbor. J. Antibiot. 2020, 73, 141–151. [Google Scholar] [CrossRef]
- Lipun, K.; Chantavorakit, T.; Mingma, R.; Duangmal, K. Streptomyces acidicola sp. nov., isolated from a peat swamp forest in Thailand. J. Antibiot. 2020, 73, 435–440. [Google Scholar] [CrossRef]
- Luo, X.X.; Gao, G.B.; Xia, Z.F.; Chen, Z.J.; Wan, C.X.; Zhang, L.L. Streptomyces salilacus sp. nov., an actinomycete isolated from a salt lake. Int. J. Syst. Evol. Microbiol. 2018, 68, 1514–1518. [Google Scholar] [CrossRef]
- Saygin, H.; Ay, H.; Guven, K.; Cetin, D.; Sahin, N. Streptomyces cahuitamycinicus sp. nov., isolated from desert soil and reclassification of Streptomyces galilaeus as a later heterotypic synonym of Streptomyces bobili. Int. J. Syst. Evol. Microbiol. 2020, 70, 2750–2759. [Google Scholar] [CrossRef]
- Zhang, R.; Han, X.; Xia, Z.; Luo, X.; Wan, C.; Zhang, L. Streptomyces luozhongensis sp. nov., a novel actinomycete with antifungal activity and antibacterial activity. Antonie Van Leeuwenhoek 2017, 110, 195–203. [Google Scholar] [CrossRef]
- Amin, A.; Ahmed, I.; Khalid, N.; Osman, G.; Khan, I.U.; Xiao, M.; Li, W.J. Streptomyces caldifontis sp. nov., isolated from a hot water spring of Tatta Pani, Kotli, Pakistan. Antonie Van Leeuwenhoek 2017, 110, 77–86. [Google Scholar] [CrossRef]
- Pan, T.; He, H.; Li, C.; Zhao, J.; Zhang, Y.; Li, J.; Wang, X.; Liu, C.; Zhang, J.; Xiang, W. Streptomyces daqingensis sp. nov., isolated from saline-alkaline soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1358–1363. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, L.Y.; Yang, Z.W.; Asem, M.D.; Fang, B.Z.; Salam, N.; Alkhalifah, D.H.M.; Hozzein, W.N.; Nie, G.X.; Li, W.J. Streptomyces desertarenae sp. nov., a novel actinobacterium isolated from a desert sample. Antonie Van Leeuwenhoek 2019, 112, 367–374. [Google Scholar] [CrossRef]
- Ay, H.; Nouioui, I.; Del Carmen Montero-Calasanz, M.; Klenk, H.P.; Isik, K.; Cetin, D.; Sahin, N. Streptomyces sediminis sp. nov. isolated from crater lake sediment. Antonie Van Leeuwenhoek 2018, 111, 493–500. [Google Scholar] [CrossRef] [PubMed]
- Idris, H.; Labeda, D.P.; Nouioui, I.; Castro, J.F.; Del Carmen Montero-Calasanz, M.; Bull, A.T.; Asenjo, J.A.; Goodfellow, M. Streptomyces aridus sp. nov., isolated from a high altitude Atacama Desert soil and emended description of Streptomyces noboritoensis Isono et al. 1957. Antonie Van Leeuwenhoek 2017, 110, 705–717. [Google Scholar] [CrossRef] [Green Version]
- Kamjam, M.; Nopnakorn, P.; Zhang, L.; Peng, F.; Deng, Z.; Hong, K. Streptomyces polaris sp. nov. and Streptomyces septentrionalis sp. nov., isolated from frozen soil. Antonie Van Leeuwenhoek 2019, 112, 375–387. [Google Scholar] [CrossRef] [PubMed]
- Li, K.; Tang, X.; Zhao, J.; Guo, Y.; Tang, Y.; Gao, J. Streptomyces cadmiisoli sp. nov., a novel actinomycete isolated from cadmium-contaminated soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 1024–1029. [Google Scholar] [CrossRef]
- Mo, P.; Yu, Y.Z.; Zhao, J.R.; Gao, J. Streptomyces xiangtanensis sp. nov., isolated from a manganese-contaminated soil. Antonie Van Leeuwenhoek 2017, 110, 297–304. [Google Scholar] [CrossRef]
- Mo, P.; Zhao, J.; Li, K.; Tang, X.; Gao, J. Streptomyces manganisoli sp. nov., a novel actinomycete isolated from manganese-contaminated soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 1890–1895. [Google Scholar] [CrossRef]
- Tang, X.; Zhao, J.; Li, K.; Chen, Z.; Sun, Y.; Gao, J. Streptomyces cyaneochromogenes sp. nov., a blue pigment-producing actinomycete from manganese-contaminated soil. Int. J. Syst. Evol. Microbiol. 2019, 69, 2202–2207. [Google Scholar] [CrossRef]
- Cortes-Albayay, C.; Dorador, C.; Schumann, P.; Schniete, J.K.; Herron, P.; Andrews, B.; Asenjo, J.; Nouioui, I. Streptomyces altiplanensis sp. nov., an alkalitolerant species isolated from Chilean Altiplano soil, and emended description of Streptomyces chryseus (Krasil’nikov et al. 1965) Pridham 1970. Int. J. Syst. Evol. Microbiol. 2019, 69, 2498–2505. [Google Scholar] [CrossRef]
- Li, J.; Wang, L.; Ye, Z.; Lu, L.; Li, Y. Streptomyces tibetensis sp. nov., an actinomycete isolated from the Tibetan Plateau. Antonie Van Leeuwenhoek 2020, 113, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Wang, L.W.; Bao, J. Streptomyces dengpaensis sp. nov., an actinomycete isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3322–3326. [Google Scholar] [CrossRef] [PubMed]
- Tanasupawat, S.; Phongsopitanun, W.; Suwanborirux, K.; Ohkuma, M.; Kudo, T. Streptomyces actinomycinicus sp. nov., isolated from soil of a peat swamp forest. Int. J. Syst. Evol. Microbiol. 2016, 66, 290–295. [Google Scholar] [CrossRef]
- Yu, Y.; Fu, Y.; Guo, X.; Yan, R.; Wang, H.; Zhao, J.; Wang, X.; Zhang, J.; Xiang, W. Streptomyces durbertensis sp. nov., isolated from saline-alkali soil. Int. J. Syst. Evol. Microbiol. 2018, 68, 3635–3640. [Google Scholar] [CrossRef] [PubMed]
- Bobek, J.; Smidova, K.; Cihak, M. A Waking Review: Old and Novel Insights into the Spore Germination in Streptomyces. Front. Microbiol. 2017, 8, 2205. [Google Scholar] [CrossRef] [Green Version]
- Rottig, A.; Atasayar, E.; Meier-Kolthoff, J.P.; Sproer, C.; Schumann, P.; Schauer, J.; Steinbuchel, A. Streptomyces jeddahensis sp. nov., an oleaginous bacterium isolated from desert soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 1676–1682. [Google Scholar] [CrossRef]
- Xie, Y.X.; Han, X.X.; Luo, X.X.; Xia, Z.F.; Wan, C.X.; Zhang, L.L. Streptomyces canalis sp. nov., an actinomycete isolated from an alkali-removing canal. Int. J. Syst. Evol. Microbiol. 2016, 66, 3219–3223. [Google Scholar] [CrossRef]
- Goodfellow, M.; Busarakam, K.; Idris, H.; Labeda, D.P.; Nouioui, I.; Brown, R.; Kim, B.Y.; Del Carmen Montero-Calasanz, M.; Andrews, B.A.; Bull, A.T. Streptomyces asenjonii sp. nov., isolated from hyper-arid Atacama Desert soils and emended description of Streptomyces viridosporus Pridham et al. 1958. Antonie Van Leeuwenhoek 2017, 110, 1133–1148. [Google Scholar] [CrossRef]
- Kusuma, A.B.; Nouioui, I.; Klenk, H.P.; Goodfellow, M. Streptomyces harenosi sp. nov., a home for a gifted strain isolated from Indonesian sand dune soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 4874–4882. [Google Scholar] [CrossRef]
- Zhang, L.; Ruan, C.; Peng, F.; Deng, Z.; Hong, K. Streptomyces arcticus sp. nov., isolated from frozen soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1482–1487. [Google Scholar] [CrossRef]
- Akhwale, J.K.; Goker, M.; Rohde, M.; Sproer, C.; Schumann, P.; Klenk, H.P.; Boga, H.I. Streptomyces alkaliphilus sp. nov., isolated from sediments of Lake Elmenteita in the Kenyan Rift Valley. Antonie Van Leeuwenhoek 2015, 107, 1249–1259. [Google Scholar] [CrossRef]
- Adnani, N.; Rajski, S.R.; Bugni, T.S. Symbiosis-inspired approaches to antibiotic discovery. Nat. Prod. Rep. 2017, 34, 784–814. [Google Scholar] [CrossRef] [PubMed]
- Seipke, R.F.; Barke, J.; Brearley, C.; Hill, L.; Yu, D.W.; Goss, R.J.; Hutchings, M.I. A single Streptomyces symbiont makes multiple antifungals to support the fungus farming ant Acromyrmex octospinosus. PLoS ONE 2011, 6, e22028. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanchanasin, P.; Yuki, M.; Kudo, T.; Ohkuma, M.; Kuncharoen, N.; Phongsopitanun, W.; Tanasupawat, S. Streptomyces bauhiniae sp. nov., isolated from tree bark of Bauhinia variegata Linn. in Thailand. Int. J. Syst. Evol. Microbiol. 2020, 70, 228–233. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Whang, K.S. Streptomyces fuscigenes sp. nov., isolated from bamboo (Sasa borealis) litter. Int. J. Syst. Evol. Microbiol. 2018, 68, 1541–1545. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Tian, J.; Li, X.; Gan, L.; He, L.; Chu, Y.; Tian, Y. Streptomyces dioscori sp. nov., a Novel Endophytic Actinobacterium Isolated from Bulbil of Dioscorea bulbifera L. Curr. Microbiol. 2018, 75, 1384–1390. [Google Scholar] [CrossRef]
- Guo, S.; Liu, C.; Liu, S.; Guan, X.; Guo, L.; Jia, F.; Wang, X.; Xiang, W. Streptomyces polygonati sp. nov., an endophytic actinomycete isolated from a root of Polygonatum odoratum (Mill.). Int. J. Syst. Evol. Microbiol. 2016, 66, 1488–1493. [Google Scholar] [CrossRef]
- Li, X.; Lai, X.; Gan, L.; Long, X.; Hou, Y.; Zhang, Y.; Tian, Y. Streptomyces geranii sp. nov., a novel endophytic actinobacterium isolated from root of Geranium carolinianum L. Int. J. Syst. Evol. Microbiol. 2018, 68, 2562–2567. [Google Scholar] [CrossRef]
- Wang, Y.; Xia, Z.; Liu, Z.; Wan, C.; Luo, X.; Zhang, L. Streptomyces carminius sp. nov., a novel actinomycete isolated from Sophora alopecuroides in Xinjiang, China. Antonie Van Leeuwenhoek 2018, 111, 1807–1814. [Google Scholar] [CrossRef]
- Kaewkla, O.; Franco, C.M.M. Streptomyces roietensis sp. nov., an endophytic actinobacterium isolated from the surface-sterilized stem of jasmine rice, Oryza sativa KDML 105. Int. J. Syst. Evol. Microbiol. 2017, 67, 4868–4872. [Google Scholar] [CrossRef]
- Klykleung, N.; Phongsopitanun, W.; Pittayakhajonwut, P.; Ohkuma, M.; Kudo, T.; Tanasupawat, S. Streptomyces phyllanthi sp. nov., isolated from the stem of Phyllanthus amarus. Int. J. Syst. Evol. Microbiol. 2016, 66, 3923–3928. [Google Scholar] [CrossRef] [PubMed]
- Wang, Z.; Jiang, B.; Li, X.; Gan, L.; Long, X.; Zhang, Y.; Tian, Y. Streptomyces populi sp. nov., a novel endophytic actinobacterium isolated from stem of Populus adenopoda Maxim. Int. J. Syst. Evol. Microbiol. 2018, 68, 2568–2573. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.F.; Li, Q.L.; Xiao, M.; Zhang, Y.G.; Zhou, X.K.; Narsing Rao, M.P.; Duan, Y.Q.; Li, W.J. Streptomyces capparidis sp. nov., a novel endophytic actinobacterium isolated from fruits of Capparis spinosa L. Int. J. Syst. Evol. Microbiol. 2017, 67, 133–137. [Google Scholar] [CrossRef] [PubMed]
- Yan, X.; Li, Y.; Wang, N.; Chen, Y.; Huang, L.L. Streptomyces ginkgonis sp. nov., an endophyte from Ginkgo biloba. Antonie Van Leeuwenhoek 2018, 111, 891–896. [Google Scholar] [CrossRef]
- Madhaiyan, M.; Poonguzhali, S.; Saravanan, V.S.; Duraipandiyan, V.; Al-Dhabi, N.A.; Pragatheswari, D.; Santhanakrishnan, P.; Kim, S.J.; Weon, H.Y.; Kwon, S.W. Streptomyces pini sp. nov., an actinomycete isolated from phylloplane of pine (Pinus sylvestris L.) needle-like leaves. Int. J. Syst. Evol. Microbiol. 2016, 66, 4204–4210. [Google Scholar] [CrossRef]
- Li, C.; Jin, P.; Liu, C.; Ma, Z.; Zhao, J.; Li, J.; Wang, X.; Xiang, W. Streptomyces bryophytorum sp. nov., an endophytic actinomycete isolated from moss (Bryophyta). Antonie Van Leeuwenhoek 2016, 109, 1209–1215. [Google Scholar] [CrossRef]
- Constant, P.; Poissant, L.; Villemur, R. Isolation of Streptomyces sp. PCB7, the first microorganism demonstrating high-affinity uptake of tropospheric H2. ISME J. 2008, 2, 1066–1076. [Google Scholar] [CrossRef]
- Bai, L.; Liu, C.; Guo, L.; Piao, C.; Li, Z.; Li, J.; Jia, F.; Wang, X.; Xiang, W. Streptomyces formicae sp. nov., a novel actinomycete isolated from the head of Camponotus japonicus Mayr. Antonie Van Leeuwenhoek 2016, 109, 253–261. [Google Scholar] [CrossRef]
- Hayakawa, M.; Kajiura, T.; Nonomura, H. New methods for the highly selective Isolation of Streptosporangium and Dactylosporangium from soil. J. Ferment. Bioeng. 1991, 72, 327–333. [Google Scholar] [CrossRef]
- Quecine, M.C.; Araujo, W.L.; Marcon, J.; Gai, C.S.; Azevedo, J.L.; Pizzirani-Kleiner, A.A. Chitinolytic activity of endophytic Streptomyces and potential for biocontrol. Lett. Appl. Microbiol. 2008, 47, 486–491. [Google Scholar] [CrossRef]
- Frey-Klett, P.; Garbaye, J.; Tarkka, M. The mycorrhiza helper bacteria revisited. New Phytol. 2007, 176, 22–36. [Google Scholar] [CrossRef] [PubMed]
- Saeng-In, P.; Phongsopitanun, W.; Savarajara, A.; Tanasupawat, S. Streptomyces lichenis sp. nov., isolated from lichen. Int. J. Syst. Evol. Microbiol. 2018, 68, 3641–3646. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.Q.; Chen, B.; Li, X.; Li, B.B.; Li, C.H.; Huang, Q.H.; Zhang, Q.H.; Dai, W.H.; Jiang, Y.J. Streptomyces tremellae sp. nov., isolated from a culture of the mushroom Tremella fuciformis. Int. J. Syst. Evol. Microbiol. 2016, 66, 5028–5033. [Google Scholar] [CrossRef]
- Chevrette, M.G.; Carlson, C.M.; Ortega, H.E.; Thomas, C.; Ananiev, G.E.; Barns, K.J.; Book, A.J.; Cagnazzo, J.; Carlos, C.; Flanigan, W.; et al. The antimicrobial potential of Streptomyces from insect microbiomes. Nat. Commun. 2019, 10, 516. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rohland, J.; Meyers, P.R. Streptomyces fractus sp. nov., a novel streptomycete isolated from the gut of a South African termite. Antonie Van Leeuwenhoek 2015, 107, 1127–1134. [Google Scholar] [CrossRef]
- Schwitalla, J.W.; Benndorf, R.; Martin, K.; Vollmers, J.; Kaster, A.K.; de Beer, Z.W.; Poulsen, M.; Beemelmanns, C. Streptomyces smaragdinus sp. nov., isolated from the gut of the fungus growing-termite Macrotermes natalensis. Int. J. Syst. Evol. Microbiol. 2020, 70, 5806–5811. [Google Scholar] [CrossRef]
- Piao, C.; Zheng, W.; Li, Y.; Liu, C.; Jin, L.; Song, W.; Yan, K.; Wang, X.; Xiang, W. Two new species of the genus Streptomyces: Streptomyces camponoti sp. nov. and Streptomyces cuticulae sp. nov. isolated from the cuticle of Camponotus japonicus Mayr. Arch. Microbiol. 2017, 199, 963–970. [Google Scholar] [CrossRef]
- Cao, T.; Mu, S.; Lu, C.; Zhao, S.; Li, D.; Yan, K.; Xiang, W.; Liu, C. Streptomyces amphotericinicus sp. nov., an amphotericin-producing actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int. J. Syst. Evol. Microbiol. 2017, 67, 4967–4973. [Google Scholar] [CrossRef]
- Jiang, S.; Piao, C.; Yu, Y.; Cao, P.; Li, C.; Yang, F.; Li, M.; Xiang, W.; Liu, C. Streptomyces capitiformicae sp. nov., a novel actinomycete producing angucyclinone antibiotics isolated from the head of Camponotus japonicus Mayr. Int. J. Syst. Evol. Microbiol. 2018, 68, 118–124. [Google Scholar] [CrossRef]
- Li, Y.; Ye, L.; Wang, X.; Zhao, J.; Ma, Z.; Yan, K.; Xiang, W.; Liu, C. Streptomyces camponoticapitis sp. nov., an actinomycete isolated from the head of an ant (Camponotus japonicus Mayr). Int. J. Syst. Evol. Microbiol. 2016, 66, 3855–3859. [Google Scholar] [CrossRef]
- Ye, L.; Zhao, S.; Li, Y.; Jiang, S.; Zhao, Y.; Li, J.; Yan, K.; Wang, X.; Xiang, W.; Liu, C. Streptomyces lasiicapitis sp. nov., an actinomycete that produces kanchanamycin, isolated from the head of an ant (Lasius fuliginosus L.). Int. J. Syst. Evol. Microbiol. 2017, 67, 1529–1534. [Google Scholar] [CrossRef] [PubMed]
- Poulsen, M.; Oh, D.C.; Clardy, J.; Currie, C.R. Chemical analyses of wasp-associated Streptomyces bacteria reveal a prolific potential for natural products discovery. PLoS ONE 2011, 6, e16763. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Hamm, P.S.; Dunlap, C.A.; Mullowney, M.W.; Caimi, N.A.; Kelleher, N.L.; Thomson, R.J.; Porras-Alfaro, A.; Northup, D.E. Streptomyces buecherae sp. nov., an actinomycete isolated from multiple bat species. Antonie Van Leeuwenhoek 2020, 113, 2213–2221. [Google Scholar] [CrossRef] [PubMed]
- Hamm, P.S.; Caimi, N.A.; Northup, D.E.; Valdez, E.W.; Buecher, D.C.; Dunlap, C.A.; Labeda, D.P.; Porras-Alfaro, A. Streptomyces corynorhini sp. nov., isolated from Townsend’s big-eared bats (Corynorhinus townsendii). Antonie Van Leeuwenhoek 2019, 112, 1297–1305. [Google Scholar] [CrossRef]
- Liu, C.; Ye, L.; Li, Y.; Jiang, S.; Liu, H.; Yan, K.; Xiang, W.; Wang, X. Streptomyces kronopolitis sp. nov., an actinomycete that produces phoslactomycins isolated from a millipede (Kronopolites svenhedind Verhoeff). Int. J. Syst. Evol. Microbiol. 2016, 66, 5352–5357. [Google Scholar] [CrossRef]
- Olanrewaju, O.S.; Babalola, O.O. Streptomyces: Implications and interactions in plant growth promotion. Appl. Microbiol. Biotechnol. 2019, 103, 1179–1188. [Google Scholar] [CrossRef] [Green Version]
- Adams, A.S.; Jordan, M.S.; Adams, S.M.; Suen, G.; Goodwin, L.A.; Davenport, K.W.; Currie, C.R.; Raffa, K.F. Cellulose-degrading bacteria associated with the invasive woodwasp Sirex noctilio. ISME J. 2011, 5, 1323–1331. [Google Scholar] [CrossRef]
- Amore, A.; Pepe, O.; Ventorino, V.; Birolo, L.; Giangrande, C.; Vincenza, F. Cloning and recombinant expression of a cellulase from the cellulytic strain Streptomyces sp. G12 isolated from compost. Microb. Cell Factories 2012, 11, 164. [Google Scholar] [CrossRef] [Green Version]
- Li, X.; Li, E.; Zhu, Y.; Teng, C.; Sun, B.; Song, H.; Yang, R. A typical endo-xylanase from Streptomyces rameus L2001 and its unique characteristics in xylooligosaccharide production. Carbohydr. Res. 2012, 359, 30–36. [Google Scholar] [CrossRef]
- Bontemps, C.; Toussaint, M.; Revol, P.V.; Hotel, L.; Jeanbille, M.; Uroz, S.; Turpault, M.P.; Blaudez, D.; Leblond, P. Taxonomic and functional diversity of Streptomyces in a forest soil. FEMS Microbiol. Lett. 2013, 342, 157–167. [Google Scholar] [CrossRef] [Green Version]
- Tang, B.; Yu, Y.; Zhi, X.; Yang, L.; Cen, X.; Zhao, G.; Ding, X. Streptomyces yangpuensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 1224–1229. [Google Scholar] [CrossRef] [PubMed]
- Abdulla, H. Bioweathering and biotransformation of granitic rock minerals by actinomycetes. Microb. Ecol. 2009, 58, 753–761. [Google Scholar] [CrossRef] [PubMed]
- Jin, L.; Zhao, Y.; Song, W.; Duan, L.; Jiang, S.; Wang, X.; Zhao, J.; Xiang, W. Streptomyces inhibens sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int. J. Syst. Evol. Microbiol. 2019, 69, 688–695. [Google Scholar] [CrossRef]
- Vargas Hoyos, H.A.; Nobre Santos, S.; Da Silva, L.J.; Paulino Silva, F.S.; Bonaldo Genuario, D.; Domingues Zucchi, T.; Melo, I.S. Streptomyces rhizosphaericola sp. nov., an actinobacterium isolated from the wheat rhizosphere. Int. J. Syst. Evol. Microbiol. 2019, 69, 2431–2439. [Google Scholar] [CrossRef]
- Zhao, J.; Shi, L.; Li, W.; Wang, J.; Wang, H.; Tian, Y.; Xiang, W.; Wang, X. Streptomyces tritici sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int. J. Syst. Evol. Microbiol. 2018, 68, 492–497. [Google Scholar] [CrossRef] [PubMed]
- Kanchanasin, P.; Moonmangmee, D.; Phongsopitanun, W.; Tanasupawat, S.; Moonmangmee, S. Streptomyces cerasinus sp. nov., isolated from soil in Thailand. Int. J. Syst. Evol. Microbiol. 2017, 67, 3854–3859. [Google Scholar] [CrossRef]
- Luo, X.X.; Kai, L.; Wang, Y.; Wan, C.X.; Zhang, L.L. Streptomyces luteus sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 543–547. [Google Scholar] [CrossRef]
- Take, A.; Inahashi, Y.; Omura, S.; Takahashi, Y.; Matsumoto, A. Streptomyces boninensis sp. nov., isolated from soil from a limestone cave in the Ogasawara Islands. Int. J. Syst. Evol. Microbiol. 2018, 68, 1795–1799. [Google Scholar] [CrossRef]
- Han, X.; Zheng, J.; Xin, D.; Xin, Y.; Wei, X.; Zhang, J. Streptomyces albiflavescens sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65 Pt 5, 1467–1473. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kim, J. Antifungal and antibacterial activities of Streptomyces polymachus sp. nov. isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 2385–2390. [Google Scholar] [CrossRef] [Green Version]
- Zhou, S.; Yang, X.; Huang, D.; Huang, X. Streptomyces solisilvae sp. nov., isolated from tropical forest soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 3553–3558. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.L.; Wang, H.; Xue, Z.L.; Li, J.S.; Qi, H.; Zhang, H.; Zhao, T.; Wang, J.D.; Xiang, W.S. Two new glutarimide antibiotics from Streptomyces sp. HS-NF-780. J. Antibiot. 2019, 72, 241–245. [Google Scholar] [CrossRef] [PubMed]
- Yu, Z.; Han, C.; Yu, B.; Zhao, J.; Yan, Y.; Huang, S.; Liu, C.; Xiang, W. Taxonomic Characterization, and Secondary Metabolite Analysis of Streptomyces triticiradicis sp. nov.: A Novel Actinomycete with Antifungal Activity. Microorganisms 2020, 8, 77. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Saygin, H.; Veyisoglu, A.; Tatar, D.; Nigiz, C.; Tokatli, A.; Sahin, N. Streptomyces coryli sp. nov., isolated from hazelnut orchard soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 4791–4797. [Google Scholar] [CrossRef]
- Zhao, J.; Tang, X.; Li, K.; Guo, Y.; Feng, M.; Gao, J. Streptomyces paludis sp. nov., isolated from an alpine wetland soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 773–778. [Google Scholar] [CrossRef]
- Tokatli, A.; Idil, O.; Veyisoglu, A.; Saygin, H.; Guven, K.; Cetin, D.; Sahin, N. Streptomyces boluensis sp. nov., isolated from lake sediment. Arch. Microbiol. 2020, 202, 2303–2309. [Google Scholar] [CrossRef]
- Zhang, Q.Y.; Liu, C.F.; Wang, Y.; Xia, Z.F.; Huang, Y.J.; Luo, X.X. Streptomyces roseicoloratus sp. nov., isolated from cotton soil. Int. J. Syst. Evol. Microbiol. 2020, 70, 738–743. [Google Scholar] [CrossRef]
- Xing, J.; Jiang, X.; Kong, D.; Zhou, Y.; Li, M.; Han, X.; Ma, Q.; Tan, H.; Ruan, Z. Streptomyces soli sp. nov., isolated from birch forest soil. Arch. Microbiol. 2020, 202, 1687–1692. [Google Scholar] [CrossRef]
- Sun, B.; Yuan, L.; Xia, Z.; Wan, C.; Zhang, L. Streptomyces albicerus sp. nov., a novel actinomycete isolated from the sediments of the Tailan River in Xinjiang, China. Arch. Microbiol. 2020, 202, 1639–1646. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, S.; Yang, R.; Chen, X.; Zhang, D.; Zhang, W.; Li, S.; Chen, T.; Liu, G.; Dyson, P. Streptomyces dangxiongensis sp. nov., isolated from soil of Qinghai-Tibet Plateau. Int. J. Syst. Evol. Microbiol. 2019, 69, 2729–2734. [Google Scholar] [CrossRef]
- Zhao, J.; Han, L.; Yu, M.; Cao, P.; Li, D.; Guo, X.; Liu, Y.; Wang, X.; Xiang, W. Characterization of Streptomyces sporangiiformans sp. nov., a Novel Soil Actinomycete with Antibacterial Activity against Ralstonia solanacearum. Microorganisms 2019, 7, 360. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Li, D.; Han, L.; Zhao, J.; Ju, H.; Jiang, S.; Guo, X.; Wang, X.; Xiang, W. Streptomyces monticola sp. nov., a novel actinomycete isolated from soil. Antonie Van Leeuwenhoek 2019, 112, 451–460. [Google Scholar] [CrossRef] [PubMed]
- Sujarit, K.; Kudo, T.; Ohkuma, M.; Pathom-Aree, W.; Lumyong, S. Streptomyces venetus sp. nov., an actinomycete with a blue aerial mycelium. Int. J. Syst. Evol. Microbiol. 2018, 68, 3333–3339. [Google Scholar] [CrossRef] [PubMed]
- Zhao, J.; Li, D.; Jiang, H.; Han, L.; Jiang, S.; Guo, X.; Wang, X.; Xiang, W. Streptomyces xiangluensis sp. nov., a novel actinomycete isolated from soil. Antonie Van Leeuwenhoek 2018, 111, 2249–2256. [Google Scholar] [CrossRef] [PubMed]
- Piao, C.; Ling, L.; Zhao, J.; Jin, L.; Jiang, S.; Guo, X.; Wang, X.; Xiang, W. Streptomyces urticae sp. nov., isolated from rhizosphere soil of Urtica urens L. Antonie Van Leeuwenhoek 2018, 111, 1835–1843. [Google Scholar] [CrossRef] [PubMed]
- Ayed, A.; Slama, N.; Mankai, H.; Bachkouel, S.; El Kahoui, S.; Tabbene, O.; Limam, F. Streptomyces tunisialbus sp. nov., a novel Streptomyces species with antimicrobial activity. Antonie Van Leeuwenhoek 2018, 111, 1571–1581. [Google Scholar] [CrossRef] [PubMed]
- Cao, T.; Shen, Y.; Zhao, J.; Liu, C.; Zhao, X.; Jin, L.; Li, Y.; Wang, X.; Xiang, W. Streptomyces flavalbus sp. nov., an actinobacterium isolated from rhizosphere of maize (Zea mays L.). Antonie Van Leeuwenhoek 2018, 111, 1047–1054. [Google Scholar] [CrossRef]
- Shen, Y.; Sun, T.; Jiang, S.; Mu, S.; Li, D.; Guo, X.; Zhang, J.; Zhao, J.; Xiang, W. Streptomyces lutosisoli sp. nov., a novel actinomycete isolated from muddy soil. Antonie Van Leeuwenhoek 2018, 111, 2403–2412. [Google Scholar] [CrossRef]
- Tian, Y.; Han, C.; Zhao, J.; Shi, H.; Hu, J.; Jiang, S.; Han, X.; Wang, X.; Xiang, W. Streptomyces triticisoli sp. nov., a novel actinomycete isolated from rhizosphere soil of wheat (Triticum aestivum L.). Int. J. Syst. Evol. Microbiol. 2018, 68, 3327–3332. [Google Scholar] [CrossRef]
- Wu, H.; Liu, B.; Ou, X.; Pan, S.; Shao, Y.; Huang, F. Streptomyces thermoalkaliphilus sp. nov., an alkaline cellulase producing thermophilic actinomycete isolated from tropical rainforest soil. Antonie Van Leeuwenhoek 2018, 111, 413–422. [Google Scholar] [CrossRef]
- le Roes-Hill, M.; Prins, A.; Meyers, P.R. Streptomyces swartbergensis sp. nov., a novel tyrosinase and antibiotic producing actinobacterium. Antonie Van Leeuwenhoek 2018, 111, 589–600. [Google Scholar] [CrossRef]
- Moonmangmee, D.; Kanchanasin, P.; Phongsopitanun, W.; Tanasupawat, S.; Moonmangmee, S. Streptomyces xylanilyticus sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 4189–4194. [Google Scholar] [CrossRef] [PubMed]
- Pereira, P.H.F.; Macrae, A.; Reinert, F.; de Souza, R.F.; Coelho, R.R.R.; Potter, G.; Klenk, H.P.; Labeda, D.P. Streptomyces odonnellii sp. nov., a proteolytic streptomycete isolated from soil under cerrado (savanna) vegetation cover. Int. J. Syst. Evol. Microbiol. 2017, 67, 5211–5215. [Google Scholar] [CrossRef] [PubMed]
- Zhang, H.; Zheng, J.; Zhuang, J.; Xin, Y.; Zheng, X.; Zhang, J. Streptomyces fuscichromogenes sp. nov., an actinomycete from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 77–81. [Google Scholar] [CrossRef]
- Sripreechasak, P.; Phongsopitanun, W.; Tamura, T.; Tanasupawat, S. Streptomyces krungchingensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2017, 67, 50–54. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Whang, K.S. Streptomyces rhizosphaerihabitans sp. nov. and Streptomyces adustus sp. nov., isolated from bamboo forest soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 3573–3578. [Google Scholar] [CrossRef]
- Luo, X.; Sun, Y.; Xie, S.; Wan, C.; Zhang, L. Streptomyces indoligenes sp. nov., isolated from rhizosphere soil of Populus euphratica. Int. J. Syst. Evol. Microbiol. 2016, 66, 2424–2428. [Google Scholar] [CrossRef]
- Cheng, C.; Li, Y.Q.; Asem, M.D.; Lu, C.Y.; Shi, X.H.; Chu, X.; Zhang, W.Q.; Di An, D.; Li, W.J. Streptomyces xinjiangensis sp. nov., an actinomycete isolated from Lop Nur region. Arch. Microbiol. 2016, 198, 785–791. [Google Scholar] [CrossRef]
- She, W.; Sun, Z.; Yi, L.; Zhao, S.; Liang, Y. Streptomyces alfalfae sp. nov. and comparisons with its closest taxa Streptomyces silaceus, Streptomyces flavofungini and Streptomyces intermedius. Int. J. Syst. Evol. Microbiol. 2016, 66, 44–49. [Google Scholar] [CrossRef]
- Sujarit, K.; Kudo, T.; Ohkuma, M.; Pathom-Aree, W.; Lumyong, S. Streptomyces palmae sp. nov., isolated from oil palm (Elaeis guineensis) rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 3983–3988. [Google Scholar] [CrossRef]
- Zhao, S.; Ye, L.; Liu, C.; Abagana, A.Y.; Zheng, W.; Sun, P.; Li, J.; Xiang, W.; Wang, X. Streptomyces gamaensis sp. nov., a novel actinomycete with antifungal activity isolated from soil in Gama, Chad. Antonie Van Leeuwenhoek 2017, 110, 471–477. [Google Scholar] [CrossRef] [PubMed]
- Sripreechasak, P.; Tamura, T.; Shibata, C.; Suwanborirux, K.; Tanasupawat, S. Streptomyces andamanensis sp. nov., isolated from soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 2030–2034. [Google Scholar] [CrossRef] [PubMed]
- Zhang, B.; Tang, S.; Chen, X.; Zhang, L.; Zhang, G.; Zhang, W.; Liu, G.; Chen, T.; Li, S.; Dyson, P. Streptomyces lacrimifluminis sp. nov., a novel actinobacterium that produces antibacterial compounds, isolated from soil. Int. J. Syst. Evol. Microbiol. 2016, 66, 4981–4986. [Google Scholar] [CrossRef] [PubMed]
- Nguyen, T.M.; Kim, J. Streptomyces olivicoloratus sp. nov., an antibiotic-producing bacterium isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3262–3270. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, L.; Liu, C.; Sun, P.; Li, J.; Li, W.; Xiang, W.; Wang, X. Streptomonospora halotolerans sp. nov., an actinomycete isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3183–3189. [Google Scholar] [CrossRef]
- Zhao, J.; Guo, L.; Liu, C.; Bai, L.; Han, C.; Li, J.; Xiang, W.; Wang, X. Streptomyces tyrosinilyticus sp. nov., a novel actinomycete isolated from river sediment. Int. J. Syst. Evol. Microbiol. 2015, 65, 3091–3096. [Google Scholar] [CrossRef]
- Guan, X.; Liu, C.; Zhao, J.; Fang, B.; Zhang, Y.; Li, L.; Jin, P.; Wang, X.; Xiang, W. Streptomyces maoxianensis sp. nov., a novel actinomycete isolated from soil in Maoxian, China. Antonie Van Leeuwenhoek 2015, 107, 1119–1126. [Google Scholar] [CrossRef]
- Guo, X.; Zhang, L.; Li, X.; Gao, Y.; Ruan, J.; Huang, Y. Streptomyces rubrisoli sp. nov., neutrotolerant acidophilic actinomycetes isolated from red soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3103–3108. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kim, J. Streptomyces gilvifuscus sp. nov., an actinomycete that produces antibacterial compounds isolated from soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3493–3500. [Google Scholar] [CrossRef]
- Zhang, B.H.; Cheng, J.; Chen, W.; Li, H.Q.; Yang, J.Y.; Park, D.J.; Kim, C.J.; Shen, R.; Duan, Y.Q.; Li, W.J. Streptomyces lushanensis sp. nov., a novel actinomycete with anti-cyanobacterial activity. J. Antibiot. 2015, 68, 5–8. [Google Scholar] [CrossRef]
- Nguyen, T.M.; Kim, J. Streptomyces bambusae sp. nov., Showing Antifungal and Antibacterial Activities, Isolated from Bamboo (Bambuseae) Rhizosphere Soil Using a Modified Culture Method. Curr. Microbiol. 2015, 71, 658–668. [Google Scholar] [CrossRef] [PubMed]
- Lee, H.J.; Whang, K.S. Streptomyces sasae sp. nov., isolated from bamboo (Sasa borealis) rhizosphere soil. Int. J. Syst. Evol. Microbiol. 2015, 65, 3547–3551. [Google Scholar] [CrossRef] [PubMed]
- Williams, S.T.; Shameemullah, M.; Watson, E.T.; Mayfield, C.I. Studies on the ecology of actinomycetes in soil—VI. The influence of moisture tension on growth and survival. Soil Biol. Biochem. 1972, 4, 215–225. [Google Scholar] [CrossRef]
- Dharmaraj, S.; Ashokkumar, B.; Dhevendaran, K. Isolation of marine Streptomyces and the evaluation of its bioactive molecules. Afr. J. Microbiol. Res. 2010, 4, 240–248. [Google Scholar]
- Abdelmohsen, U.R.; Bayer, K.; Hentschel, U. Diversity, abundance and natural products of marine sponge-associated actinomycetes. Nat. Prod. Rep. 2014, 31, 381–399. [Google Scholar] [CrossRef]
- Moran, M.A.; Rutherford, L.T.; Hodson, R.E. Evidence for indegenous Streptomyces population in a marine environment determined with a 16S rRNA probe. Appl. Environ. Microbiol. 1995, 61, 3695–3700. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, K.; Gupta, R.K. Diversity and isolation of rare actinomycetes: An overview. Crit. Rev. Microbiol. 2013, 39, 256–294. [Google Scholar] [CrossRef]
- Bhawsar, S. Scuba Diving for Marine Microbiologists. Available online: https://www.biotecharticles.com/Careers-Article/Scuba-Diving-For-Marine-Microbiologists-1400.html (accessed on 1 September 2021).
- Dharmaraj, S. Marine Streptomyces as a novel source of bioactive substances. World J. Microbiol. Biotechnol. 2010, 26, 2123–2139. [Google Scholar] [CrossRef]
- Maldonado, L.A.; Fenical, W.; Jensen, P.R.; Kauffman, C.A.; Mincer, T.J.; Ward, A.C.; Bull, A.T.; Goodfellow, M. Salinispora arenicola gen. nov., sp. nov. and Salinispora tropica sp. nov., obligate marine actinomycetes belonging to the family Micromonosporaceae. Int. J. Syst. Evol. Microbiol. 2005, 55 Pt 5, 1759–1766. [Google Scholar] [CrossRef] [Green Version]
- Mudryk, Z.; Donderski, W. Effect of sodium chloride on the metabolic activity of halophillic bacteria isolated from the lake Gardno estuary. Estuaries 1991, 14, 495–498. [Google Scholar] [CrossRef]
- Barka, E.A.; Vatsa, P.; Sanchez, L.; Gaveau-Vaillant, N.; Jacquard, C.; Meier-Kolthoff, J.P.; Klenk, H.P.; Clement, C.; Ouhdouch, Y.; van Wezel, G.P. Taxonomy, Physiology, and Natural Products of Actinobacteria. Microbiol. Mol. Biol. Rev. 2016, 80, 1–43. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Blockley, A.; Elliott, D.; Roberts, A.; Sweet, M. Symbiotic Microbes from Marine Invertebrates: Driving a New Era of Natural Product Drug Discovery. Diversity 2017, 9, 49. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Kong, F.; Zhou, S.; Huang, D.; Zheng, J.; Zhu, W. Streptomyces tirandamycinicus sp. nov., a Novel Marine Sponge-Derived Actinobacterium with Antibacterial Potential against Streptococcus agalactiae. Front. Microbiol. 2019, 10, 482. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Huang, X.; Zhou, S.; Huang, D.; Chen, J.; Zhu, W. Streptomyces spongiicola sp. nov., an actinomycete derived from marine sponge. Int. J. Syst. Evol. Microbiol. 2016, 66, 738–743. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Dhaneesha, M.; Hasin, O.; Sivakumar, K.C.; Ravinesh, R.; Naman, C.B.; Carmeli, S.; Sajeevan, T.P. DNA Binding and Molecular Dynamic Studies of Polycyclic Tetramate Macrolactams (PTM) with Potential Anticancer Activity Isolated from a Sponge-Associated Streptomyces zhaozhouensis subsp. mycale subsp. nov. Mar. Biotechnol. 2019, 21, 124–137. [Google Scholar] [CrossRef]
- Li, L.; Wang, J.; Zhou, Y.J.; Lin, H.W.; Lu, Y.H. Streptomyces reniochalinae sp. nov. and Streptomyces diacarni sp. nov., from marine sponges. Int. J. Syst. Evol. Microbiol. 2019, 69, 99–104. [Google Scholar] [CrossRef]
- Harunari, E.; Hamada, M.; Shibata, C.; Tamura, T.; Komaki, H.; Imada, C.; Igarashi, Y. Streptomyces hyaluromycini sp. nov., isolated from a tunicate (Molgula manhattensis). J. Antibiot. 2016, 69, 159–163. [Google Scholar] [CrossRef]
- Silva, F.S.; Souza, D.T.; Zucchi, T.D.; Pansa, C.C.; de Figueiredo Vasconcellos, R.L.; Crevelin, E.J.; de Moraes, L.A.; Melo, I.S. Streptomyces atlanticus sp. nov., a novel actinomycete isolated from marine sponge Aplysina fulva (Pallas, 1766). Antonie Van Leeuwenhoek 2016, 109, 1467–1474. [Google Scholar] [CrossRef]
- Ng, Y.K.; Hodson, M.P.; Hewavitharana, A.K.; Bose, U.; Shaw, P.N.; Fuerst, J.A. Effects of salinity on antibiotic production in sponge-derived Salinispora actinobacteria. J. Appl. Microbiol. 2014, 117, 109–125. [Google Scholar] [CrossRef]
- Davidson, P.M.; Taylor, T.M.; Schmidt, S.E. Chemical Preservatives and Natural Antimicrobial Compounds. In Food Microbiology: Fundamentals and Frontiers, 4th ed.; American Society for Microbiology Press: Washington, DC, USA, 2012; pp. 765–801. [Google Scholar]
- Pang, H.; Xin, X.; He, J.; Cui, B.; Guo, D.; Liu, S.; Yan, Z.; Liu, C.; Wang, X.; Nan, J. Effect of NaCl Concentration on Microbiological Properties in NaCl Assistant Anaerobic Fermentation: Hydrolase Activity and Microbial Community Distribution. Front. Microbiol. 2020, 11, 589222. [Google Scholar] [CrossRef]
- Chipley, J.R. Sodium Benzoate and Benzoic Acid. In Antimicrobials in Food; Davidson, P.M., Sofos, J.N., Branen, A.L., Eds.; Tailor and Francis Group: Boca Raton, FL, USA, 2005; pp. 11–48. [Google Scholar]
- Pan, H.Q.; Cheng, J.; Zhang, D.F.; Yu, S.Y.; Khieu, T.N.; Son, C.K.; Jiang, Z.; Hu, J.C.; Li, W.J. Streptomyces bohaiensis sp. nov., a novel actinomycete isolated from Scomberomorus niphonius in the Bohai Sea. J. Antibiot. 2015, 68, 246–252. [Google Scholar] [CrossRef] [PubMed]
- Stach, J.E.; Maldonado, L.A.; Masson, D.G.; Ward, A.C.; Goodfellow, M.; Bull, A.T. Statistical approaches for estimating actinobacterial diversity in marine sediments. Appl. Environ. Microbiol. 2003, 69, 6189–6200. [Google Scholar] [CrossRef] [Green Version]
- Tamelander, T.; Spilling, K.; Winder, M. Organic matter export to the seafloor in the Baltic Sea: Drivers of change and future projections. Ambio 2017, 46, 842–851. [Google Scholar] [CrossRef] [PubMed]
- Snelgrove, P.V.R. Getting to the bottom of Marine biodiversity: Sedimentary habitats. BioScience 1999, 49, 129–138. [Google Scholar] [CrossRef] [Green Version]
- Hoshino, T.; Doi, H.; Uramoto, G.I.; Wormer, L.; Adhikari, R.R.; Xiao, N.; Morono, Y.; D’Hondt, S.; Hinrichs, K.U.; Inagaki, F. Global diversity of microbial communities in marine sediment. Proc. Natl. Acad. Sci. USA 2020, 117, 27587–27597. [Google Scholar] [CrossRef]
- Kallmeyer, J.; Pockalny, R.; Adhikari, R.R.; Smith, D.C.; D’Hondt, S. Global distribution of microbial abundance and biomass in subseafloor sediment. Proc. Natl. Acad. Sci. USA 2012, 109, 16213–16216. [Google Scholar] [CrossRef] [Green Version]
- Ma, G.Q.; Xia, Z.F.; Wan, C.X.; Zhang, Y.; Luo, X.X.; Zhang, L.L. Streptomyces kalpinensis sp. nov., an actinomycete isolated from a salt water beach. Int. J. Syst. Evol. Microbiol. 2017, 67, 4892–4896. [Google Scholar] [CrossRef]
- Iniyan, A.M.; Wink, J.; Landwehr, W.; Ramprasad, E.V.V.; Sasikala, C.; Ramana, C.V.; Schumann, P.; Sproer, C.; Bunk, B.; Joseph, F.R.S.; et al. Streptomyces marianii sp. nov., a novel marine actinomycete from southern coast of India. J. Antibiot. 2021, 74, 59–69. [Google Scholar] [CrossRef]
- Zhang, B.; Tang, S.; Chen, X.; Zhang, G.; Zhang, W.; Chen, T.; Liu, G.; Li, S.; Dos Santos, L.T.; Castro, H.C.; et al. Streptomyces qaidamensis sp. nov., isolated from sand in the Qaidam Basin, China. J. Antibiot. 2018, 71, 880–886. [Google Scholar] [CrossRef] [Green Version]
- Biswas, K.; Choudhury, J.D.; Mahansaria, R.; Saha, M.; Mukherjee, J. Streptomyces euryhalinus sp. nov., a new actinomycete isolated from a mangrove forest. J. Antibiot. 2017, 70, 747–753. [Google Scholar] [CrossRef]
- Huang, H.; Liu, M.; Zhong, W.; Mo, K.; Zhu, J.; Zou, X.; Hu, Y.; Bao, S. Streptomyces caeni sp. nov., isolated from mangrove mud. Int. J. Syst. Evol. Microbiol. 2018, 68, 3080–3083. [Google Scholar] [CrossRef] [PubMed]
- Das, S.; Mangwani, N. Ocean acidification and marine microorganisms: Responses and consequences. Oceanologia 2015, 57, 349–361. [Google Scholar] [CrossRef] [Green Version]
- Law, J.W.; Ser, H.L.; Duangjai, A.; Saokaew, S.; Bukhari, S.I.; Khan, T.M.; Ab Mutalib, N.S.; Chan, K.G.; Goh, B.H.; Lee, L.H. Streptomyces colonosanans sp. nov., A Novel Actinobacterium Isolated from Malaysia Mangrove Soil Exhibiting Antioxidative Activity and Cytotoxic Potential against Human Colon Cancer Cell Lines. Front. Microbiol. 2017, 8, 877. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.L.; Palanisamy, U.D.; Yin, W.F.; Chan, K.G.; Goh, B.H.; Lee, L.H. Streptomyces malaysiense sp. nov.: A novel Malaysian mangrove soil actinobacterium with antioxidative activity and cytotoxic potential against human cancer cell lines. Sci. Rep. 2016, 6, 24247. [Google Scholar] [CrossRef] [Green Version]
- Ser, H.L.; Zainal, N.; Palanisamy, U.D.; Goh, B.H.; Yin, W.F.; Chan, K.G.; Lee, L.H. Streptomyces gilvigriseus sp. nov., a novel actinobacterium isolated from mangrove forest soil. Antonie Van Leeuwenhoek 2015, 107, 1369–1378. [Google Scholar] [CrossRef]
- Veyisoglu, A.; Cetin, D.; Inan Bektas, K.; Guven, K.; Sahin, N. Streptomyces ovatisporus sp. nov., isolated from deep marine sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 4856–4863. [Google Scholar] [CrossRef]
- Terahara, T.; Naemura, T.; Nampo, Y.; Kobayashi, T.; Imada, C.; Hamada, M.; Tamura, T. Streptomyces otsuchiensis sp. nov., a biosurfactant-producing actinobacterium isolated from marine sediment. Int. J. Syst. Evol. Microbiol. 2019, 69, 3740–3744. [Google Scholar] [CrossRef]
- Zainal, N.; Ser, H.L.; Yin, W.F.; Tee, K.K.; Lee, L.H.; Chan, K.G. Streptomyces humi sp. nov., an actinobacterium isolated from soil of a mangrove forest. Antonie Van Leeuwenhoek 2016, 109, 467–474. [Google Scholar] [CrossRef]
- Phongsopitanun, W.; Kudo, T.; Ohkuma, M.; Pittayakhajonwut, P.; Suwanborirux, K.; Tanasupawat, S. Streptomyces verrucosisporus sp. nov., isolated from marine sediments. Int. J. Syst. Evol. Microbiol. 2016, 66, 3607–3613. [Google Scholar] [CrossRef]
- Ray, L.; Mishra, S.R.; Panda, A.N.; Das, S.; Rastogi, G.; Pattanaik, A.K.; Adhya, T.K.; Suar, M.; Raina, V. Streptomyces chitinivorans sp. nov., a chitinolytic strain isolated from estuarine lake sediment. Int. J. Syst. Evol. Microbiol. 2016, 66, 3241–3248. [Google Scholar] [CrossRef]
- Ser, H.L.; Palanisamy, U.D.; Yin, W.F.; Abd Malek, S.N.; Chan, K.G.; Goh, B.H.; Lee, L.H. Presence of antioxidative agent, Pyrrolo[1,2-a]pyrazine-1,4-dione, hexahydro- in newly isolated Streptomyces mangrovisoli sp. nov. Front. Microbiol. 2015, 6, 854. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Yousif, G.; Busarakam, K.; Kim, B.Y.; Goodfellow, M. Streptomyces mangrovi sp. nov., isolated from mangrove forest sediment. Antonie Van Leeuwenhoek 2015, 108, 783–791. [Google Scholar] [CrossRef] [PubMed]
- Chen, C.; Ye, Y.; Wang, R.; Zhang, Y.; Wu, C.; Debnath, S.C.; Ma, Z.; Wang, J.; Wu, M. Streptomyces nigra sp. nov. Is a Novel Actinobacterium Isolated From Mangrove Soil and Exerts a Potent Antitumor Activity In Vitro. Front. Microbiol. 2018, 9, 1587. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ma, G.Q.; Xia, Z.F.; Zhang, Y.; Wan, C.X.; Luo, X.X.; Zhang, L.L. Streptomyces litoralis sp. nov., isolated from a salt water beach. Int. J. Syst. Evol. Microbiol. 2016, 66, 5051–5055. [Google Scholar] [CrossRef] [PubMed]
- Ser, H.L.; Tan, L.T.; Palanisamy, U.D.; Abd Malek, S.N.; Yin, W.F.; Chan, K.G.; Goh, B.H.; Lee, L.H. Streptomyces antioxidans sp. nov., a Novel Mangrove Soil Actinobacterium with Antioxidative and Neuroprotective Potentials. Front. Microbiol. 2016, 7, 899. [Google Scholar] [CrossRef] [Green Version]
- Chater, K.F. Recent advances in understanding Streptomyces. F1000Research 2016, 5, 2795. [Google Scholar] [CrossRef] [Green Version]
- Waksman, S.A.; Woodruff, H.B. Bacteriostatic and Bactericidal Substances Produced by a Soil Actinomyces. Proc. Soc. Exp. Biol. Med. 1940, 45, 609–614. [Google Scholar] [CrossRef]
- Waksman, S.A.; Woodruff, H.B. Selective antibiotic action of various substances of microbial origin. J. Bacteriol. 1942, 44, 373–384. [Google Scholar] [CrossRef] [Green Version]
- Schatz, A.; Waksman, S.S. Effect of Streptomycin and Other Antibiotic Substances upon Mycobacterium tuberculosis and Related Organisms. Proc. Soc. Exp. Biol. Med. 2015, 57, 244–248. [Google Scholar] [CrossRef]
- Manteca, A.; Sanchez, J. Streptomyces development in colonies and soils. Appl. Environ. Microbiol. 2009, 75, 2920–2924. [Google Scholar] [CrossRef] [Green Version]
- Tiwari, K.; Gupta, R.K. Rare actinomycetes: A potential storehouse for novel antibiotics. Crit. Rev. Biotechnol. 2012, 32, 108–132. [Google Scholar] [CrossRef] [PubMed]
- Nair, S.; Abraham, J. Natural Products from Actinobacteria for Drug Discovery. In Advances in Pharmaceutical Biotechnology; Springer: Singapore, 2020; pp. 333–363. [Google Scholar]
- Voser, T.M.; Campell, M.D.; Carroll, A.R. How diffrent are rare marine microbial natural products compared to their terrestrial counterparts. Nat. Prod. Rep. 2022, 39, 7–19. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Hu, X.; Li, L.; Hu, X.; Wang, J.; Hu, X.; Liu, H.; Yu, L.; You, X.; Jiang, B.; et al. 1-hydroxy-7-oxolavanducyanin and Delta(7″,8″)-6″-hydroxynaphthomevalin from Streptomyces sp. CPCC 203577. J. Antibiot. 2020, 73, 324–328. [Google Scholar] [CrossRef]
- Perez-Bonilla, M.; Oves-Costales, D.; Gonzalez, I.; de la Cruz, M.; Martin, J.; Vicente, F.; Genilloud, O.; Reyes, F. Krisynomycins, Imipenem Potentiators against Methicillin-Resistant Staphylococcus aureus, Produced by Streptomyces canus. J. Nat. Prod. 2020, 83, 2597–2606. [Google Scholar] [CrossRef] [PubMed]
- Menegatti, C.; Lourenzon, V.B.; Rodriguez-Hernandez, D.; da Paixao Melo, W.G.; Ferreira, L.L.G.; Andricopulo, A.D.; do Nascimento, F.S.; Pupo, M.T. Meliponamycins: Antimicrobials from Stingless Bee-Associated Streptomyces sp. J. Nat. Prod. 2020, 83, 610–616. [Google Scholar] [CrossRef]
- Maiti, P.K.; Das, S.; Sahoo, P.; Mandal, S. Streptomyces sp. SM01 isolated from Indian soil produces a novel antibiotic picolinamycin effective against multi drug resistant bacterial strains. Sci. Rep. 2020, 10, 10092. [Google Scholar] [CrossRef]
- Wang, X.; Elshahawi, S.I.; Ponomareva, L.V.; Ye, Q.; Liu, Y.; Copley, G.C.; Hower, J.C.; Hatcher, B.E.; Kharel, M.K.; Van Lanen, S.G.; et al. Structure Determination, Functional Characterization, and Biosynthetic Implications of Nybomycin Metabolites from a Mining Reclamation Site-Associated Streptomyces. J. Nat. Prod. 2019, 82, 3469–3476. [Google Scholar] [CrossRef]
- Kaweewan, I.; Hemmi, H.; Komaki, H.; Kodani, S. Isolation and structure determination of a new antibacterial peptide pentaminomycin C from Streptomyces cacaoi subsp. cacaoi. J. Antibiot. 2020, 73, 224–229. [Google Scholar] [CrossRef]
- Arora, N.; Kumar, S.; Satti, N.K.; Ali, A.; Gupta, P.; Katoch, M. A strain of Streptomyces sp. isolated from rhizospheric soil of Crataegus oxycantha producing nalidixic acid, a synthetic antibiotic. J. Appl. Microbiol. 2018, 124, 1393–1400. [Google Scholar] [CrossRef]
- Yang, Z.; Shao, L.; Wang, M.; Rao, M.; Ge, M.; Xu, Y. Two novel quinomycins discovered by UPLC-MS from Streptomyces sp. HCCB11876. J. Antibiot. 2019, 72, 164–168. [Google Scholar] [CrossRef]
- Abbas, M.; Elshahawi, S.I.; Wang, X.; Ponomareva, L.V.; Sajid, I.; Shaaban, K.A.; Thorson, J.S. Puromycins B-E, Naturally Occurring Amino-Nucleosides Produced by the Himalayan Isolate Streptomyces sp. PU-14G. J. Nat. Prod. 2018, 81, 2560–2566. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Elshahawi, S.I.; Cai, W.; Zhang, Y.; Ponomareva, L.V.; Chen, X.; Copley, G.C.; Hower, J.C.; Zhan, C.G.; Parkin, S.; et al. Bi- and Tetracyclic Spirotetronates from the Coal Mine Fire Isolate Streptomyces sp. LC-6-2. J. Nat. Prod. 2017, 80, 1141–1149. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tian, H.; Shafi, J.; Ji, M.; Bi, Y.; Yu, Z. Antimicrobial Metabolites from Streptomyces sp. SN0280. J. Nat. Prod. 2017, 80, 1015–1019. [Google Scholar] [CrossRef]
- Abdelkader, M.S.A.; Philippon, T.; Asenjo, J.A.; Bull, A.T.; Goodfellow, M.; Ebel, R.; Jaspars, M.; Rateb, M.E. Asenjonamides A-C, antibacterial metabolites isolated from Streptomyces asenjonii strain KNN 42.f from an extreme-hyper arid Atacama Desert soil. J. Antibiot. 2018, 71, 425–431. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Park, H.B.; Park, J.S.; Lee, S.I.; Shin, B.; Oh, D.C.; Kwon, H.C. Gordonic Acid, a Polyketide Glycoside Derived from Bacterial Coculture of Streptomyces and Gordonia Species. J. Nat. Prod. 2017, 80, 2542–2546. [Google Scholar] [CrossRef]
- Son, S.; Hong, Y.S.; Jang, M.; Heo, K.T.; Lee, B.; Jang, J.P.; Kim, J.W.; Ryoo, I.J.; Kim, W.G.; Ko, S.K.; et al. Genomics-Driven Discovery of Chlorinated Cyclic Hexapeptides Ulleungmycins A and B from a Streptomyces Species. J. Nat. Prod. 2017, 80, 3025–3031. [Google Scholar] [CrossRef]
- Wu, C.; Du, C.; Ichinose, K.; Choi, Y.H.; van Wezel, G.P. Discovery of C-Glycosylpyranonaphthoquinones in Streptomyces sp. MBT76 by a Combined NMR-Based Metabolomics and Bioinformatics Workflow. J. Nat. Prod. 2017, 80, 269–277. [Google Scholar] [CrossRef] [Green Version]
- Cai, W.; Wang, X.; Elshahawi, S.I.; Ponomareva, L.V.; Liu, X.; McErlean, M.R.; Cui, Z.; Arlinghaus, A.L.; Thorson, J.S.; Van Lanen, S.G. Antibacterial and Cytotoxic Actinomycins Y6-Y9 and Zp from Streptomyces sp. Strain Go-GS12. J. Nat. Prod. 2016, 79, 2731–2739. [Google Scholar] [CrossRef] [Green Version]
- Yekkour, A.; Meklat, A.; Bijani, C.; Toumatia, O.; Errakhi, R.; Lebrihi, A.; Mathieu, F.; Zitouni, A.; Sabaou, N. A novel hydroxamic acid-containing antibiotic produced by a Saharan soil-living Streptomyces strain. Lett. Appl. Microbiol. 2015, 60, 589–596. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Rateb, M.E.; Teng, Q.; Yang, D.; Rudolf, J.D.; Zhu, X.; Huang, Y.; Zhao, L.X.; Jiang, Y.; Li, X.; et al. Angucyclines and Angucyclinones from Streptomyces sp. CB01913 Featuring C-Ring Cleavage and Expansion. J. Nat. Prod. 2015, 78, 2471–2480. [Google Scholar] [CrossRef] [Green Version]
- Noomnual, S.; Thasana, N.; Sungkeeree, P.; Mongkolsuk, S.; Loprasert, S. Streptanoate, a new anticancer butanoate from Streptomyces sp. DC3. J. Antibiot. 2016, 69, 124–127. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Z.K.; Guo, L.; Chen, C.; Liu, S.W.; Zhang, L.; Dai, S.J.; He, Q.Y.; You, X.F.; Hu, X.X.; Tuo, L.; et al. Xiakemycin A, a novel pyranonaphthoquinone antibiotic, produced by the Streptomyces sp. CC8-201 from the soil of a karst cave. J. Antibiot. 2015, 68, 771–774. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sheng, Y.; Lam, P.W.; Shahab, S.; Santosa, D.A.; Proteau, P.J.; Zabriskie, T.M.; Mahmud, T. Identification of Elaiophylin Skeletal Variants from the Indonesian Streptomyces sp. ICBB 9297. J. Nat. Prod. 2015, 78, 2768–2775. [Google Scholar] [CrossRef] [PubMed]
- Wu, C.; Du, C.; Gubbens, J.; Choi, Y.H.; van Wezel, G.P. Metabolomics-Driven Discovery of a Prenylated Isatin Antibiotic Produced by Streptomyces Species MBT28. J. Nat. Prod. 2015, 78, 2355–2363. [Google Scholar] [CrossRef] [PubMed]
- Kim, M.C.; Cullum, R.; Hebishy, A.M.S.; Mohamed, H.A.; Faraag, A.H.I.; Salah, N.M.; Abdelfattah, M.S.; Fenical, W. Mersaquinone, A New Tetracene Derivative from the Marine-Derived Streptomyces sp. EG1 Exhibiting Activity against Methicillin-Resistant Staphylococcus aureus (MRSA). Antibiotics 2020, 9, 252. [Google Scholar] [CrossRef]
- Song, Y.; Yang, J.; Yu, J.; Li, J.; Yuan, J.; Wong, N.K.; Ju, J. Chlorinated bis-indole alkaloids from deep-sea derived Streptomyces sp. SCSIO 11791 with antibacterial and cytotoxic activities. J. Antibiot. 2020, 73, 542–547. [Google Scholar] [CrossRef]
- Zhang, D.; Yi, W.; Ge, H.; Zhang, Z.; Wu, B. Bioactive Streptoglutarimides A–J from the Marine-Derived Streptomyces sp. ZZ741. J. Nat. Prod. 2019, 82, 2800–2808. [Google Scholar] [CrossRef]
- Chen, S.; Zhang, D.; Chen, M.; Zhang, Z.; Lian, X.Y. A rare diketopiperazine glycoside from marine-sourced Streptomyces sp. ZZ446. Nat. Prod. Res. 2020, 34, 1046–1050. [Google Scholar] [CrossRef]
- Jiang, Y.J.; Zhang, D.S.; Zhang, H.J.; Li, J.Q.; Ding, W.J.; Xu, C.D.; Ma, Z.J. Medermycin-Type Naphthoquinones from the Marine-Derived Streptomyces sp. XMA39. J. Nat. Prod. 2018, 81, 2120–2124. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, M.; Wu, C.; Tan, Y.; Li, J.; Hao, X.; Duan, Y.; Guan, Y.; Shang, X.; Wang, Y.; et al. Identification and Proposed Relative and Absolute Configurations of Niphimycins C-E from the Marine-Derived Streptomyces sp. IMB7-145 by Genomic Analysis. J. Nat. Prod. 2018, 81, 178–187. [Google Scholar] [CrossRef]
- Kitani, S.; Ueguchi, T.; Igarashi, Y.; Leetanasaksakul, K.; Thamchaipenet, A.; Nihira, T. Rakicidin F, a new antibacterial cyclic depsipeptide from a marine sponge-derived Streptomyces sp. J. Antibiot. 2017, 71, 139–141. [Google Scholar] [CrossRef] [PubMed]
- Iniyan, A.M.; Sudarman, E.; Wink, J.; Kannan, R.R.; Vincent, S.G.P. Ala-geninthiocin, a new broad spectrum thiopeptide antibiotic, produced by a marine Streptomyces sp. ICN19. J. Antibiot. 2019, 72, 99–105. [Google Scholar] [CrossRef]
- Takehana, Y.; Umekita, M.; Hatano, M.; Kato, C.; Sawa, R.; Igarashi, M. Fradiamine A, a new siderophore from the deep-sea actinomycete Streptomyces fradiae MM456M-mF7. J. Antibiot. 2017, 70, 611–615. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brana, A.F.; Sarmiento-Vizcaino, A.; Osset, M.; Perez-Victoria, I.; Martin, J.; de Pedro, N.; de la Cruz, M.; Diaz, C.; Vicente, F.; Reyes, F.; et al. Lobophorin K, a New Natural Product with Cytotoxic Activity Produced by Streptomyces sp. M-207 Associated with the Deep-Sea Coral Lophelia pertusa. Mar. Drugs 2017, 15, 144. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Nong, X.H.; Wei, X.Y.; Qi, S.H. Pteridic acids C-G spirocyclic polyketides from the marine-derived Streptomyces sp. SCSGAA 0027. J. Antibiot. 2017, 70, 1047–1052. [Google Scholar] [CrossRef]
- Wang, Q.; Zhang, Y.; Wang, M.; Tan, Y.; Hu, X.; He, H.; Xiao, C.; You, X.; Wang, Y.; Gan, M. Neo-actinomycins A and B, natural actinomycins bearing the 5H-oxazolo[4,5-b]phenoxazine chromophore, from the marine-derived Streptomyces sp. IMB094. Sci. Rep. 2017, 7, 3591. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Paulus, C.; Rebets, Y.; Tokovenko, B.; Nadmid, S.; Terekhova, L.P.; Myronovskyi, M.; Zotchev, S.B.; Ruckert, C.; Braig, S.; Zahler, S.; et al. New natural products identified by combined genomics-metabolomics profiling of marine Streptomyces sp. MP131-18. Sci. Rep. 2017, 7, 42382. [Google Scholar] [CrossRef]
- Sun, C.; Liu, Z.; Zhu, X.; Fan, Z.; Huang, X.; Wu, Q.; Zheng, X.; Qin, X.; Zhang, T.; Zhang, H.; et al. Antitubercular Ilamycins from Marine-Derived Streptomyces atratus SCSIO ZH16 DeltailaR. J. Nat. Prod. 2020, 83, 1646–1657. [Google Scholar] [CrossRef]
- Xu, J.H.; Gu, K.B.; Zhang, D.J.; Li, Y.G.; Tian, L. Ghanamycins A and B, two novel gamma-butyrolactones from marine-derived Streptomyces ghanaensis TXC6-16. J. Antibiot. 2017, 70, 733–736. [Google Scholar] [CrossRef]
- Zhou, S.Y.; Zou, Y.L.; Wang, G.W.; Liao, Z.H.; Chen, M. Two new compounds from a marine-derived Streptomyces sp. J. Asian Nat. Prod. Res. 2017, 19, 1172–1176. [Google Scholar] [CrossRef]
- Wang, C.X.; Ding, R.; Jiang, S.T.; Tang, J.S.; Hu, D.; Chen, G.D.; Lin, F.; Hong, K.; Yao, X.S.; Gao, H. Aldgamycins J–O, 16-Membered Macrolides with a Branched Octose Unit from Streptomycetes sp. and Their Antibacterial Activities. J. Nat. Prod. 2016, 79, 2446–2454. [Google Scholar] [CrossRef] [PubMed]
- Wibowo, M.; Gotfredsen, C.H.; Sassetti, E.; Melchiorsen, J.; Clausen, M.H.; Gram, L.; Ding, L. Azodyrecins A–C: Azoxides from a Soil-Derived Streptomyces Species. J. Nat. Prod. 2020, 83, 3519–3525. [Google Scholar] [CrossRef] [PubMed]
- Lee, B.; Son, S.; Lee, J.K.; Jang, M.; Heo, K.T.; Ko, S.K.; Park, D.J.; Park, C.S.; Kim, C.J.; Ahn, J.S.; et al. Isolation of new streptimidone derivatives, glutarimide antibiotics from Streptomyces sp. W3002 using LC-MS-guided screening. J. Antibiot. 2020, 73, 184–188. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Abbas, M.; Zhang, Y.; Elshahawi, S.I.; Ponomareva, L.V.; Cui, Z.; Van Lanen, S.G.; Sajid, I.; Voss, S.R.; Shaaban, K.A.; et al. Baraphenazines A–G, Divergent Fused Phenazine-Based Metabolites from a Himalayan Streptomyces. J. Nat. Prod. 2019, 82, 1686–1693. [Google Scholar] [CrossRef]
- He, X.; Wang, Y.; Luo, R.H.; Yang, L.M.; Wang, L.; Guo, D.; Yang, J.; Deng, Y.; Zheng, Y.T.; Huang, S.X. Dimeric Pyranonaphthoquinone Glycosides with Anti-HIV and Cytotoxic Activities from a Soil-Derived Streptomyces. J. Nat. Prod. 2019, 82, 1813–1819. [Google Scholar] [CrossRef]
- Gui, C.; Yuan, J.; Mo, X.; Huang, H.; Zhang, S.; Gu, Y.C.; Ju, J. Cytotoxic Anthracycline Metabolites from a Recombinant Streptomyces. J. Nat. Prod. 2018, 81, 1278–1289. [Google Scholar] [CrossRef]
- Son, S.; Jang, M.; Lee, B.; Hong, Y.S.; Ko, S.K.; Jang, J.H.; Ahn, J.S. Ulleungdin, a Lasso Peptide with Cancer Cell Migration Inhibitory Activity Discovered by the Genome Mining Approach. J. Nat. Prod. 2018, 81, 2205–2211. [Google Scholar] [CrossRef]
- Lu, C.; Zhao, Y.; Jia, W.Q.; Zhang, H.; Qi, H.; Xiang, W.S.; Wang, J.D.; Wang, X.J. A new anthracycline-type metabolite from Streptomyces sp. NEAU-L3. J. Antibiot. 2017, 70, 1026–1028. [Google Scholar] [CrossRef]
- Gao, M.Y.; Qi, H.; Li, J.S.; Zhang, H.; Zhang, J.; Wang, J.D.; Xiang, W.S. A new polysubstituted cyclopentene derivative from Streptomyces sp. HS-NF-1046. J. Antibiot. 2017, 70, 216–218. [Google Scholar] [CrossRef]
- Chen, J.J.; Rateb, M.E.; Love, M.S.; Xu, Z.; Yang, D.; Zhu, X.; Huang, Y.; Zhao, L.X.; Jiang, Y.; Duan, Y.; et al. Herbicidins from Streptomyces sp. CB01388 Showing Anti-Cryptosporidium Activity. J. Nat. Prod. 2018, 81, 791–797. [Google Scholar] [CrossRef]
- Liu, S.H.; Xu, M.D.; Zhang, H.; Qi, H.; Zhang, J.; Liu, C.X.; Wang, J.D.; Xiang, W.S.; Wang, X.J. New cytotoxic spectinabilin derivative from ant-associated Streptomyces sp. 1H-GS5. J. Antibiot. 2016, 69, 128–131. [Google Scholar] [CrossRef] [PubMed]
- Sun, P.; Zhao, Q.; Wu, Z.; Zhang, W.; Liu, W. 1,19-seco-Avermectin analogues from a DeltaaveCDE mutant Streptomyces avermectinius strain. J. Nat. Prod. 2015, 78, 301–305. [Google Scholar] [CrossRef] [PubMed]
- Han, X.; Liu, Z.; Zhang, Z.; Zhang, X.; Zhu, T.; Gu, Q.; Li, W.; Che, Q.; Li, D. Geranylpyrrol A and Piericidin F from Streptomyces sp. CHQ-64 DeltardmF. J. Nat. Prod. 2017, 80, 1684–1687. [Google Scholar] [CrossRef] [PubMed]
- Kim, S.H.; Shin, Y.; Lee, S.H.; Oh, K.B.; Lee, S.K.; Shin, J.; Oh, D.C. Salternamides A–D from a Halophilic Streptomyces sp. Actinobacterium. J. Nat. Prod. 2015, 78, 836–843. [Google Scholar] [CrossRef]
- Bae, M.; An, J.S.; Hong, S.H.; Bae, E.S.; Chung, B.; Kwon, Y.; Hong, S.; Oh, K.B.; Shin, J.; Lee, S.K.; et al. Donghaecyclinones A-C: New Cytotoxic Rearranged Angucyclinones from a Volcanic Island-Derived Marine Streptomyces sp. Mar. Drugs 2020, 18, 121. [Google Scholar] [CrossRef] [Green Version]
- Wang, J.; Wang, H.; Sun, C.; Li, F.; Wu, Y.; Zhang, G.; Gu, Q.; Zhu, T.; Li, D.; Che, Q. Dimeric Tetrahydroanthracene Regioisomers and Their Monomeric Precursor Produced by Streptomyces fumigatiscleroticus HDN10255. J. Nat. Prod. 2020, 83, 2797–2802. [Google Scholar] [CrossRef]
- Ma, L.F.; Chen, M.J.; Liang, D.E.; Shi, L.M.; Ying, Y.M.; Shan, W.G.; Li, G.Q.; Zhan, Z.J. Streptomyces albogriseolus SY67903 Produces Eunicellin Diterpenoids Structurally Similar to Terpenes of the Gorgonian Muricella sibogae, the Bacterial Source. J. Nat. Prod. 2020, 83, 1641–1645. [Google Scholar] [CrossRef]
- Fukuda, T.; Nagai, K.; Kanamoto, A.; Tomoda, H. 2-Epi-anthracimycin, a new cytotoxic agent from the marine-derived actinomycete Streptomyces sp. OPMA00631. J. Antibiot. 2020, 73, 548–553. [Google Scholar] [CrossRef]
- Zhou, B.; Jiang, Y.J.; Ji, Y.Y.; Zhang, H.J.; Shen, L. Lactoquinomycin C and D, two new medermycin derivatives from the marine-derived Streptomyces sp. SS17A. Nat. Prod. Res. 2020, 34, 1213–1218. [Google Scholar] [CrossRef]
- Liu, W.; Zhang, W.; Jin, H.; Zhang, Q.; Chen, Y.; Jiang, X.; Zhang, G.; Zhang, L.; Zhang, W.; She, Z.; et al. Genome Mining of Marine-Derived Streptomyces sp. SCSIO 40010 Leads to Cytotoxic New Polycyclic Tetramate Macrolactams. Mar. Drugs 2019, 17, 663. [Google Scholar] [CrossRef] [Green Version]
- Kawahara, T.; Izumikawa, M.; Kozone, I.; Hashimoto, J.; Kagaya, N.; Koiwai, H.; Komatsu, M.; Fujie, M.; Sato, N.; Ikeda, H.; et al. Neothioviridamide, a Polythioamide Compound Produced by Heterologous Expression of a Streptomyces sp. Cryptic RiPP Biosynthetic Gene Cluster. J. Nat. Prod. 2018, 81, 264–269. [Google Scholar] [CrossRef] [PubMed]
- Chen, M.; Chai, W.; Song, T.; Ma, M.; Lian, X.Y.; Zhang, Z. Anti-glioma Natural Products Downregulating Tumor Glycolytic Enzymes from Marine Actinomycete Streptomyces sp. ZZ406. Sci. Rep. 2018, 8, 72. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chen, L.; Chai, W.; Wang, W.; Song, T.; Lian, X.Y.; Zhang, Z. Cytotoxic Bagremycins from Mangrove-Derived Streptomyces sp. Q22. J. Nat. Prod. 2017, 80, 1450–1456. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.J.; Li, J.Q.; Zhang, H.J.; Ding, W.J.; Ma, Z.J. Cyclizidine-Type Alkaloids from Streptomyces sp. HNA39. J. Nat. Prod. 2018, 81, 394–399. [Google Scholar] [CrossRef]
- Wang, J.N.; Zhang, H.J.; Li, J.Q.; Ding, W.J.; Ma, Z.J. Bioactive Indolocarbazoles from the Marine-Derived Streptomyces sp. DT-A61. J. Nat. Prod. 2018, 81, 949–956. [Google Scholar] [CrossRef] [PubMed]
- Zhang, W.; Che, Q.; Tan, H.; Qi, X.; Li, J.; Li, D.; Gu, Q.; Zhu, T.; Liu, M. Marine Streptomyces sp. derived antimycin analogues suppress HeLa cells via depletion HPV E6/E7 mediated by ROS-dependent ubiquitin-proteasome system. Sci. Rep. 2017, 7, 42180. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Che, Q.; Li, J.; Li, D.; Gu, Q.; Zhu, T. Structure and absolute configuration of drimentine I, an alkaloid from Streptomyces sp. CHQ-64. J. Antibiot. 2016, 69, 467–469. [Google Scholar] [CrossRef] [PubMed]
- Son, S.; Jang, M.; Lee, B.; Jang, J.P.; Hong, Y.S.; Kim, B.Y.; Ko, S.K.; Jang, J.H.; Ahn, J.S. A pipecolic acid-rich branched cyclic depsipeptide ulleungamide C from a Streptomyces species induces G0/G1 cell cycle arrest in promyelocytic leukemia cells. J. Antibiot. 2021, 74, 181–189. [Google Scholar] [CrossRef]
- An, J.S.; Lee, J.Y.; Kim, E.; Ahn, H.; Jang, Y.J.; Shin, B.; Hwang, S.; Shin, J.; Yoon, Y.J.; Lee, S.K.; et al. Formicolides A and B, Antioxidative and Antiangiogenic 20-Membered Macrolides from a Wood Ant Gut Bacterium. J. Nat. Prod. 2020, 83, 2776–2784. [Google Scholar] [CrossRef]
- Huo, C.; Zheng, Z.; Xu, Y.; Ding, Y.; Zheng, H.; Mu, Y.; Niu, Y.; Gao, J.; Lu, X. Naphthacemycins from a Streptomyces sp. as Protein-Tyrosine Phosphatase Inhibitors. J. Nat. Prod. 2020, 83, 1394–1399. [Google Scholar] [CrossRef]
- Song, Y.J.; Zheng, H.B.; Peng, A.H.; Ma, J.H.; Lu, D.D.; Li, X.; Zhang, H.Y.; Xie, W.D. Strepantibins A–C: Hexokinase II Inhibitors from a Mud Dauber Wasp Associated Streptomyces sp. J. Nat. Prod. 2019, 82, 1114–1119. [Google Scholar] [CrossRef]
- Yang, L.; Li, H.; Wu, P.; Mahal, A.; Xue, J.; Xu, L.; Wei, X. Dinghupeptins A–D, Chymotrypsin Inhibitory Cyclodepsipeptides Produced by a Soil-Derived Streptomyces. J. Nat. Prod. 2018, 81, 1928–1936. [Google Scholar] [CrossRef]
- Bae, M.; Moon, K.; Kim, J.; Park, H.J.; Lee, S.K.; Shin, J.; Oh, D.C. Mohangic Acids A–E, p-Aminoacetophenonic Acids from a Marine-Mudflat-Derived Streptomyces sp. J. Nat. Prod. 2016, 79, 332–339. [Google Scholar] [CrossRef]
- Cowen, L.E.; Sanglard, D.; Howard, S.J.; Rogers, P.D.; Perlin, D.S. Mechanisms of Antifungal Drug Resistance. Cold Spring Harb. Perspect. Med. 2014, 5, a019752. [Google Scholar] [CrossRef]
- Jeon, B.J.; Kim, J.D.; Han, J.W.; Kim, B.S. Antifungal activity of rimocidin and a new rimocidin derivative BU16 produced by Streptomyces mauvecolor BU16 and their effects on pepper anthracnose. J. Appl. Microbiol. 2016, 120, 1219–1228. [Google Scholar] [CrossRef] [Green Version]
- Munro, C.A. Fungal echinocandin resistance. F1000 Biol. Rep. 2010, 2, 66. [Google Scholar] [CrossRef] [Green Version]
- Zarrin, M.; Faramarzi, S. Study of Azole-Resistant and Cyp51a Gene in Aspergillus fumigatus. Open Access Maced. J. Med. Sci. 2018, 6, 747–750. [Google Scholar] [CrossRef]
- Yang, J.; Yang, Z.; Yin, Y.; Rao, M.; Liang, Y.; Ge, M. Three novel polyene macrolides isolated from cultures of Streptomyces lavenduligriseus. J. Antibiot. 2016, 69, 62–65. [Google Scholar] [CrossRef]
- Wang, W.; Song, T.; Chai, W.; Chen, L.; Chen, L.; Lian, X.Y.; Zhang, Z. Rare Polyene-polyol Macrolides from Mangrove-derived Streptomyces sp. ZQ4BG. Sci. Rep. 2017, 7, 1703. [Google Scholar] [CrossRef] [Green Version]
- Yang, F.X.; Huang, J.P.; Liu, Z.; Wang, Z.; Yang, J.; Tang, J.; Yu, Z.; Yan, Y.; Kai, G.; Huang, S.X. Benwamycins A–G, Trialkyl-Substituted Benzene Derivatives from a Soil-Derived Streptomyces. J. Nat. Prod. 2020, 83, 111–117. [Google Scholar] [CrossRef]
- Shin, B.; Ahn, S.; Noh, M.; Shin, J.; Oh, D.C. Suncheonosides A–D, Benzothioate Glycosides from a Marine-Derived Streptomyces sp. J. Nat. Prod. 2015, 78, 1390–1396. [Google Scholar] [CrossRef]
- Tang, D.; Liu, L.L.; He, Q.R.; Yan, W.; Li, D.; Gao, J.M. Ansamycins with Antiproliferative and Antineuroinflammatory Activity from Moss-Soil-Derived Streptomyces cacaoi subsp. asoensis H2S5. J. Nat. Prod. 2018, 81, 1984–1991. [Google Scholar] [CrossRef]
- Nong, X.H.; Zhang, X.Y.; Xu, X.Y.; Wang, J.; Qi, S.H. Nahuoic Acids B–E, Polyhydroxy Polyketides from the Marine-Derived Streptomyces sp. SCSGAA 0027. J. Nat. Prod. 2016, 79, 141–148. [Google Scholar] [CrossRef]
- Bauermeister, A.; Pereira, F.; Grilo, I.R.; Godinho, C.C.; Paulino, M.; Almeida, V.; Gobbo-Neto, L.; Prieto-Davo, A.; Sobral, R.G.; Lopes, N.P.; et al. Intra-clade metabolomic profiling of MAR4 Streptomyces from the Macaronesia Atlantic region reveals a source of anti-biofilm metabolites. Environ. Microbiol. 2019, 21, 1099–1112. [Google Scholar] [CrossRef]
- Hong, S.H.; Ban, Y.H.; Byun, W.S.; Kim, D.; Jang, Y.J.; An, J.S.; Shin, B.; Lee, S.K.; Shin, J.; Yoon, Y.J.; et al. Camporidines A and B: Antimetastatic and Anti-inflammatory Polyketide Alkaloids from a Gut Bacterium of Camponotus kiusiuensis. J. Nat. Prod. 2019, 82, 903–910. [Google Scholar] [CrossRef]
- Li, L.; Li, S.; Jiang, B.; Zhang, M.; Zhang, J.; Yang, B.; Li, L.; Yu, L.; Liu, H.; You, X.; et al. Isarubrolones Containing a Pyridooxazinium Unit from Streptomyces as Autophagy Activators. J. Nat. Prod. 2019, 82, 1149–1154. [Google Scholar] [CrossRef]
- Jang, J.P.; Hwang, G.J.; Jang, M.; Takahashi, S.; Ko, S.K.; Osada, H.; Jang, J.H.; Ahn, J.S. Aturanosides A and B, Glycosylated Anthraquinones with Antiangiogenic Activity from a Soil-Derived Streptomyces Species. J. Nat. Prod. 2018, 81, 2004–2009. [Google Scholar] [CrossRef]
- Igarashi, Y.; Kyoso, T.; Kim, Y.; Oikawa, T. Simamycin (5′-O-geranyluridine): A new prenylated nucleoside from Streptomyces sp. J. Antibiot. 2017, 70, 607–610. [Google Scholar] [CrossRef] [Green Version]
- Shaaban, K.A.; Saunders, M.A.; Zhang, Y.; Tran, T.; Elshahawi, S.I.; Ponomareva, L.V.; Wang, X.; Zhang, J.; Copley, G.C.; Sunkara, M.; et al. Spoxazomicin D and Oxachelin C, Potent Neuroprotective Carboxamides from the Appalachian Coal Fire-Associated Isolate Streptomyces sp. RM-14-6. J. Nat. Prod. 2017, 80, 2–11. [Google Scholar] [CrossRef] [Green Version]
- Abdelfattah, M.S.; Ishikawa, N.; Karmakar, U.K.; Yamaku, K.; Ishibashi, M. New phenazine analogues from Streptomyces sp. IFM 11694 with TRAIL resistance-overcoming activities. J. Antibiot. 2016, 69, 446–450. [Google Scholar] [CrossRef]
- Arai, M.A.; Koryudzu, K.; Ishibashi, M. Inubosins A, B, and C are acridine alkaloids isolated from a culture of Streptomyces sp. IFM 11440 with Ngn2 promoter activity. J. Nat. Prod. 2015, 78, 311–314. [Google Scholar] [CrossRef]


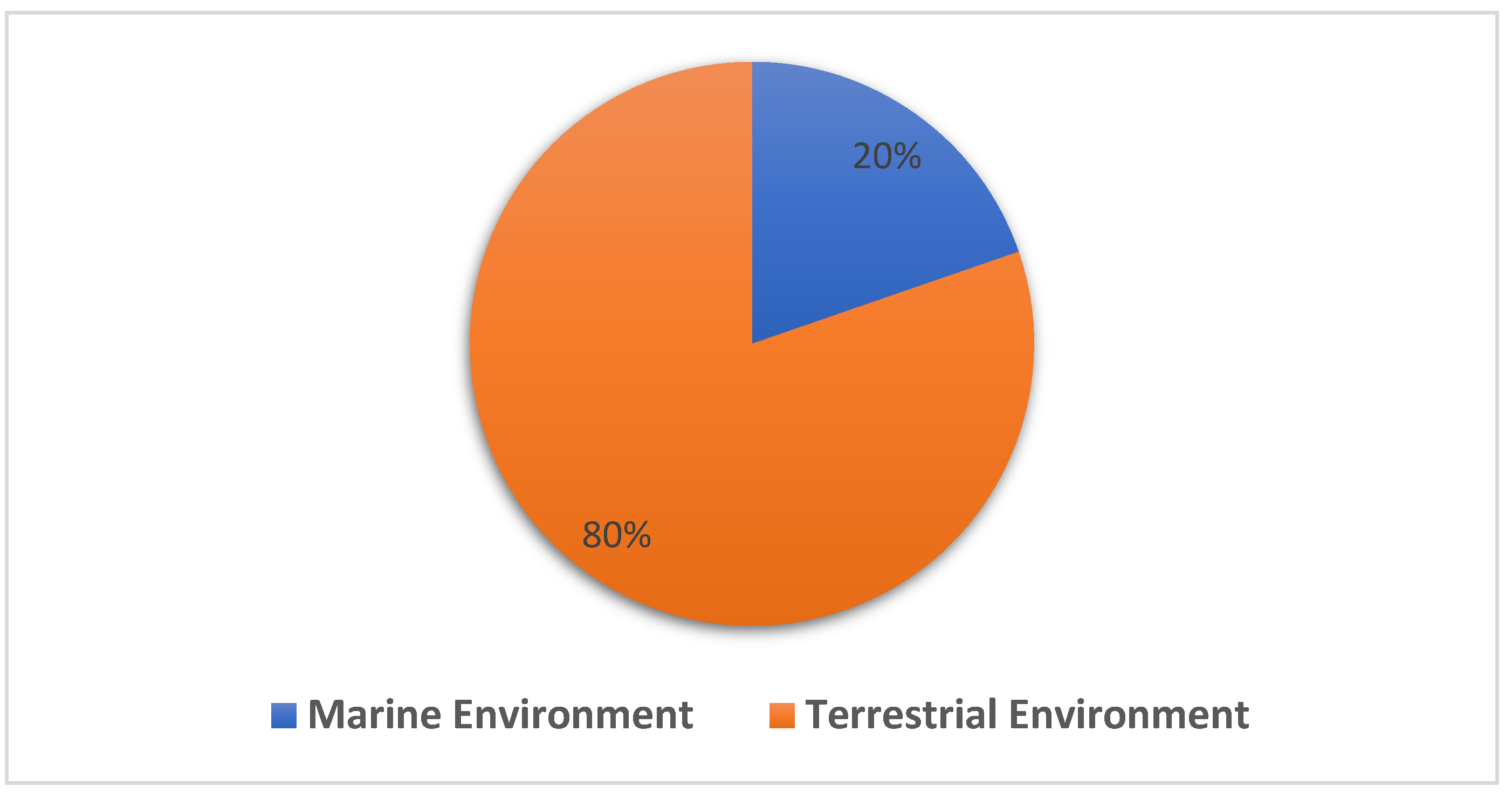









| Pre-Treatment | Terrestrial Source | Isolation Medium | Incubation Time/Temperature | References |
|---|---|---|---|---|
| Heat Treatment | ||||
| Heated at 120 °C for 15 min | Arid, non-saline soil sample (sand dunes) | Actinomycete isolation agar (HiMedia), pH 7.3 | 45 °C for up to 14 days | [74] |
| One gram of soil was suspended in 1.5% (w/v) phenol solution and incubated at room temperature for 30 min | Soil from peat swamp forest | Humic acid vitamin (HV) agar supplemented with nalidixic acid (25 μg mL−1) and nystatin (50 μg mL−1) | 30 °C for 14 days | [51] |
| Air-dried at room temperature for 14 days and suspended with strength Ringer’s solution | Soil sample from a commercial hazelnutorchard | Stevenson’s medium no. 3 supplemented with cycloheximide (50 μg mL−1), nalidixic acid (10 μg mL−1), nystatin (50 μg mL−1) and novobiocin (10 μg mL−1) | 28 °C for 21 days. | [129] |
| Heating at 55 °C for 6 min in a thermo-regulated bath | Arid soil samples | Starch Casein agar within the pH range of 7.0–7.2 | 28 °C for 14–21 days | [48] |
| Atacama desert soil | −1 cycloheximide | 28 °C for 14 days | [65] | |
| Heated at 60 °C for 20 min | Acidic mine area soil | Acidified (pH 5) Starch-Casein Agar supplemented withcycloheximide and nystatin, each at 50 μg mL−1 | 30 °C for 14 days | [49] |
| Heated at 85 °C for 15 min | Rock soil sample | Humic acid vitamin agar | 28 °C for three weeks | [50] |
| Wet heat (20 min, 60 °C) | Crater lake sediments | ISP 2 medium supplemented with 10 mg/L tetracycline, nystatin (50 μg/mL) and rifampicin (5 mg mL−1) | 28 °C for 14 days | [58] |
| Heat treated at 120 °C for 1 h | Soil from the banks of Gamka river | MC agar adjusted to pH 7.4 | 30 °C for 21 days | [146] |
| Pre-heated at 55 °C for 20 min, was incubated at 28 °C for 21 days | Hot water spring soil | Starch casein agar medium within the pH range of 7.0–7.2 supplemented with 25 μg mL−1 nystatin | 28 °C for 2 weeks | [55] |
| Pre-heated suspension 60 °C for 20 min | Lake sediment | M1 agar supplemented with filter-sterilized cycloheximide (50 mg mL−1) and rifampicin (5 mg mL−1) | 28 °C for 21 days | [131] |
| Dried at 55 °C for 48 hrs. | Soil sample near Xiangtan manganese mine | Gause’s synthetic medium 1 adjusted to pH 7.2 and supplemented with 2.0–3.0 mL of K2Cr2O7 solution (1.775 g L−1) in a 100 mL medium | 30 °C after incubation for 7–12 days | [62] |
| Heated at 55 °C in a water bath for 5 min | Soil | Starch casein nitrate agar plates (HiMedia) supplemented with 25 mg mL−1 nystatin | 28 °C for 14 days | [157] |
| Air-dried at room temperature for 14 days | Desert soil | Minimal medium within the pH range of 7.5–8.0 supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (10 μg mL−1) | 28 days at 28 °C | [53] |
| Soil from mount Song | Sodium succinate-asparagine agar pH 7.2 supplemented with cycloheximide (50 mg L−1) and Nalidixic acid (20 mg L−1). | 28 °C for 21 days | [137] | |
| Soil collected from mount Song | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 28 days | [136] | |
| Air-dried at room temperature for 7 days | Soil of a peat swamp forest | Humic acid vitamin (HV) agar supplemented with nalidixic acid (25 mg mL−1) and cycloheximide (50 mg mL−1) | 30 °C for 4 days | [68] |
| Soil | Humic acid vitamin (HV) agarsupplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1), | 30 °C for 3 weeks. | [121] | |
| Soil | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1) | 30 °C for 3 weeks | [147] | |
| Tree bark of Bauhinia variegata Linn | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (25 μg mL−1) | 30 °C for 3 weeks | [79] | |
| Air-dried at room temperature for a week | Soil sampled at a manganese-contaminated field | Gause’s synthetic medium 1 supplemented with 0.04 g K2Cr2O7 | 28 °C for 7–14 days | [64] |
| Alpine wetland soil | Gause’s synthetic medium 2 adjusted to pH 7.2 | 28 °C for 21 days | [130] | |
| Air-dried at room temperature for 48 h | Root of Geranium carolinianum Linn | Water–yeast extract agar supplemented with actidione (50 mg L−1) and nalidixic acid (25 mg L−1) | 28 °C for 2–6 weeks | [83] |
| Air-dried | Soil of a tropical rainforest | Humic acid vitamin (HV) agar | 50 °C in the dark for 5 days | [145] |
| Air-dried at room temperature | Forestsoil | Humic acid vitamin (HV) agar | 28 °C for 3 weeks | [164] |
| Air-dried for 72 hrs and then incubated at 40 °C for 16 hrs. | Forest soil | Humic acid vitamin (HV) agar | 28 °C for 3 weeks | [125] |
| Heated to 40 °C for 16 hrs. | Forest soil | HV agar supplemented with 50 mgL−1 cycloheximide at pH 7.2 and starch-casein agar at pH 7.2 and supplemented with 50 mg mL−1 filter-sterilized cycloheximide, 50 mg mL−1 nystatin and 0.5 mg mL−1 rifampicin | 28 °C for 3 weeks | [159] |
| Physical treatment | ||||
| Shaken at 250 r.p.m. in 100 mL of sterile water with glass beads for 30 min at 20 °C | Rhizosphere soil of wheat (Triticum aestivum L.) | Cellulose-proline agar (CPA) supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 3 weeks | [128] |
| Mixed on a tumble shaker for an hour | Alkaline soil adjacent to a meteoric alkaline soda lake | Starch casein agar adjusted to pH 8.5 with 1N NaOH and supplemented with 5% (w/v) NaCl and cycloheximide and nystatin (each at 50 μg mL−1) | 28 °C for 4 weeks | [37] |
| Shaken at 180 r.p.m. overnight | Desert | Gauze’s No. 1 medium 1 supplemented with Nystatin (100 mg mL−1) and nalidixic acid (50 mg mL−1) which had been filter sterilized (0.22 μm pore) before being added to 45 °C molten agar | 28 °C for 21 days | [40] |
| 1 g of soil was diluted in 50 m of 1 g L−1 3-morpholinopropanesulfoinc acid solution with 0.2 g l-1 CaCO3. The resulting soil suspension was shaken at 180 r.p.m. min−1 at 30 °C for 1 hr | Acid sandy soil | C supplemented with an inhibitor solution containing K2Cr2O7 (25 mg mL−1), calcium propionate (30 mg mL−1) and cycloheximide (50 mg mL−1) | 30 °C for 14 days | [66] |
| Sonic oscillator (Sonics Vibra-Cell VCX750) for 40 s at 30 W in 9 mL sterilized water | Bamboo (Sasa borealis) litter | Bennett’s Agar supplemented with cycloheximide (50 μg mL−1) and nalidixic acid (20 μg mL−1) at pH 5.5 | 28 °C for 2 weeks | [80] |
| Shaken at 180 r.p.m. at 30 °C for 1 hr | Desert soil | (25 mg mL−1), calcium propionate (30 mg mL−1) and cycloheximide (50 mg mL−1) | 30 °C for 7 days | [67] |
| , 200 r.p.m. for 1 hr. | Desert Soil | Reasoner’s 2A (R2A; BD) agar adjusted to pH 7.0. | 37 °C for 7 days | [57] |
| Suspended in distilled water (2 mL) followed by an ultrasonic treatment (160 W) for 3 min + soil suspension was incubated at 28 °C and 250 r.p.m. on a rotary shaker for 30 min | Rhizosphere soil of Urtica urens L. | Cellulose proline agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1). | 28 °C for 21 days | [140] |
| Ultrasonic treatment (160W) for 3 min followed by incubation of soil sample at 28 °C and 250 r.p.m. on a rotary shaker for 20 min | Saline–alkali soil | CMKA medium supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 14 days | [69] |
| Shaking on a rotary shaker at 180 r.p.m. at 28 °C for 30 min | Cuticle of Camponotus japonicus Mayr | Gause’s synthetic agar no. 1 adjusted to pH 7.2 and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1). | 28 °C for 21 days | [102] |
| Head of an ant | sodium succinate-asparagine agar at pH 7.2 and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1). | [103] | ||
| Head of an ant (Camponotus japonicus Mayr) | sodium succinate-asparagine agar at pH 7.2 and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | [104] | ||
| Head of an ant (Camponotus japonicus Mayr) | Tap Water Yeast Extract Agar (TWYE)) supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | [105] | ||
| Head of an ant (Lasius fuliginous L.) | Humic acid vitamin (HV) agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 21 days | [106] | |
| Kept in an orbital shaker (28 °C, 180 r.p.m.) for 1 hr. | Soil | 28 °C for 14 days | [153] | |
| Shaken on a rotary shaker at 250 r.p.m. at 28 °C for 30 min | Millipede (Kronopolites svenhedind Verhoeff) | Gause’s Synthetic Agar No. 1 at pH 7.2. supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 21 days | [110] |
| Rhizosphere soil of wheat | [144] | |||
| Ultrasonic treatment (160 W) for 3 min + incubation at 28 °C and 250 r.p.m. on a rotary shaker for 20 min | Saline–alkaline soil | CMKA medium prepared with 10 % (w/v) NaCl and supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 14 days | [56] |
| l sterile water with shaking on a rotary shaker at 180 r.p.m at 28 °C for 30 min | Head of Camponotus japonicus Mayr ant | Gause’s synthetic agar no. 1 2 supplemented with cycloheximide(50 mg L−1) and nalidixic acid (20 mg L−1). | 28 °C for 21 days | [93] |
| Water bath sonicator for 2 min at 30 °C | Red soil | Modified mineral-medium agar containing 0.5 % sorbitol supplemented with cycloheximide, nystatin, nalidixic acid (each at 50 μg mL−1), and novobiocin (at 25 μg mL−1) | 28 °C for 3–4 weeks | [163] |
| Orbital shaking at 120 r.p.m. for 2 weeks | Bamboo rhizosphere soil | Humic acid vitamin agar (HV agar) at pH 7.2 and starch-casein agar supplemented with filter-sterilized μg mL−1 cycloheximide (50 μg mL−1), nystatin (50 μg mL−1), and rifampicin (0.5 μg mL−1) | 28 °C for 2 weeks | [166] |
| Chemical Treatment | ||||
| Lodewyckx pretreatment method | Bulbil of Dioscorea bulbifera L. | Humic acid vitamin (HV) agar containing cycloheximide (50 mg L−1) and nalidixic acid (25 mg L−1) | 28 °C for 2–6 weeks | [81] |
| 3% NaCl | Saltern soil | Starch Casein agar within the pH range of 7.0–7.2 supplemented with filter-sterilized cycloheximide (50 μg mL−1) and 3% NaCl | 28 °C for 30 days | [38] |
| 1.5% (w/v) NaCl | Salt lake sediment | ISP (International Streptomyces Project) medium 4 prepared with 1.5% (w/v) NaCl | 28 °C for 5 days | [52] |
| One gram of soil was suspended in 9 mL 1.5% (v/v) Phenol for 30 min | Rhizosphere soil of an oil palm (Elaeis guineensis) | g mL−1) and cycloheximide (50 μg mL−1) | 30 °C for 14 days | [138] |
| Suspended and diluted with a solution [0.38% K2HPO4, 0.12% KH2PO4, 0.51% MgSO4.H2O, 0.25% NaCl, 0.005% Fe2(SO4)3.H2O, 0.005% MnSO4.5H2O] | Soil | Humic acid vitamin (HV) agar supplemented benlate [final conc. 25 μg mL−1 (w/v)] and nalidixic acid [final conc. 25 μg mL−1 (w/v)] | 27 °C for 3 weeks | [123] |
| 2–3 cm stem section excised with sterile scalpel, washed in 20% (1.05% for roots) hydrogen peroxide (10 min) and rinsed 4x in sterile 0.02 M potassium phosphate buffer | Stem of Populus adenopoda | Humic acid vitamin (HV) agar containing nalidixic acid (25 mg L−1) and cycloheximide (50 mg L−1) | 28 °C for 2–6 weeks | [87] |
| Seeds were surface sterilized withserial washes of 75% ethanol for 1 min, 10% sodium hypochlorite for 5 min and several rinses with distilled water | Aril of a seed of Ginkgo biloba | mL−1) and actidione (50 μg mL−1) | 28 °C for 21 days | [89] |
| 5% NaCl | Subsurface soil sample of Atacama desert | Glucose-yeast extract agar (HiMedia) supplemented with cycloheximide and nystatin (each at 25 μg mL−1) | 28 °C for 14 days | [59] |
| Suspended in 1 mL sterile saline (0.9% NaCl) | Desert soil | Mineral salt medium (MSM) agar containing Nile Red (0.5 μg mL−1) | 30 °C for five days | [71] |
| 3% (w/v) NaCl | Fruits of Capparis spinosa | Tap water-yeast extract (TWYE) agarwithin the pH range of 7.0–7.2 supplemented with 3% (w/v) NaCl | 30 °C for 2–6weeks | [88] |
| Subjected to a seven-step surface sterilization procedure: a 60 s wash in sterile tap water containing cycloheximide (100 mg L−1) and nalidixic acid (20 mg L−1), followed by a wash in sterile water, a 5 min wash in 5% (v/v) NaCl, a 10 min wash in 2.5% (w/v) Na2S2O3, a 5 min wash in 75% (v/v) ethanol, a wash in sterile water and a final rinse in 10% (w/v) NaHCO3 for 10 min, and then the rinsed root sample was Dried at 100 °C for 15 min. | Root of Polygonatumodoratum (Mill.) | Humic acid-vitamin agar supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C fro 14 days | [82] |
| 1.5% (w/v) NaCl | Hypersaline soil sample | B7 medium prepared with 1.5% (w/v) NaCl | 37 °C for 10 days | [72] |
| 10% (w/v) NaCl | Saline alkaline soil | CMKA medium supplemented with cycloheximide (50 mg L−1) and nalidixic acid (20 mg L−1) | 28 °C for 14 days | [56] |
| Surface sterilized in 70% ethanol for 2 min before being washed twice in sterile distilled H2O | Gut of a South African termite | Medium II at pH 7.0 supplemented 50 μg mL−1 cycloheximide and 10 μg mL−1 nalidixic acid | 30 °C 4 weeks | [100] |
| Strain | Nature of Sample | Isolation Medium | Country | Reference |
|---|---|---|---|---|
| Streptomyces reniochalinae sp. nov. | LHW50302T from Reniochalina stalagmitis LHW51701T from Diacarnus megaspinorhabdosa | Streptomyces Isolation Medium agar plates containing 3% sea salt (w/v), 50 mg L−1 cycloheximide and 25 mg L−1 nalidixic acid. | China | [182] |
| Streptomyces diacarni sp. nov. | ||||
| Streptomyces tirandamycinicus sp. nov. | Marine sponge | Humic acid vitamin (HV) agar prepared with 50% (v/v) seawater and supplemented with K2Cr2O7 (100 mg L−1) | China | [179] |
| Streptomyces zhaozhouensis subsp. mycale subsp. nov. | Marine sponge (Mycale sp.) | Actinomycetes Isolation Agar (HiMedia) supplemented with 1% sponge extract and gentamycin (2 mg 100 mL−1), cycloheximide (2.5 mg 100 mL−1), and amphotericin B (1 mg 100 mL−1). | India | [181] |
| Streptomyces atlanticus sp. nov. | Marine sponge (Aplysina fulva) | g mL−1) and nalidixic acid (50 μg mL−1) | Brazil | [184] |
| Streptomyces hyaluromycini sp. nov. | Tunicate (Molgula manhattensis) | Inorganic salts−starch agar (ISP 4) supplemented with cycloheximide (25 mg mL−1), potassium dichromate (50 mg mL−1) and nystatin (50 mg mL−1) supplemented with nalidixic acid (20 mg L−1) and cycloheximide (50 mg L−1) | Japan | [183] |
| Streptomyces bohaiensis sp. nov. | Young Scomberomorus niphonius in (long, slender, laterally flattened, pelagic fish with longitudinal dark spots on the sides and ~ 15 cm in fork length) | Oatmeal agar international Streptomyces project (ISP 3) (HiMedia) containing nalidixic acid (25 mg L−1) and cycloheximide (50 mg L−1) | China | [189] |
| Streptomyces spongiicola sp. nov. | Marine sponge | Starch casein nitrate agar at pH 7.0–7.2, prepared with 50% (v/v) seawater and supplemented with actidione (50 mg mL−1), nystatin (50 mg mL−1) and nalidixic acid (20 mg mL−1). | China | [180] |
| Pre-Treatment | Marine Source | Isolation Medium | Incubation Time/Temperature | References |
|---|---|---|---|---|
| Heat Treatment | ||||
for 6 min, then suspended in 100 mL sterile aged seawaterand stirred for 30 min. | Mangrove mud | that had been made with 70% aged seawater in distilled water (instead of pure distilled water), and supplemented with cycloheximide (25 mg mL−1), potassium dichromate (50 mg mL−1) and nystatin (50 mg mL−1) | for 7 days | [199] |
| Sediment in mangrove soil | g mL−1) | for 14 days. | [201] | |
| Wet heat in sterilized ) using a water bath | Mangrove sediment | g mL−1) | for 7–14 days | [206] |
| Brackish sediment of a fish dumping yard in Chilika lake | Colloidal Chitin agar (CCA) medium supplemented with nystatin (50 mg L−1) | for 7 days | [208] | |
| Air-dried at room temperature for 7 days | Marine sediments | −1−1) | for 2–3 weeks | [207] |
| ) | Mangrove Forest soil | g mL−1) | for 14 days | [213] |
| [209] | ||||
| Mangrove sediments | mL−1) | for 14 days | [203] | |
| for 15 min) | Sediments around the mangrove plant Avicennia mariana | g mL−1) and supplemented with sterile seawater (3.3%, w/v) | for 4 weeks | [210] |
| Chemical Treatment | ||||
| 3.0% (w/v) NaCl solution | Marine sediment | for 5 h and 3.0% (w/v) NaCl | for 2 weeks | [205] |
| Marine sponge (Aplysina fulva) | g mL−1) | for 21 days | [184] | |
| Novel/New Antibacterial Compound | Chemical Class | Antibacterial Activity | Sample Environment | Ref. |
|---|---|---|---|---|
| Terrestrial Source | ||||
| 1-Hydroxy-7-oxolavanucyanin and Δ (7″,8″)-6″-hydroxynaphthomevalin | Phenazine/terpene hybrid | G+ | Soil | [222] |
| Krisynomycin B and C | Cyclic Depsipeptide | G+ | Desert Sand | [223] |
| Meliponamycin A and B | Cyclic Hexadepsipeptide | G+ | Bees | [224] |
| Picolinamycin | Pyrimidine alkaloid | G+ | Soil | [225] |
| Nybomycin D | Quinoline | G+ | Acid mine soil | [226] |
| Pentaminomycin C–E | Cyclic pentapeptide | G+ | Fungi | [227] |
| Nalidixic acid 1 | Quinolone | G- | Rhizospheric soil | [228] |
| Quinomycin I and J | Cyclic depsipeptide | G+ | Mount soil | [229] |
| Puromycin B–E | Amino-nucleoside | G+ | Soil | [230] |
| Abyssomicin M−X | Spirotetronate polyketide | G+ | Soil | [231] |
| Streptoone A | Linear polyketide | G+ | Soil | [232] |
| Asenjonamide A–C | β-diketone | G+ & G- | Desert soil | [233] |
| Gordonic acid | polyketide glycoside | G+ | Acid mine drainage soil | [234] |
| Ulleungmycin A and B | Non-ribosmal peptide | G+ | Volcanic soil | [235] |
| Quinomycin A–C | Pyranonapthaquinone | G+ | Mountain Soil | [236] |
| Actinomycin Y6–Y9 | Bi-cyclic chromopeptide lactone | G+ | Soil | [237] |
| 2-amino-N-(2-amino-3-phenylpropanoyl)-N-hydroxy-3-phenylpropanamide | Hydroxamic acid | G+ & G- | Desert soil | [238] |
| Angucyclines and angucyclinones | Benz[a]anthracene polyketide | G+ | Soil | [239] |
| Streptanoate | Amide ester | G+ | Soil | [240] |
| Xiakemycin A | Pyranonaphthoquinone | G+ | Soil | [241] |
| Methyl ealaiophylins | Macrodiolide | G+ | Soil | [242] |
| 7-Prenylisatin | Isatin | G+ | Mountain soils | [243] |
| Marine Source | ||||
| Mersaquinone 1 | Tetracene | MRSA | Marine sediment | [244] |
| Dionemycin 1 | Chlorinated bis-indole alkaloid | MRSA & G+ | Marine sediment | [245] |
| Streptoglutarimide A–J | Glutarimide | MRSA | Marine mud | [246] |
| Maculosin-O-α-L-rhamnopyranoside | Diketopiperazine glycoside | MRSA, G+ & G- | Coastal soil | [247] |
| Strepoxepinmycin A−D | Naphthoquinone | MRSA, G+ & G- | Marine-derived | [248] |
| Niphimycin C−E | Macrolide | G+, MRSA &VRE | Marine sediment | [249] |
| Rakicidin F | Cyclic depsipeptide | G+ & G- | Marine sponge | [250] |
| Ala-geninthiocin 1 | Thiopeptide | G+ | Marine sediment | [251] |
| Fradiamine A | Hydroxamic acid siderophore | G+ | Deep-sea sediment | [252] |
| Lobophorin K | Spirotetronate glycoside | G+ | Deep-sea coral | [253] |
| Pteridic acid C–G | Spiroketal polyketide | G+ & G- | coral | [254] |
| Neo-actinomycin A and B | Phenoxazine | MRSA & VRE | Marine sediment | [255] |
| Spiroindimicin E and F | Chlorinated bis-indole alkaloid | G+ | Marine sediment | [256] |
| Ilamycin P | Non-ribosmal peptide | G+ | Marine sediment | [257] |
| Ghanamycin A & B | γ-Butyrolactone | Phytopathogens (G+ & G-) | Saltcedar from intertidal zone | [258] |
| (2E, 6E)-3,7,11- trimethyldodeca-2,6-dienedioic acid (2) | Unsaturdated fatty Acid | G+ | Marine sediment | [259] |
| Aldgamycin J−O | Macrolide | G+& G- | Marine sediment | [260] |
| Novel/New Anticancer Compound | Chemical Class | Sample Environment | Ref. |
|---|---|---|---|
| Terrestrial Source | |||
| Azodyrecin B | Azoxide fatty acid | Soil | [261] |
| Streptimidone 1 & 3 | Glutarimide | Soil | [262] |
| Ilamycin G−R | Non-ribosmal peptide | Soil | [257] |
| 9-Methylstreptimidone 2-α-D-glucopyranoside and hydroxyiso-9-methylstreptimidone | Glutarimide | Soil | [127] |
| Baraphenazine E | Phenazine | Soil | [263] |
| Naquihexcin C, E & I | Pyranonaphthoquinone glycoside | Soil | [264] |
| Nalidixic acid | Quinolone | Rhizospheric soil | [228] |
| Quinomycin 1 & 3 | Cyclic depsipeptide | Mount soil | [229] |
| ε-Rhodomycinone 1, 4 & 8 and β-Rhodomycinone 2, 3, 5–7 & 9−12 | Anthracycline | Soil | [265] |
| Ulleungdin | Lasso peptide | Soil | [266] |
| Tetracenoquinocin A | Anthracycline | Soil | [267] |
| Hisunic acid 1 | Cyclic polyketide | Soil | [268] |
| Herbicidin L | Adenosine-nucleoside | Soil | [269] |
| Actinomycin 2−5 | Bicyclic chromopeptide lactone | Soil | [237] |
| Spectinabilin 1 | Linear polyene | Head of ant (Camponotus japonicas Mayr) | [270] |
| Angucycline | Benz[a]anthracene polyketide | Soil | [239] |
| Streptanoate | Amide ester | Soil | [240] |
| 1,19-Seco-avermectin 3−5 | Macrolide | Soil | [271] |
| Marine Source | |||
| Piericidin F | Pyridine-containing linear polyketide | Mangrove soil | [272] |
| Salternamide A | Cyclohexenone-containing linear polyketide | Saltern soil | [273] |
| Donghaecyclinone A–C | Benz[a]anthracene polyketide | Volcanic island marine sediment | [274] |
| Tetrahydroanthracene derivative 4 | Dimeric tetrahydroanthracene | Mairne sponge | [275] |
| Microeunicellol A | Terpene | Marine sediment | [276] |
| Dionemycin 1 | Chlorinated bis-indole alkaloid | Marine sediment | [245] |
| 2-epi-Anthracimycin 2 | Macrolide | Marine sediment | [277] |
| Lactoquinomycin C & D | Napthaquinone | Marine sediment | [278] |
| 10-epi-HSAF, 10-epi-deOH-HAS, d 10-epi-maltophilin, 10-epi-xanthobaccin C & 10-epi-hydroxymaltophilin | Polycyclic tetramate macrolactam | Mangrove sediment | [279] |
| Neothioviridamide | Polythioamide | Mangrove soil | [280] |
| 1-hydroxymethyl-8-hydroxy-anthraquinone-3-carboxylic acid | Anthraquinone | Fresh sea anemone (H. lineata) | [281] |
| Bagremycin C | para-hydroxybenzoic acid ester | Mangrove soils | [282] |
| Strepoxepinmycin C & D | Naphthoquinone | Marine sediment | [248] |
| Cyclizidine C | Indolizidine alkaloid | Marine sediment | [283] |
| 9-HydroxyK252c, 3-hydroxy-3′-Nacetylholyrine A, 3- hydroxyholyrine A, streptocarbazole E | Indolocarbazole | Marine sediment | [284] |
| Geninthiocin 1 | Macrocyclic peptide | Subtidal marine sediment | [251] |
| Deformylated antimycin 6 & 7 | Diester alkaloid | Mangrove sediment | [285] |
| Lobophorin K | Spirotetronate glycoside | Deep sea coral | [253] |
| Neo-actinomycin A | Bi-cyclic chromopeptide lactone | Marine sediment | [255] |
| Drimentine I | Hybrid isoprenoid-diketopiperazine | Marine sediment | [286] |
| Novel/New Antifungal Compound | Chemical Class | Enzyme Modulatory Activity | Sample Environment | Ref. |
|---|---|---|---|---|
| Terrestrial Source | ||||
| Ulleungamide C | Cyclic depsipeptide | Inhibitor and inducer | Soil | [287] |
| Formicolide A and B | Macrolide | Inducer | Ant gut (Formica yessensis) | [288] |
| Naphthacemycin B5-B13 | Naphthacene | Inhibitor | Medicinal plant Senecio scandens | [289] |
| Strepantibin A−C | Terphenyl | Inhibitor | Larvae of mud dauber wasp (Sceliphron madraspatanum) | [290] |
| Dinghupeptin A & B | Cyclodepsipeptide | Inhibitor | Soil | [291] |
| Lorneic acid F & I | Trisubstituted aromatic acid | Inhibitor | Bark of Betula mandshurica Nakai | [280] |
| Marine Source | ||||
| Mohangic acid E | Linear polyene | Inducer | Marine mud flat | [292] |
| Salternamide A and D | Cyclohexenone-containing linear polyketide | Inhibitor | Saltern soil | [273] |
| Strepoxepinmycin D | Naphthoquinone | Inhibitor | Marine sediment | [248] |
| Cyclizidine C, F, H & I | Indolizidine alkaloid | Inhibitor | Marine sediment | [283] |
| 3-hydroxy-K252c | Indolocarbazole | Inhibitor | Marine sediment | [284] |
| Novel/New Antifungal Compound | Chemical Class | Sample Environment | Ref. |
|---|---|---|---|
| Terrestrial Source | |||
| Picolinamycin | Pyrimidine alkaloid | Soil | [225] |
| Baraphenazine E | Phenazine | Soil | [263] |
| 2-amino-N-(2-amino-3-phenylpropanoyl)-N-hydroxy-3-phenylpropanamide | Hydroxamic acid | Desert soil | [238] |
| Rimocidin derivative BU16 | Macrolide | Soil | [294] |
| Filipin III, 15-Glycidylfilipin III, 16α, 17α-Epoxyfilipin V &16β, 17β-Epoxyfilipin V | Macrolide | Soil | [297] |
| Streptoone B | Linear polyketide | Soil | [232] |
| Abyssomicin M−X | Spirotetronate polyketide | Creek soil | [231] |
| Marine Source | |||
| Streptoglutarimide A−J | Glutarimide | Marine mud | [246] |
| Flavofungin I and II | Macrolide | Mangrove soil | [298] |
| Novel/New Bioactive Compound | Chemical Class | Biological Activity | Sample Environment | Ref. |
|---|---|---|---|---|
| Naphthacemycin B5-B13 | Naphthacene | Antidiabetic | Medicinal plant Senecio scandens | [289] |
| Benwamycin 2 & 6 | Trialkyl-substituted polyketide | Antiproliferative | Soil | [299] |
| Suncheonoside A, B, and D | Benzothioate glycosides | Antidiabetic | Marine sediment | [300] |
| Strepantibin A and B | Terpenyl | Antiproliferative | Larvae of mud dauber wasp Sceliphron madraspatanum | [290] |
| Trienomycin J | Macrolide | Antiproliferative | Moss soil-derived | [301] |
| Streptovitacin A | Glutarimide | Antiproliferative | Marine mud | [246] |
| Nahuoic acid B−E | Polyol polyketide | Antibiofilm | Marine sediment | [302] |
| Napyradiomycin SF2415B3 | Hybrid isoprenoid | Marine sediment | [303] | |
| Camporidine A | Prenylated naphthoquinone | Anti-inflammatory | Gut of carpenter ant Camponotus kiusiuensis | [304] |
| Formicolide A and B | Macrolide | Antiangiogenic | Gut bacterial strain of the wood ant (Formica yessensis) | [288] |
| Meliponamycin A & B | Cyclic hexadepsipeptide | Entomopathogenic | Melipona scutellaris nurse bees | [211] |
| Isarubrolone 3 & 4 | Polycyclic tropoloalkaloid | Autophagy inducer | Soil | [305] |
| Abyssomicin I | Spirotetronate polyketide | Inhibits tumor cell invasion | Rock soil | [50] |
| Naquihexcin C | Pyranonaphthoquinone glycoside | HIV-1 inhibitor | Rhizospheric soil | [264] |
| Camporidine A | Prenylated naphthoquinone | Antimetastatic | Gut of carpenter ant (Camponotus kiusiuensis) | [304] |
| Aturanoside A & B | Anthraquinone glycoside | Suppresses vascular endothelial growth factor (VEGF) | Soil | [306] |
| Trienomycin J−L | Macrolide | Inhibited nitric oxide production | Soil moss | [301] |
| Herbicidin L | Adenosine-nucleoside | Antiparasitic | Soil | [269] |
| Simamycin | Prenylated nucleoside | Induces differentiation of preadipocytes into matured adipocytes | Soil | [307] |
| Oxachelin CSpoxazomicin D | Oxazoline carboxamide, peptide | Potent neuroprotectives | Soil | [308] |
| Aotaphenazine | Phenazine | Overcome tumor necrosisFactor-related apoptosis-inducing ligand (TRAIL). | Soil | [309] |
| Aotaphenazine | Phenazine | Enhances the levels of apoptosis inducing proteins | Soil | [309] |
| Inubosin B | Acridine alkaloid | Ngn2 promoter activity and induces mRNA expression of genes related to neural stem cell differentiation. | Soil | [310] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Donald, L.; Pipite, A.; Subramani, R.; Owen, J.; Keyzers, R.A.; Taufa, T. Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective. Microbiol. Res. 2022, 13, 418-465. https://doi.org/10.3390/microbiolres13030031
Donald L, Pipite A, Subramani R, Owen J, Keyzers RA, Taufa T. Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective. Microbiology Research. 2022; 13(3):418-465. https://doi.org/10.3390/microbiolres13030031
Chicago/Turabian StyleDonald, Lavinia, Atanas Pipite, Ramesh Subramani, Jeremy Owen, Robert A. Keyzers, and Taitusi Taufa. 2022. "Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective" Microbiology Research 13, no. 3: 418-465. https://doi.org/10.3390/microbiolres13030031
APA StyleDonald, L., Pipite, A., Subramani, R., Owen, J., Keyzers, R. A., & Taufa, T. (2022). Streptomyces: Still the Biggest Producer of New Natural Secondary Metabolites, a Current Perspective. Microbiology Research, 13(3), 418-465. https://doi.org/10.3390/microbiolres13030031







