Development of a Real-Time PCR Method for the Detection of European and Siberian Subtypes of Tick-Borne Encephalitis Virus
Abstract
:1. Introduction
2. Materials and Methods
2.1. Primer Design
2.2. Optimization of Primer Concentration
2.3. Sequencing
2.4. Positive Control and Virus Strains
3. Results
3.1. Primer Tm Analysis and Design Refinement
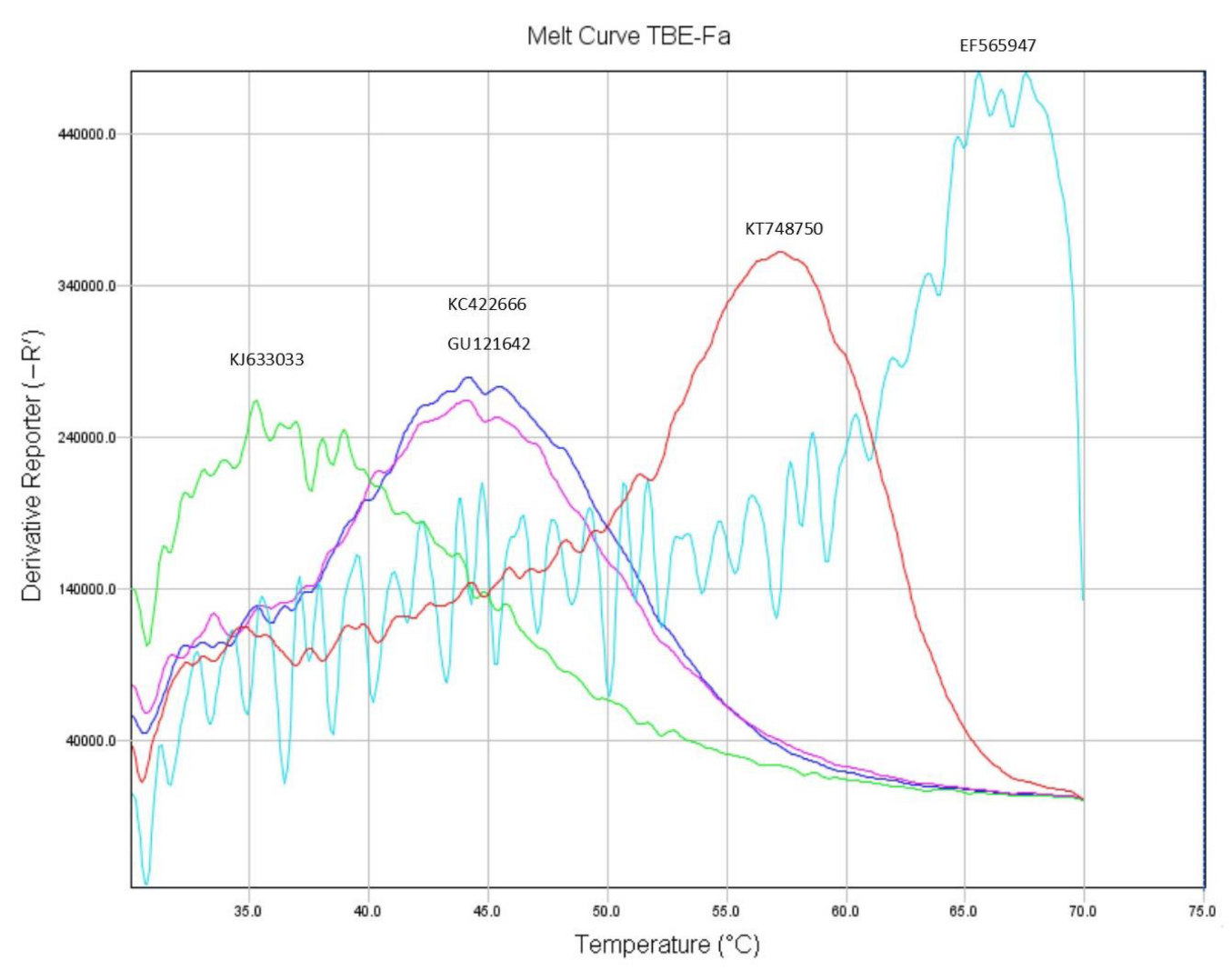
3.2. Detection of TBEV
3.3. Sensitivity and Efficiency of the Real-Time PCR
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
Appendix A
| Subtype | TBE-R16G4T | Tm | |
|---|---|---|---|
| 5′GACTCTGCACAACAAGGACA3′ | |||
| EF565947 | Eu | 3′CTGGGACGTGTTGTTTCTGT5′ | 57 °C |
| KT749573 | Sib | 3′CTGAGACGTGTTGTTCCTGT5′ | 66 °C |
| KT748750 | Sib | 3′CTGAGACGTGTAGTTCCTGT5′ | 60 °C |
| AB022296 | FE | 3′ATGAGACGTATTGTTCCTGT5′ | 55 °C |
| KJ633033 | Bai | 3′CTGAGACGTGTTATCCCTGT5′ | 51 °C |
| JX315851 | Sib | 3′TTGAGACGTATTGTTCCAGT5′ | 50 °C |
| KU761569 | FE | 3′ATGAGACGTATTGTCCCTAT5′ | 41 °C |
| KC422666 | FE | 3′ATGAGCCGTGTTATTCCTGT5′ | 46 °C |
| GU121642 | FE | 3′ATGAGCTGTGTTGTTCCTGT5′ | 54 °C |
| AF231807 | FE | 3′TTGAGACGTATCGTTCCTAT5′ | 40 °C |
References
- Pulkkinen, L.I.A.; Butcher, S.J.; Anastasina, M. Tick-Borne Encephalitis Virus: A Structural View. Viruses 2018, 10, 350. [Google Scholar] [CrossRef] [PubMed]
- Ecker, M.; Allison, S.L.; Meixner, T.; Heinz, F.X. Sequence analysis and genetic classification of tick-borne encephalitis viruses from Europe and Asia. J. Gen. Virol. 1999, 80 Pt 1, 179–185. [Google Scholar] [CrossRef]
- Adelshin, R.V.; Sidorova, E.A.; Bondaryuk, A.N.; Trukhina, A.G.; Sherbakov, D.Y.; White Iii, R.A.; Andaev, E.I.; Balakhonov, S.V. “886-84-like” tick-borne encephalitis virus strains: Intraspecific status elucidated by comparative genomics. Ticks Tick-Borne Dis. 2019, 10, 1168–1172. [Google Scholar] [CrossRef] [PubMed]
- Dai, X.; Shang, G.; Lu, S.; Yang, J.; Xu, J. A new subtype of eastern tick-borne encephalitis virus discovered in Qinghai-Tibet Plateau, China. Emerg. Microbes Infect. 2018, 7, 74. [Google Scholar] [CrossRef]
- Demina, T.V.; Dzhioev, Y.P.; Verkhozina, M.M.; Kozlova, I.V.; Tkachev, S.E.; Plyusnin, A.; Doroshchenko, E.K.; Lisak, O.V.; Zlobin, V.I. Genotyping and characterization of the geographical distribution of tick-borne encephalitis virus variants with a set of molecular probes. J. Med. Virol. 2010, 82, 965–976. [Google Scholar] [CrossRef] [PubMed]
- Dobler, G.; Gniel, D.; Petermann, R.; Pfeffer, M. Epidemiology and distribution of tick-borne encephalitis. Wien. Med. Wochenschr. 2012, 162, 230–238. [Google Scholar] [CrossRef]
- Jaaskelainen, A.E.; Tonteri, E.; Pieninkeroinen, I.; Sironen, T.; Voutilainen, L.; Kuusi, M.; Vaheri, A.; Vapalahti, O. Siberian subtype tick-borne encephalitis virus in Ixodes ricinus in a newly emerged focus, Finland. Ticks Tick-Borne Dis. 2016, 7, 216–223. [Google Scholar] [CrossRef]
- Bogovic, P.; Strle, F. Tick-borne encephalitis: A review of epidemiology, clinical characteristics, and management. World J. Clin. Cases 2015, 3, 430–441. [Google Scholar] [CrossRef]
- Andreassen, A.; Jore, S.; Cuber, P.; Dudman, S.; Tengs, T.; Isaksen, K.; Hygen, H.O.; Viljugrein, H.; Anestad, G.; Ottesen, P.; et al. Prevalence of tick borne encephalitis virus in tick nymphs in relation to climatic factors on the southern coast of Norway. Parasites Vectors 2012, 5, 177. [Google Scholar] [CrossRef]
- Vikse, R.; Paulsen, K.M.; Edgar, K.S.; Pettersson, J.H.O.; Ottesen, P.S.; Okbaldet, Y.B.; Kiran, N.; Lamsal, A.; Lindstedt, H.E.H.; Pedersen, B.N.; et al. Geographical distribution and prevalence of tick-borne encephalitis virus in questing Ixodes ricinus ticks and phylogeographic structure of the Ixodes ricinus vector in Norway. Zoonoses Public Health 2020, 67, 370–381. [Google Scholar] [CrossRef]
- Norwegian Surveillance System for Communicable Diseases (MSIS), Norwegian Institute of Public Health. 2022. Available online: https://msis.no/ (accessed on 10 August 2022).
- Mehl, R. The distribution and host relations of Norwegian ticks (Acari, Ixodides). Fauna Norwgica Ser. B 1983, 30, 46–51. [Google Scholar]
- Soleng, A.; Edgar, K.S.; Paulsen, K.M.; Pedersen, B.N.; Okbaldet, Y.B.; Skjetne, I.E.B.; Gurung, D.; Vikse, R.; Andreassen, A.K. Distribution of Ixodes ricinus ticks and prevalence of tick-borne encephalitis virus among questing ticks in the Arctic Circle region of northern Norway. Ticks Tick-Borne Dis. 2018, 9, 97–103. [Google Scholar] [CrossRef] [PubMed]
- Hvidsten, D.; Frafjord, K.; Gray, J.S.; Henningsson, A.J.; Jenkins, A.; Kristiansen, B.E.; Lager, M.; Rognerud, B.; Slåtsve, A.M.; Stordal, F.; et al. The distribution limit of the common tick, Ixodes ricinus, and some associated pathogens in north-western Europe. Ticks Tick-Borne Dis. 2020, 11, 101388. [Google Scholar] [CrossRef] [PubMed]
- Kjær, L.J.; Soleng, A.; Edgar, K.S.; Lindstedt, H.E.H.; Paulsen, K.M.; Andreassen, Å.K.; Korslund, L.; Kjelland, V.; Slettan, A.; Stuen, S.; et al. A large-scale screening for the taiga tick, Ixodes persulcatus, and the meadow tick, Dermacentor reticulatus, in southern Scandinavia, 2016. Parasites Vectors 2019, 12, 338. [Google Scholar] [CrossRef]
- Jaenson, T.G.T.; Wilhelmsson, P. First records of tick-borne pathogens in populations of the taiga tick Ixodes persulcatus in Sweden. Parasites Vectors 2019, 12, 559. [Google Scholar] [CrossRef]
- Laaksonen, M.; Sajanti, E.; Sormunen, J.J.; Penttinen, R.; Hanninen, J.; Ruohomaki, K.; Saaksjarvi, I.; Vesterinen, E.J.; Vuorinen, I.; Hytonen, J.; et al. Crowdsourcing-based nationwide tick collection reveals the distribution of Ixodes ricinus and I. persulcatus and associated pathogens in Finland. Emerg. Microbes Infect. 2017, 6, e31. [Google Scholar] [CrossRef]
- Jaaskelainen, A.E.; Tonteri, E.; Sironen, T.; Pakarinen, L.; Vaheri, A.; Vapalahti, O. European subtype tick-borne encephalitis virus in Ixodes persulcatus ticks. Emerg. Infect. Dis. 2011, 17, 323–325. [Google Scholar] [CrossRef]
- Kuivanen, S.; Smura, T.; Rantanen, K.; Kamppi, L.; Kantonen, J.; Kero, M.; Jaaskelainen, A.; Jaaskelainen, A.J.; Sane, J.; Myllykangas, L.; et al. Fatal Tick-Borne Encephalitis Virus Infections Caused by Siberian and European Subtypes, Finland, 2015. Emerg. Infect. Dis. 2018, 24, 946–948. [Google Scholar] [CrossRef]
- Hasle, G.; Bjune, G.; Edvardsen, E.; Jakobsen, C.; Linnehol, B.; Røer, J.E.; Mehl, R.; Røed, K.H.; Pedersen, J.; Leinaas, H.P. Transport of ticks by migratory passerine birds to Norway. J. Parasitol. 2009, 95, 1342–1351. [Google Scholar] [CrossRef]
- Klitgaard, K.; Chriél, M.; Isbrand, A.; Jensen, T.K.; Bødker, R. Identification of Dermacentor reticulatus Ticks Carrying Rickettsia raoultii on Migrating Jackal, Denmark. Emerg. Infect. Dis. 2017, 23, 2072–2074. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer—New capabilities and interfaces. Nucleic Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Ye, J.; Coulouris, G.; Zaretskaya, I.; Cutcutache, I.; Rozen, S.; Madden, T.L. Primer-BLAST: A tool to design target-specific primers for polymerase chain reaction. BMC Bioinform. 2012, 13, 134. [Google Scholar] [CrossRef] [PubMed]
- Katoh, K.; Standley, D.M. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013, 30, 772–780. [Google Scholar] [CrossRef]
- Waterhouse, A.M.; Procter, J.B.; Martin, D.M.; Clamp, M.; Barton, G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics 2009, 25, 1189–1191. [Google Scholar] [CrossRef] [PubMed]
- Bustin, S.A.; Benes, V.; Garson, J.A.; Hellemans, J.; Huggett, J.; Kubista, M.; Mueller, R.; Nolan, T.; Pfaffl, M.W.; Shipley, G.L.; et al. The MIQE guidelines: Minimum information for publication of quantitative real-time PCR experiments. Clin Chem 2009, 55, 611–622. [Google Scholar] [CrossRef]
- Reusken, C.; Baronti, C.; Mögling, R.; Papa, A.; Leitmeyer, K.; Charrel, R.N. Toscana, West Nile, Usutu and tick-borne encephalitis viruses: External quality assessment for molecular detection of emerging neurotropic viruses in Europe, 2017. Euro Surveill. 2019, 24. [Google Scholar] [CrossRef]
- You, Y.; Moreira, B.G.; Behlke, M.A.; Owczarzy, R. Design of LNA probes that improve mismatch discrimination. Nucleic Acids Res. 2006, 34, e60. [Google Scholar] [CrossRef]
- Wang, Q.Y.; Li, F.; Shen, X.X.; Fu, S.H.; He, Y.; Lei, W.W.; Liang, G.D.; Wang, H.Y.; Ma, X.J. A Reverse-transcription Recombinase-aided Amplification Assay for the Rapid Detection of the Far-Eastern Subtype of Tick-borne Encephalitis Virus. Biomed. Environ. Sci. BES 2019, 32, 357–362. [Google Scholar] [CrossRef]
- Schwaiger, M.; Cassinotti, P. Development of a quantitative real-time RT-PCR assay with internal control for the laboratory detection of tick borne encephalitis virus (TBEV) RNA. J. Clin. Virol. Off. Publ. Pan Am. Soc. Clin. Virol. 2003, 27, 136–145. [Google Scholar] [CrossRef]
- Gäumann, R.; Mühlemann, K.; Strasser, M.; Beuret, C.M. High-throughput procedure for tick surveys of tick-borne encephalitis virus and its application in a national surveillance study in Switzerland. Appl. Env. Microbiol. 2010, 76, 4241–4249. [Google Scholar] [CrossRef]
- Lindblom, P.; Wilhelmsson, P.; Fryland, L.; Sjöwall, J.; Haglund, M.; Matussek, A.; Ernerudh, J.; Vene, S.; Nyman, D.; Andreassen, A.; et al. Tick-borne encephalitis virus in ticks detached from humans and follow-up of serological and clinical response. Ticks Tick-Borne Dis. 2014, 5, 21–28. [Google Scholar] [CrossRef] [PubMed]
| Primer | Sequence 5′–3′ | |
|---|---|---|
| Prototype primer set | TBE-Fa | ATGTGTACGACGCCAACAAA |
| TBE-Fg | ATGTGTACGACGCCAACAAG | |
| TBE-R | GACCCTGCACAACAAIGACA | |
| Refined primer set | TBE-Fa8T | ATGTGTATGACGCCAACAAA |
| TBE-Fg8T | ATGTGTATGACGCCAACAAG | |
| TBE-R16G | GACCCTGCACAACAAGGACA | |
| TBE-R16G4T 1 | GACTCTGCACAACAAGGACA |
| Virus Strain | Virus/Subtype | Material | Accession Number |
|---|---|---|---|
| 1993/783 | TBEV-Eu | Culture supernatant | MT311860 |
| Absettarov | TBEV-Eu | Supplied as RNA | KJ000002 |
| Hochosterwitz | TBEV-Eu | Culture supernatant | MT311861 |
| Hypr | TBEV-Eu | Supplied as RNA/EQA | U39292 |
| Neudörfl 1 | TBEV-Eu | Vaccine | U27495 |
| Sokoup | TBEV-Eu | Supplied as RNA | NA |
| Latvia 1-96 | TBEV-Sib | Culture supernatant | GU183382 |
| Vasilchenko | TBEV-Sib | Supplied as RNA | AF069066 |
| Sofjin | TBEV-FE | Supplied as RNA | AB062064 |
| LI/NOR | Louping ill virus | Culture supernatant | D12936 |
| - | Zika | EQA | NA |
| - | Dengue type 1 | EQA | NA |
| - | Dengue type 2 | EQA | NA |
| - | Dengue type 3 | EQA | NA |
| - | Dengue type 4 | EQA | NA |
| - | Yellow fever | EQA | NA |
| - | Toscana lineage A | EQA | NA |
| - | Toscana lineage B | EQA | NA |
| - | West Nile lineage 1 | EQA | NA |
| - | West Nile lineage 2 | EQA | NA |
| - | Utsu virus | EQA | NA |
| Accession No. | Sub- Type | TBE-Fa | Tm | ΔTm | TBE-Fg | Tm | ΔTm | TBE-R | Tm | ΔTm |
|---|---|---|---|---|---|---|---|---|---|---|
| 5′ATGTGTACGACGCCAACAAA 3′ | 5′ATGTGTACGACGCCAACAAG3′ | 5′GACCCTGCACAACAAIGACA3′ | ||||||||
| EF565947 | EU | 3′TACACATGCTGCGGTTGTTT5′ 1 | 66 °C | 3′TACACATGCTGCGGTTGTTT5′ | 66 °C | 3′CTGGGACGTGTTGTTTCTGT5′ 2 | 60 °C | |||
| KT749573 | Sib | 3′TACACATACTACGGTTGTCT5′ | 43 °C | 23 °C | 3′TACACATACTACGGTTGTCT5′ | 42 °C | 24 °C | 3′CTGAGACGTGTTGTTCCTGT5′ 3,4 | 56 °C | 4 °C |
| KT748750 | Sib | 3′TACACATACTGCGGTTGTTT5′ | 56 °C | 10 °C | 3′TACACATACTGCGGTTGTTT5′ | 56 °C | 10 °C | 3′CTGAGACGTGTAGTTCCTGT5′ | 50 °C | 10 °C |
| AB022296 | FE | 3′TGCACATACTGCGTTTGTTT5′ | 43 °C | 23 °C | 3′TGCACATACTGCGTTTGTTT5′ | 42 °C | 24 °C | 3′ATGAGACGTATTGTTCCTGT5′ | 44 °C | 16 °C |
| KJ633033 | Bai | 3′TACATATACTGCGTTTGTTC5′ | 37 °C | 29 °C | 3′TACATATACTGCGTTTGTTC5′ | 39 °C | 27 °C | 3′CTGAGACGTGTTATCCCTGT5′ | 38 °C | 22 °C |
| JX315851 | Sib | 3′TACACATACTGCGGTTGTTC5′ 3 | 55 °C | 11 °C | 3′TACACATACTGCGGTTGTTC5′ | 57 °C | 9 °C | 3′TTGAGACGTATTGTTCCAGT5′ | 38 °C | 22 °C |
| KU761569 | FE | 3′TACACATACTGCGATTGTTT5′ 4,5 | 46 °C | 20 °C | 3′TACACATACTGCGATTGTTT5′ | 45 °C | 21 °C | 3′ATGAGACGTATTGTCCCTAT5′ | <35 °C | - |
| KC422666 | FE | 3′TACACATACTGCGATTGTTC5′ | 43 °C | 23 °C | 3′TACACATACTGCGATTGTTC5′ | 46 °C | 20 °C | 3′ATGAGCCGTGTTATTCCTGT5′ 5 | 36 °C | 24 °C |
| GU121642 | FE | 3′TGCACATACTACGGTTGTTT5′ | 44 °C | 22 °C | 3′TGCACATACTACGGTTGTTT5′ | 42 °C | 24 °C | 3′ATGAGCTGTGTTGTTCCTGT5′ | 46 °C | 14 °C |
| AF231807 | Sib | 3′TACACATACTGCGGTTGTTC5′ | 57 °C | 9 °C | 3′TACACATACTGCGGTTGTTC5′ | 58 °C | 8 °C | 3′TTGAGACGTATCGTTCCTAT5′ | <35 °C | - |
| Accession no. | Subtype | TBE-Fa8T | Tm | ΔTm | TBE-R16G | Tm |
|---|---|---|---|---|---|---|
| 5′ATGTGTATGACGCCAACAAA3′ | 5′GACCCTGCACAACAAGGACA3′ | |||||
| EF565947 | EU | 3′TACACATGCTGCGGTTGTTT5′ | 61 °C | 4 °C | 3′CTGGGACGTGTTGTTTCTGT5′ | 61 °C |
| KT749573 | Sib | 3′TACACATACTACGGTTGTCT5′ | 53 °C | 12 °C | 3′CTGAGACGTGTTGTTCCTGT5′ | 61 °C |
| KT748750 | Sib | 3′TACACATACTGCGGTTGTTT5′ | 65 °C | 0 °C | 3′CTGAGACGTGTAGTTCCTGT5′ | 55 °C |
| AB022296 | FE | 3′TGCACATACTGCGTTTGTTT5′ | 55 °C | 10 °C | 3′ATGAGACGTATTGTTCCTGT5′ | 50 °C |
| KJ633033 | Bai | 3′TACATATACTGCGTTTGTTC5′ | 47 °C | 18 °C | 3′CTGAGACGTGTTATCCCTGT5′ | 43 °C |
| JX315851 | Sib | 3′TACACATACTGCGGTTGTTC5′ | 65 °C | 0 °C | 3′TTGAGACGTATTGTTCCAGT5′ | 42 °C |
| KU761569 | FE | 3′TACACATACTGCGATTGTTT5′ | 56 °C | 9 °C | 3′ATGAGACGTATTGTCCCTAT5′ | <35 °C |
| KC422666 | FE | 3′TACACATACTGCGATTGTTC5′ | 56 °C | 9 °C | 3′ATGAGCCGTGTTATTCCTGT5′ | 42 °C |
| GU121642 | FE | 3′TGCACATACTACGGTTGTTT5′ | 54 °C | 11 °C | 3′ATGAGCTGTGTTGTTCCTGT5′ | 54 °C |
| AF231807 | FE | 3′TACACATACTGCGGTTGTTC5′ | 64 °C | 1 °C | 3′TTGAGACGTATCGTTCCTAT5′ | <35 °C |
| Virus Strain | Real-Time PCR Result | Sequencing |
|---|---|---|
| 1993/783 | Positive | Confirmed |
| Absettarov | Positive | Confirmed |
| Hochosterwitz | Positive | Confirmed |
| Hypr | Positive | Not confirmed |
| Neudörfl 1 | Positive | Confirmed |
| Sokoup | Positive | Confirmed (TBEV-Eu) |
| Latvia-1-96 | Positive | Confirmed |
| Vasilchenko | Positive | Confirmed |
| Sofijn | Positive | Confirmed |
| LI/NOR | Negative | Not sequenced |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pedersen, B.N.; Jenkins, A.; Paulsen, K.M.; Basset, C.; Andreassen, Å.K. Development of a Real-Time PCR Method for the Detection of European and Siberian Subtypes of Tick-Borne Encephalitis Virus. Microbiol. Res. 2023, 14, 1545-1558. https://doi.org/10.3390/microbiolres14040106
Pedersen BN, Jenkins A, Paulsen KM, Basset C, Andreassen ÅK. Development of a Real-Time PCR Method for the Detection of European and Siberian Subtypes of Tick-Borne Encephalitis Virus. Microbiology Research. 2023; 14(4):1545-1558. https://doi.org/10.3390/microbiolres14040106
Chicago/Turabian StylePedersen, Benedikte N., Andrew Jenkins, Katrine M. Paulsen, Coraline Basset, and Åshild K. Andreassen. 2023. "Development of a Real-Time PCR Method for the Detection of European and Siberian Subtypes of Tick-Borne Encephalitis Virus" Microbiology Research 14, no. 4: 1545-1558. https://doi.org/10.3390/microbiolres14040106
APA StylePedersen, B. N., Jenkins, A., Paulsen, K. M., Basset, C., & Andreassen, Å. K. (2023). Development of a Real-Time PCR Method for the Detection of European and Siberian Subtypes of Tick-Borne Encephalitis Virus. Microbiology Research, 14(4), 1545-1558. https://doi.org/10.3390/microbiolres14040106






