A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces
Abstract
:1. Introduction
2. Biofilm Formation
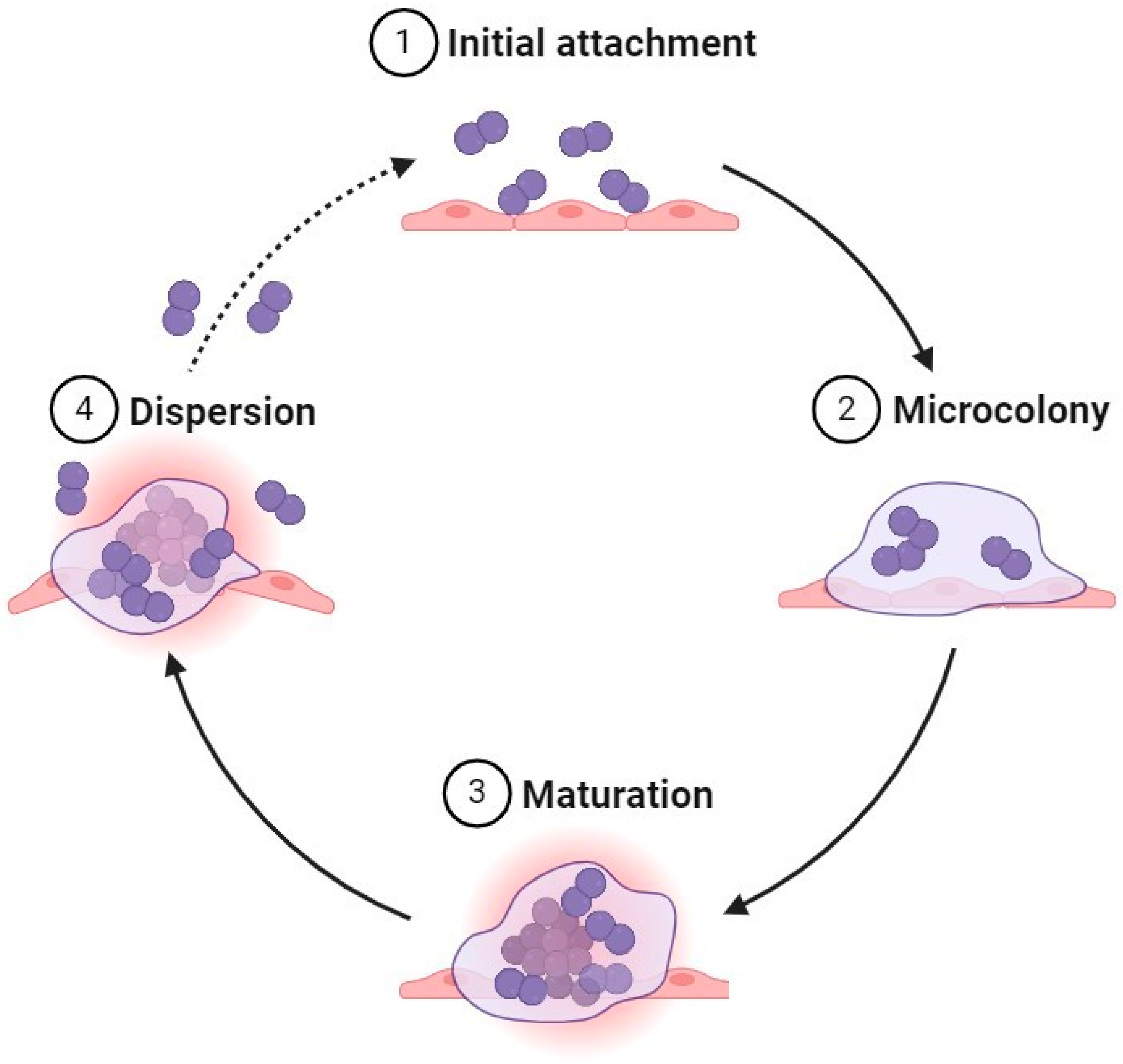
2.1. Formation Steps
2.1.1. Initial Attachment
2.1.2. Microcolony
2.1.3. Maturation
2.1.4. Dispersion
3. The Biofilm Matrix Components
3.1. Polysaccharides
3.2. Proteins
3.3. Lipids and Biosurfactants
3.4. Extracellular DNA
4. Biofilm Issues in Food Processing
5. Detection Methods of Biofilm Formation
6. Biofilm Prevention and Treatment in the Food Industry
6.1. Conventional Sanitizing Materials
6.2. Enzymes
6.3. Essential Oils
6.4. Bacteriocins
7. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Borges, A.; Meireles, A.; Mergulhão, F.; Melo, L.; Simões, M. Biofilm control with enzymes. In Recent Trends in Biofilm Science and Technology; Elsevier: Amsterdam, The Netherlands, 2020; pp. 249–271. [Google Scholar] [CrossRef]
- Zhu, T.; Yang, C.; Bao, X.; Chen, F.; Guo, X. Strategies for controlling biofilm formation in food industry. Grain Oil Sci. Technol. 2022, 5, 179–186. [Google Scholar] [CrossRef]
- Bamford, N.C.; MacPhee, C.E.; Stanley-Wall, N.R. Microbial primer: An introduction to biofilms—What they are, why they form and their impact on built and natural environments. Microbiology 2023, 169, 8. [Google Scholar] [CrossRef] [PubMed]
- Ma, Z.; Tang, X.; Stanford, K.; Chen, X.; McAllister, T.A.; Niu, Y.D. Single- and dual-species biofilm formation by Shiga toxin-producing Escherichia coli and Salmonella, and their susceptibility to an engineered peptide WK2. Microorganisms 2021, 9, 2510. [Google Scholar] [CrossRef] [PubMed]
- Nan, Y.; Rodas-Gonzalez, A.; Stanford, K.; Nadon, C.; Yang, X.; McAllister, T.; Narváez-Bravo, C. Formation and transfer of multi-species biofilms containing E. coli O103:H2 on food contact surfaces to beef. Front. Microbiol. 2022, 13, 863778. [Google Scholar] [CrossRef]
- Wagner, E.M.; Pracser, N.; Thalguter, S.; Fischel, K.; Rammer, N.; Pospíšilová, L.; Alispahic, M.; Wagner, M.; Rychli, K. Identification of biofilm hotspots in a meat processing environment: Detection of spoilage bacteria in multi-species biofilms. Int. J. Food Microbiol. 2020, 328, 108668. [Google Scholar] [CrossRef]
- Jahid, I.K.; Ha, S.D. A review of microbial biofilms of produce: Future challenge to food safety. Food Sci. Biotechnol. 2012, 21, 299–316. [Google Scholar] [CrossRef]
- Mizan, M.F.; Jahid, I.K.; Ha, S.D. Microbial biofilms in seafood: A food-hygiene challenge. Food Microbiol. 2015, 49, 41–55. [Google Scholar] [CrossRef]
- Carrascosa, C.; Raheem, D.; Ramos, F.; Saraiva, A.; Raposo, A. Microbial biofilms in the food industry—A comprehensive review. Int. J. Environ. Res. Public Health 2021, 18, 2014. [Google Scholar] [CrossRef]
- Lequette, Y.; Boels, G.; Clarisse, M.; Faille, C. Using enzymes to remove biofilms of bacterial isolates sampled in the food industry. Biofouling 2010, 26, 421–431. [Google Scholar] [CrossRef]
- Giaouris, E.E.; Simões, M.V. Pathogenic biofilm formation in the food industry and alternative control strategies. In Foodborne Diseases; Academic Press: New York, NY, USA, 2018; pp. 309–377. [Google Scholar] [CrossRef]
- Penesyan, A.; Paulsen, I.T.; Kjelleberg, S.; Gillings, M.R. Three faces of biofilms: A microbial lifestyle, a nascent multicellular organism, and an incubator for diversity. npj Biofilms Microbiomes 2021, 7, 80. [Google Scholar] [CrossRef]
- Klausen, M.; Aaes-Jørgensen, A.; Molin, S.; Tolker-Nielsen, T. Involvement of bacterial migration in the development of complex multicellular structures in Pseudomonas aeruginosa biofilms. Mol. Microbiol. 2003, 50, 61–68. [Google Scholar] [CrossRef] [PubMed]
- Beuchat, L.R.; Kim, H.; Gurtler, J.B.; Lin, L.; Ryu, J.H.; Richards, G.M. Cronobacter sakazakii in foods and factors affecting its survival, growth, and inactivation. Int. J. Food Microbiol. 2009, 136, 204–213. [Google Scholar] [CrossRef] [PubMed]
- Merino, L.E.; Procura, F.; Trejo, F.M.; Bueno, D.J.; Golowczyc, M.A. Biofilm formation by Salmonella sp. in the poultry industry: Detection, control and eradication strategies. Food Res. Int. 2019, 119, 530–540. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.; Wingender, J.; Szewzyk, U.; Steinberg, P.D.; Rice, S.A.; Kjelleberg, S. Biofilms: An emergent form of bacterial life. Nat. Rev. Microbiol. 2016, 14, 563–575. [Google Scholar] [CrossRef]
- Stewart, P.S.; Roe, F.; Rayner, J.; Elkins, J.G.; Lewandowski, Z.; Ochsner, U.A.; Hassett, D.J. Effect of catalase on hydrogen peroxide penetration into Pseudomonas aeruginosa biofilms. Appl. Environ. Microbiol. 2000, 66, 836–838. [Google Scholar] [CrossRef]
- Stewart, P.S.; Owkes, M. Simulation of catalase-dependent tolerance of microbial biofilm to hydrogen peroxide with a biofilm computer model. npj Biofilms Microbiomes 2023, 9, 60. [Google Scholar] [CrossRef]
- Trmcic, A.; Chen, H.; Trzaskowska, M.; Tamber, S.; Wang, S. Biofilm-forming capacity of five Salmonella strains and their fate on postharvest mini cucumbers. J. Food Prot. 2018, 81, 1871–1879. [Google Scholar] [CrossRef] [PubMed]
- Ripolles-Avila, C.; Ríos-Castillo, A.G.; Fontecha-Umaña, F.; Rodríguez-Jerez, J.J. Removal of Salmonella enterica serovar Typhimurium and Cronobacter sakazakii biofilms from food contact surfaces through enzymatic catalysis. J. Food Saf. 2020, 40, 2. [Google Scholar] [CrossRef]
- Meireles, A.; Borges, A.; Giaouris, E.; Simões, M. The current knowledge on the application of anti-biofilm enzymes in the food industry. Food Res. Int. 2016, 86, 140–146. [Google Scholar] [CrossRef]
- Gómez-Sequeda, N.; Cáceres, M.; Stashenko, E.; Hidalgo, W.; Ortíz, C. Antimicrobial and antibiofilm activities of essential oils against Escherichia coli O157:H7 and methicillin-resistant Staphylococcus aureus (MRSA). Antibiotics 2020, 9, 730. [Google Scholar] [CrossRef]
- Donlan, R.M. Biofilms: Microbial life on surfaces. Emerg. Infect. Dis. 2002, 8, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Bai, X.; Nakatsu, C.H.; Bhunia, A.K. Bacterial biofilms and their implications in pathogenesis and food safety. Foods 2021, 10, 2117. [Google Scholar] [CrossRef] [PubMed]
- Tasneem, U.; Yasin, N.; Nisa, I.; Shah, F.; Rasheed, U.; Momin, F.; Zaman, S.; Qasim, M. Biofilm producing bacteria: A serious threat to public health in developing countries. J. Food Sci. Nutr. 2018, 1, 25–31. [Google Scholar] [CrossRef]
- Flemming, H.; Wingender, J. The biofilm matrix. Nat. Rev. Microbiol. 2010, 8, 623–633. [Google Scholar] [CrossRef]
- Fux, C.A.; Costerton, J.W.; Stewart, P.S.; Stoodley, P. Survival strategies of infectious biofilms. Trends Microbiol. 2005, 13, 34–40. [Google Scholar] [CrossRef] [PubMed]
- Steenackers, H.; Hermans, K.; Vanderleyden, J.; De Keersmaecker, S.C.J. Salmonella biofilms: An overview on occurrence, structure, regulation and eradication. Food Res. Int. 2012, 45, 502–531. [Google Scholar] [CrossRef]
- Costerton, J.W.; Lewandowski, Z.; Caldwell, D.E.; Korber, D.R.; Lappin-Scott, H.M. Microbial biofilms. Annu. Rev. Microbiol. 1995, 49, 711–745. [Google Scholar] [CrossRef]
- Watnick, P.; Kolter, R. Biofilm, city of microbes. J. Bacteriol. 2000, 182, 2675–2679. [Google Scholar] [CrossRef]
- Lindsay, D.; von Holy, A. Bacterial biofilms within the clinical setting: What healthcare professionals should know. J. Hosp. Infect. 2006, 64, 313–325. [Google Scholar] [CrossRef]
- Garrett, T.R.; Bhakoo, M.; Zhang, Z. Bacterial adhesion and biofilms on surfaces. Prog. Nat. Sci. Mater. Int. 2008, 18, 1049–1056. [Google Scholar] [CrossRef]
- Rabin, N.; Zheng, Y.; Opoku-Temeng, C.; Du, Y.; Bonsu, E.; Sintim, H.O. Biofilm formation mechanisms and targets for developing antibiofilm agents. Future Med. Chem. 2015, 7, 493–512. [Google Scholar] [CrossRef] [PubMed]
- Zhao, X.; Zhao, F.; Wang, J.; Zhong, N. Biofilm formation and control strategies of foodborne pathogens: Food safety perspectives. RSC Adv. 2017, 7, 36670–36683. [Google Scholar] [CrossRef]
- O’Toole, G.A.; Kaplan, H.B.; Kolter, R. Biofilm formation as microbial development. Annu. Rev. Microbiol. 2000, 54, 49–79. [Google Scholar] [CrossRef]
- Waters, C.M.; Lu, W.; Rabinowitz, J.D.; Bassler, B.L. Quorum sensing controls biofilm formation in Vibrio cholerae through modulation of cyclic di-GMP levels and repression of vpsT. J. Bacteriol. 2008, 190, 2527–2536. [Google Scholar] [CrossRef]
- Tolker-Nielsen, T.; Molin, S. Spatial organization of microbial biofilm communities. Microb. Ecol. 2000, 40, 75–84. [Google Scholar] [CrossRef]
- Lepuschitz, S.; Ruppitsch, W.; Pekard-Amenitsch, S.; Forsythe, S.J.; Cormican, M.; Mach, R.L.; Piérard, D.; Allerberger, F. Multicenter study of Cronobacter sakazakii infections in humans, Europe, 2017. Emerg. Infect. Dis. 2019, 25, 515–522. [Google Scholar] [CrossRef] [PubMed]
- Flemming, H.; Van Hullebusch, E.D.; Neu, T.R.; Nielsen, P.H.; Seviour, T.; Stoodley, P.; Wingender, J.; Wuertz, S. The biofilm matrix: Multitasking in a shared space. Nat. Rev. Microbiol. 2022, 21, 70–86. [Google Scholar] [CrossRef]
- Solomon, E.B.; Niemira, B.A.; Sapers, G.M. Annihilation of Escherichia coli O157:H7 on lettuce by electrolyzed oxidizing water. J. Food Prot. 2005, 68, 173–178. [Google Scholar] [CrossRef]
- Limoli, D.H.; Jones, C.J.; Wozniak, D.J. Bacterial extracellular polysaccharides in biofilm formation and function. Microbiol. Spectr. 2015, 3, 223–247. [Google Scholar] [CrossRef]
- Singh, S.; Datta, S.; Narayanan, K.B.; Rajnish, K.N. Bacterial exo-polysaccharides in biofilms: Role in antimicrobial resistance and treatments. J. Genet. Eng. Biotechnol. 2021, 19, 140. [Google Scholar] [CrossRef]
- Lear, G.; Lewis, G.D.; Cenci, G. Rheological characterization of the extracellular polymeric substances produced by five species of environmental aerobic biofilm-forming bacteria. Water Res. 2010, 44, 3862–3870. [Google Scholar] [CrossRef]
- Rather, M.A.; Gupta, K.; Mandal, M. Microbial biofilm: Formation, architecture, antibiotic resistance, and control strategies. Braz. J. Microbiol. 2021, 52, 1701–1718. [Google Scholar] [CrossRef] [PubMed]
- Adetunji, A.I.; Olaniran, A.O. Production and potential biotechnological applications of microbial surfactants: An overview. Saudi J. Biol. Sci. 2021, 28, 669–679. [Google Scholar] [CrossRef]
- Ibáñez de Aldecoa, A.L.; Zafra, O.; González-Pastor, J.E. Mechanisms and regulation of extracellular DNA release and its biological roles in microbial communities. Front. Microbiol. 2017, 8, 1390. [Google Scholar] [CrossRef] [PubMed]
- Uruén, C.; Chopo-Escuin, G.; Tommassen, J.; Mainar-Jaime, R.C.; Arenas, J. Biofilms as promoters of bacterial antibiotic resistance and tolerance. Antibiotics 2020, 10, 3. [Google Scholar] [CrossRef]
- Kumpel, E.; Nelson, K.L. Comparing microbial water quality in an intermittent and continuous piped water supply. Water Res. 2013, 47, 5176–5188. [Google Scholar] [CrossRef]
- Francolini, I.; Donelli, G. Prevention and control of biofilm-based medical-device-related infections. FEMS Immunol. Med. Microbiol. 2010, 59, 227–238. [Google Scholar] [CrossRef]
- Galié, S.; Garcia-Gutierrez, C.; Miguélez, E.M.; Villar, C.J.; Lombó, F. Biofilms in the food industry: Health aspects and control methods. Front. Microbiol. 2018, 9, 89. [Google Scholar] [CrossRef]
- Srey, S.; Jahid, I.K.; Ha, S. Biofilm formation in food industries: A food safety concern. Food Control 2013, 31, 572–585. [Google Scholar] [CrossRef]
- Stepanović, S.; Ćirković, I.; Ranin, L.; Švabić-Vlahović, M. Biofilm formation by Salmonella spp. and Listeria monocytogenes on plastic surface. Lett. Appl. Microbiol. 2004, 38, 428–432. [Google Scholar] [CrossRef]
- Oh, S.; Chen, P.; Kang, D. Biofilm formation by Enterobacter sakazakii grown in artificial broth and infant milk formula on plastic surface. J. Rapid Methods Autom. Microbiol. 2007, 15, 311–319. [Google Scholar] [CrossRef]
- Brooks, J.D.; Flint, S. Biofilms in the food industry: Problems and potential solutions. Int. J. Food Sci. Technol. 2008, 43, 2163–2176. [Google Scholar] [CrossRef]
- Reij, M.; Aantrekker, E.D. Recontamination as a source of pathogens in processed foods. Int. J. Food Microbiol. 2004, 91, 1–11. [Google Scholar] [CrossRef] [PubMed]
- Corcoran, M.; Morris, D.; De Lappe, N.; O’Connor, J.; Lalor, P.; Dockery, P.; Cormican, M. Commonly used disinfectants fail to eradicate Salmonella enterica biofilm from food contact surface materials. Appl. Environ. Microbiol. 2013, 80, 1507–1514. [Google Scholar] [CrossRef]
- Van Houdt, R.; Michiels, C.W. Biofilm formation and the food industry: A focus on the bacterial outer surface. J. Appl. Microbiol. 2010, 109, 1117–1131. [Google Scholar] [CrossRef] [PubMed]
- Shi, X.; Zhu, X. Biofilm formation and food safety in food industries. Trends Food Sci. Technol. 2009, 20, 407–413. [Google Scholar] [CrossRef]
- Smith, A.B.; Johnson, C.D. Investigation of a Salmonella Typhimurium outbreak associated with biofilm formation at a dairy processing plant. J. Food Saf. 2009, 29, 189–202. [Google Scholar]
- Hurrell, E.; Kucerova, E.; Loughlin, M.; Caubilla-Barron, J.; Forsythe, S. Biofilm formation on enteral feeding tubes by Cronobacter sakazakii, Salmonella serovars and other Enterobacteriaceae. Int. J. Food Microbiol. 2009, 136, 227–231. [Google Scholar] [CrossRef]
- Kim, H.; Ryu, J.-H.; Beuchat, L.R. Attachment of and biofilm formation by Enterobacter sakazakii on stainless steel and enteral feeding tubes. Appl. Environ. Microbiol. 2006, 72, 5846–5856. [Google Scholar] [CrossRef]
- Wilson, C.; Lukowicz, R.; Merchant, S.; Valquier-Flynn, H.; Caballero, J.; Sandoval, J.; Okuom, M.; Huber, C.; Brooks, T.D.; Wilson, E.; et al. Quantitative and qualitative assessment methods for biofilm growth: A mini-review. Res. Rev. J. Eng. Technol. 2017, 6, 1–9. [Google Scholar]
- Haney, E.; Trimble, M.; Cheng, J.; Vallé, Q.; Hancock, R. Critical assessment of methods to quantify biofilm growth and evaluate antibiofilm activity of host defence peptides. Biomolecules 2018, 8, 29. [Google Scholar] [CrossRef] [PubMed]
- Mandakhalikar, K.D.; Rahmat, J.N.; Chiong, E.; Neoh, K.G.; Shen, L.; Tambyah, P.A. Extraction and quantification of biofilm bacteria: Method optimized for urinary catheters. Sci. Rep. 2018, 8, 8069. [Google Scholar] [CrossRef] [PubMed]
- Koo, H. Inhibition of Streptococcus mutans biofilm accumulation and polysaccharide production by apigenin and tt-farnesol. J. Antimicrob. Chemother. 2003, 52, 782–789. [Google Scholar] [CrossRef] [PubMed]
- Ambriz-Aviña, V.; Contreras-Garduño, J.A.; Pedraza-Reyes, M. Applications of flow cytometry to characterize bacterial physiological responses. Biomed. Res. Int. 2014, 2014, 461941. [Google Scholar] [CrossRef]
- Schwartz, K.; Stephenson, R.; Hernandez, M.; Jambang, N.; Boles, B. The use of drip flow and rotating disk reactors for Staphylococcus aureus biofilm analysis. J. Vis. Exp. 2010, 46, 2470. [Google Scholar] [CrossRef]
- Adetunji, V.O.; Odetokun, I.A. Assessment of biofilm in E. coli O157:H7 and Salmonella strains: Influence of cultural conditions. Am. J. Food Technol. 2012, 7, 582–595. [Google Scholar] [CrossRef]
- Cloete, T.E.; Brözel, V.S.; Von Holy, A. Practical aspects of biofouling control in industrial water systems. Int. Biodeterior. Biodegrad. 1992, 29, 299–341. [Google Scholar] [CrossRef]
- Buchholz, F.; Harms, H.; Maskow, T. Biofilm research using calorimetry—A marriage made in heaven? Biotechnol. J. 2010, 5, 1339–1350. [Google Scholar] [CrossRef]
- Olaimat, A.N.; Ababneh, A.M.; Al-Holy, M.; Al-Nabulsi, A.; Qatatsheh, A.A.; Jaradat, Z.W.; Albiss, B.A.; Osaili, T.; Holley, R.A. Antimicrobial and antibiofilm activities of Aleppo pine essential oil and enzymes against Salmonella enterica biofilms formed on stainless steel and plastic surfaces. Food Control 2024, 164, 110587. [Google Scholar] [CrossRef]
- Relucenti, M.; Familiari, G.; Donfrancesco, O.; Taurino, M.; Li, X.; Chen, R.; Artini, M.; Papa, R.; Selan, L. Microscopy methods for biofilm imaging: Focus on SEM and VP-SEM pros and cons. Biology 2021, 10, 51. [Google Scholar] [CrossRef]
- Chen, Y.; Chauhan, S.; Gong, C.; Dayton, H.; Xu, C.; De, D.; Tsai, Y.-Y.W.; Datta, M.S.; Rosoklija, G.B.; Dwork, A.J.; et al. Low-cost and scalable projected light-sheet microscopy for the high-resolution imaging of cleared tissue and living samples. Nat. Biomed. Eng. 2024, 8, 1109–1123. [Google Scholar] [CrossRef] [PubMed]
- Bogachev, M.I.; Volkov, V.Y.; Markelov, O.A.; Trizna, E.Y.; Baydamshina, D.R.; Melnikov, V.; Murtazina, R.R.; Zelenikhin, P.V.; Sharafutdinov, I.S.; Kayumov, A.R. Fast and simple tool for the quantification of biofilm-embedded cells sub-populations from fluorescent microscopic images. PLoS ONE 2018, 13, e0193267. [Google Scholar] [CrossRef] [PubMed]
- Yokoi, N.; Yuasa, T.; Niskanen, I.; Hibino, K.; Funamizu, H.; Aizu, Y. Monitoring of the formation of biofilm inside a glass tube using light scattering patterns. Opt. Rev. 2024, 31, 225–235. [Google Scholar] [CrossRef]
- Welch, K.; Cai, Y.; Strømme, M. A method for quantitative determination of biofilm viability. J. Funct. Biomater. 2012, 3, 418–431. [Google Scholar] [CrossRef]
- Mah, T.F.C.; O’Toole, G.A. Mechanisms of biofilm resistance to antimicrobial agents. Trends Microbiol. 2001, 9, 34–39. [Google Scholar] [CrossRef]
- Gilbert, P.; Maira-Litrán, T.; McBain, A.J.; Rickard, A.H.; Whyte, F.W. The physiology and collective recalcitrance of microbial biofilm communities. In Advances in Microbial Physiology; Elsevier: Amsterdam, The Netherlands, 2002; pp. 203–256. [Google Scholar] [CrossRef]
- Aryal, M.; Muriana, P.M. Efficacy of commercial sanitizers used in food processing facilities for inactivation of Listeria monocytogenes, E. Coli O157:H7, and Salmonella biofilms. Foods 2019, 8, 639. [Google Scholar] [CrossRef]
- Meesilp, N.; Mesil, N. Effect of microbial sanitizers for reducing biofilm formation of Staphylococcus aureus and Pseudomonas aeruginosa on stainless steel by cultivation with UHT milk. Food Sci. Biotechnol. 2018, 28, 289–296. [Google Scholar] [CrossRef]
- Marriott, N.G.; Schilling, M.W.; Gravani, R.B. Principles of Food Sanitation. In Food Science Text Series; Springer International Publishing: Cham, Switzerland, 2018. [Google Scholar] [CrossRef]
- Yoon, J.; Lee, S. Review: Comparison of the effectiveness of decontaminating strategies for fresh fruits and vegetables and related limitations. Crit. Rev. Food Sci. Nutr. 2017, 58, 3189–3208. [Google Scholar] [CrossRef]
- Wirtanen, G.; Salo, S. Disinfection in food processing—Efficacy testing of disinfectants. Rev. Environ. Sci. Bio/Technol. 2003, 2, 293–306. [Google Scholar] [CrossRef]
- Arnold, W.A.; Blum, A.; Branyan, J.; Bruton, T.A.; Carignan, C.C.; Cortopassi, G.A.; Datta, S.; DeWitt, J.C.; Doherty, A.; Halden, R.U.; et al. Quaternary ammonium compounds: A chemical class of emerging concern. Environ. Sci. Technol. 2023, 57, 7645–7665. [Google Scholar] [CrossRef]
- Nahar, S.; Mizan, M.F.R.; Ha, A.J.W.; Ha, S. Advances and future prospects of enzyme-based biofilm prevention approaches in the food industry. Compr. Rev. Food Sci. Food Saf. 2018, 17, 1484–1502. [Google Scholar] [CrossRef]
- Kalpana, B.J.; Aarthy, S.; Pandian, S.K. Antibiofilm activity of α-amylase from Bacillus subtilis S8-18 against biofilm forming human bacterial pathogens. Appl. Biochem. Biotechnol. 2012, 167, 1778–1794. [Google Scholar] [CrossRef] [PubMed]
- Lentner, M.; Otto, H. Lipolytic enzymes as antibiofilm agents. In Enzymes in Action; Springer: Berlin/Heidelberg, Germany, 2013; pp. 65–87. [Google Scholar]
- Karygianni, L.; Ren, Z.; Koo, H.; Thurnheer, T. Biofilm matrixome: Extracellular components in structured microbial communities. Trends Microbiol. 2020, 28, 668–681. [Google Scholar] [CrossRef] [PubMed]
- Koo, H.; Allan, R.N.; Howlin, R.P.; Stoodley, P.; Hall-Stoodley, L. Targeting microbial biofilms: Current and prospective therapeutic strategies. Nat. Rev. Microbiol. 2017, 15, 740–755. [Google Scholar] [CrossRef] [PubMed]
- Ramakrishnan, R.; Singh, A.K.; Singh, S.; Chakravortty, D.; Das, D. Enzymatic dispersion of biofilms: An emerging biocatalytic avenue to combat biofilm-mediated microbial infections. J. Biol. Chem. 2022, 298, 102352. [Google Scholar] [CrossRef]
- Tang, X.; Flint, S.H.; Bennett, R.J.; Brooks, J.D.; Morton, R.H. Biofilm growth of individual and dual strains of Klebsiella oxytoca from the dairy industry on ultrafiltration membranes. J. Ind. Microbiol. Biotechnol. 2009, 36, 1491–1497. [Google Scholar] [CrossRef]
- Miao, L.; Wang, P.; Hou, J.; Yao, Y.; Liu, Z.; Liu, S.; Li, T. Distinct community structure and microbial functions of biofilms colonizing microplastics. Sci. Total Environ. 2019, 650, 2395–2402. [Google Scholar] [CrossRef]
- Percival, S.L.; Hill, K.E.; Malic, S.; Thomas, D.W.; Williams, D.W. Antimicrobial tolerance and the significance of persister cells in recalcitrant chronic wound biofilms. Wound Repair Regener. 2011, 19, 1–9. [Google Scholar] [CrossRef]
- Rodríguez-López, P.; Puga, C.H.; Orgaz, B.; Cabo, M.L. Quantifying the combined effects of pronase and benzalkonium chloride in removing late-stage Listeria monocytogenes–Escherichia coli dual-species biofilms. Biofouling 2017, 33, 690–702. [Google Scholar] [CrossRef]
- Wang, H.; Xing, T.; Wu, N.; Xu, X. Removal of Salmonella biofilm formed under meat processing environment by surfactant in combination with bio-enzyme. LWT 2016, 66, 298–304. [Google Scholar] [CrossRef]
- Tajkarimi, M.M.; Ibrahima, S.A.; Cliver, D.O. Antimicrobial herb and spice compounds in food. Food Control 2010, 21, 1199–1218. [Google Scholar] [CrossRef]
- Nazzaro, F.; Fratianni, F.; De Martino, L.; Coppola, R.; De Feo, V. Effect of essential oils on pathogenic bacteria. Pharmaceuticals 2013, 6, 1451–1474. [Google Scholar] [CrossRef] [PubMed]
- Rasamiravaka, T.; Labtani, Q.; Duez, P.; Jaziri, M.E. The formation of biofilms by Pseudomonas aeruginosa: A review of the natural and synthetic compounds interfering with control mechanisms. BioMed Res. Int. 2015, 2015, 759348. [Google Scholar] [CrossRef]
- Čabarkapa, I.; Čolović, R.; Ðuragić, O.; Popović, S.; Kokić, B.; Dubravka, M.; Pezo, L. Anti-biofilm activities of essential oils rich in carvacrol and thymol against Salmonella Enteritidis. Biofouling 2019, 35, 361–375. [Google Scholar] [CrossRef]
- Valeriano, C.; De Oliveira, T.L.C.; Carvalho, S.M.; Cardoso, M.D.G.; Alves, E.; Piccoli, R.H. The sanitizing action of essential oil-based solutions against Salmonella enterica serotype Enteritidis S64 biofilm formation on AISI 304 stainless steel. Food Control 2012, 25, 673–677. [Google Scholar] [CrossRef]
- Somrani, M.; Debbabi, H.; Palop, A. Antibacterial and antibiofilm activity of essential oil of clove against Listeria monocytogenes and Salmonella Enteritidis. Food Sci. Technol. Int. 2021, 28, 331–339. [Google Scholar] [CrossRef]
- Shi, C.; Yan, C.; Sui, Y.; Sun, Y.; Guo, D.; Chen, Y.; Jin, T.; Peng, X.; Ma, L.; Xia, X. Thymoquinone inhibits virulence related traits of Cronobacter sakazakii ATCC 29544 and has anti-biofilm formation potential. Front. Microbiol. 2017, 8, 2220. [Google Scholar] [CrossRef] [PubMed]
- Nuță, D.C.; Limban, C.; Chiriță, C.; Chifiriuc, M.C.; Costea, T.; Ioniță, P.; Nicolau, I.; Zarafu, I. Contribution of essential oils to the fight against microbial biofilms—A review. Processes 2021, 9, 537. [Google Scholar] [CrossRef]
- Glinel, K.; Thebault, P.; Humblot, V.; Pradier, C.M.; Jouenne, T. Antibacterial surfaces developed from bio-inspired approaches. Acta Biomater. 2012, 8, 1670–1684. [Google Scholar] [CrossRef]
- Santos, V.; Drummond, R.N.; Dias-Souza, M. Bacteriocins as antimicrobial and antibiofilm agents. In Elsevier eBooks; Elsevier: Amsterdam, The Netherlands, 2017; pp. 403–436. [Google Scholar] [CrossRef]
- Chopra, L.; Singh, G.; Jena, K.K.; Sahoo, D.K. Sonorensin: A new bacteriocin with potential of an anti-biofilm agent and a food biopreservative. Sci. Rep. 2015, 5, 13412. [Google Scholar] [CrossRef]
- Kirtonia, K.; Salauddin, M.; Bharadwaj, K.K.; Pati, S.; Dey, A.; Shariati, M.A.; Tilak, V.K.; Kuznetsova, E.; Sarkar, T. Bacteriocin: A new strategic antibiofilm agent in food industries. Biocatal. Agric. Biotechnol. 2021, 36, 102141. [Google Scholar] [CrossRef]
- Duraisamy, S.; Balakrishnan, S.; Ranjith, S.; Husain, F.; Sathyan, A.; Peter, A.S.; Prahalathan, C.; Kumarasamy, A. Bacteriocin—A potential antimicrobial peptide towards disrupting and preventing biofilm formation in the clinical and environmental locales. Environ. Sci. Pollut. Res. 2020, 27, 44922–44936. [Google Scholar] [CrossRef]
- Darbandi, A.; Asadi, A.; Mahdizade Ari, M.; Ohadi, E.; Talebi, M.; Halaj Zadeh, M.; Darb Emamie, A.; Ghanavati, R.; Kakanj, M. Bacteriocins: Properties and potential use as antimicrobials. J. Clin. Lab. Anal. 2022, 36, e24093. [Google Scholar] [CrossRef] [PubMed]
- Lamas, A.; Regal, P.; Sanjulián, L.; López-Santamarina, A.; Franco, C.M.; Cepeda, A. An Overview of Salmonella Biofilms and the Use of Bacteriocins and Bacteriophages as New Control Alternatives; IntechOpen eBooks; InTech Open: Rijeka, Croatia, 2021. [Google Scholar] [CrossRef]
- Mathur, H.; Field, D.; Rea, M.C.; Cotter, P.D.; Hill, C.; Ross, R.P. Fighting biofilms with lantibiotics and other groups of bacteriocins. npj Biofilms Microbiomes 2018, 4, 9. [Google Scholar] [CrossRef]
- Xiang, Y.-Z.; Zhang, Y.-M.; Liu, Y.-Y.; Zhang, M.; Lin, L.-B.; Zhang, Q.-L. Purification, characterization, and antibacterial and antibiofilm activity of a novel bacteriocin against Salmonella Enteritidis. Food Control 2021, 127, 108110. [Google Scholar] [CrossRef]
- Luo, L.; Yi, L.; Chen, J.; Liu, B.; Lü, X. Antibacterial mechanisms of bacteriocin BM1157 against Escherichia coli and Cronobacter sakazakii. Food Control 2021, 123, 107730. [Google Scholar] [CrossRef]
- 114. Burgos, M.J.G.; López, R.L.; del Carmen López Aguayo, M.; Pulido, R.P.; Gálvez, A. Inhibition of planktonic and sessile Salmonella enterica cells by combinations of enterocin AS-48, polymyxin B and biocides. Food Control 2013, 30, 214–221. [Google Scholar] [CrossRef]
- Kim, N.; Kim, W.J.; Kang, S. Anti-biofilm effect of crude bacteriocin derived from Lactobacillus brevis DF01 on Escherichia coli and Salmonella Typhimurium. Food Control 2019, 98, 274–280. [Google Scholar] [CrossRef]
- Pérez-Ibarreche, M.; Castellano, P.; Leclercq, A.; Vignolo, G. Control of Listeria monocytogenes biofilms on industrial surfaces by the bacteriocin-producing Lactobacillus sakei CRL1862. FEMS Microbiol. Lett. 2016, 363, 12. [Google Scholar] [CrossRef]
| Enzyme | Concentration | Microbial Biofilm | Contact Surface | Results | References |
|---|---|---|---|---|---|
| Serine protease from B. subtilis | 1% | P. fluorescens, B. mycoides, and B. cereus | stainless steel | 2.3–3.9 log CFU/cm2 reductions at 45 °C after 30 min | [10] |
| α-Amylase from B. subtilis | 50–250 μL/mL | S. aureus, V. cholerae, and P. aeruginosa | polystyrene | 51.8% to 73.1% reductions at 30 °C after 2 h | [86] |
| Lipases from marine bacterium Oceanobacillus | 150 µL/mL | P. fluorescens, E. coli, Listeria spp., B. cereus, and V. parahemolyticus | Glass | 90–95% disruption of biofilms after 1 h | [85] |
| Protease, lipase, and amylase combination | 5–10% protease, 0.5–1.0% lipase, and 2.5–5% amylase | S. Typhimurium and C. sakazakii | stainless steel and polystyrene | 27.6–61.7% biofilm detachment at 50 °C after 30 min | [20] |
| Protease, lipase, and amylase combination | 5–10% protease, 0.5–1.0% lipase, and 2.5–5% amylase | S. enterica and C. sakazakii | stainless steel and plastic | 1.2–2.2 log CFU/coupon reduction at 50 °C after 30 min | [71] |
| Essential Oil | Concentration | Microbial Biofilm | Contact Surface | Results | References |
|---|---|---|---|---|---|
| Carvacrol and thymol | 0.156 µL/mL | S. Enteritidis | polystyrene | reduce the biofilm formation by 90% | [73] |
| Peppermint and lemongrass | 7.8 µL/mL | S. Enteritidis | stainless steel | 4.0–4.2 log CFU/cm2 reductions after 10 min | [74] |
| Aleppo pine | 2000 µg/mL | S. enterica | plastic and stainless steel | 1.6 and 1.8 log CFU/coupon reductions after 30 min | [65] |
| Clove | 0.05–0.1 mg/mL | S. Enteritidis and L. monocytogenes | polystyrene | reduce the biofilm formation by 49.8–61.8% | [75] |
| Mountain pepper | 15–40 µL/mL | C. sakazakii | stainless steel | reduce the cell to below the detection level (1 CFU/cm). | [69] |
| Bacteriocins | MO | Media/Surface | Treatment Conditions | Results | Reference |
|---|---|---|---|---|---|
| Bacteriocin BM1157 | E. coli ATCC25922 | LB broth/96-well plate | 72 h/37 °C | 83% reduction in comparison with the control | [113] |
| Bacteriocin BM1157 | C. sakazakii ATCC29544 | LB broth/96-well plate | 72 h/37 °C | 80% reduction in comparison with the control | [113] |
| Enterocin AS-48 at 25 mg/L from Enterococcus faecalis | 4 strains S. Enteritidis UJ3449 | TSB broth/96-well plate | 1 h/30 °C | Reduce the number from 1 to 2 log | [114] |
| Enterocin AS-48 at 50 g/L from Enterococcus faecalis | S. Enteritidis | TSB broth/96-well plate | 1 h/30 °C | Reduce the number from 1.5 to 3.5 log | [114] |
| DF01 bacteriocin from Lactobacillus brevis DF01 | E. coli KCTC 1039 | 96-well microtiter plates | 24 h/37 °C | Inhibit the biofilm formation by 60% in comparison with control | [115] |
| DF01 bacteriocin from Lactobacillus brevis DF01 | S. Typhimurium KCTC 1925 | 96-well microtiter plates | 24 h/37 °C | Inhibit the biofilm formation by 50% in comparison with control | [115] |
| Bacteriocin AMYX6 at 36 μg/mL from B. amyloliquefaciens JDF-17 | S. Enteritidis 35 | LB medium/24-well plates | 24 h/37 °C | Reduce the biofilm formation by 52% in comparison with the control | [112] |
| Bacteriocin AMYX6 at 72 μg/mL from B. amyloliquefaciens JDF-17 | S. Enteritidis 35 | LB medium/24-well plates | 24 h/37 °C | Reduce the biofilm formation by 79% in comparison with the control | [112] |
| Bacteriocin from Lactobacillus sakei CRL1862 (800 μL) | L. monocytogenes FBUNT and Scott A | Stainless steel (SS) | 96 h/10 °C on 6-day-old biofilm | 3.1 log reduction in comparison with the control | [116] |
| Bacteriocin from Lactobacillus sakei CRL1862 (800 μL) | L. monocytogenes FBUNT and Scott A | Polytetrafluoroethylene (PTFE) | 96 h/10 °C on 6-day-old biofilm | 3.6 log reduction in comparison with the control | [116] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Olaimat, A.N.; Ababneh, A.M.; Al-Holy, M.; Al-Nabulsi, A.; Osaili, T.; Abughoush, M.; Ayyash, M.; Holley, R.A. A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces. Microbiol. Res. 2024, 15, 1973-1992. https://doi.org/10.3390/microbiolres15040132
Olaimat AN, Ababneh AM, Al-Holy M, Al-Nabulsi A, Osaili T, Abughoush M, Ayyash M, Holley RA. A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces. Microbiology Research. 2024; 15(4):1973-1992. https://doi.org/10.3390/microbiolres15040132
Chicago/Turabian StyleOlaimat, Amin N., Ahmad Mohammad Ababneh, Murad Al-Holy, Anas Al-Nabulsi, Tareq Osaili, Mahmoud Abughoush, Mutamed Ayyash, and Richard A. Holley. 2024. "A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces" Microbiology Research 15, no. 4: 1973-1992. https://doi.org/10.3390/microbiolres15040132
APA StyleOlaimat, A. N., Ababneh, A. M., Al-Holy, M., Al-Nabulsi, A., Osaili, T., Abughoush, M., Ayyash, M., & Holley, R. A. (2024). A Review of Bacterial Biofilm Components and Formation, Detection Methods, and Their Prevention and Control on Food Contact Surfaces. Microbiology Research, 15(4), 1973-1992. https://doi.org/10.3390/microbiolres15040132






