A Metabarcoding Approach for Investigating the Stomach Microbiota of the Corallivorous Snail Coralliophila meyendorffii (Muricidae, Coralliophilinae) and Its Venomous Host, the Sea-Anemone Parazoanthus axinellae (Zoanthidea, Parazoanthidae)
Abstract
1. Introduction
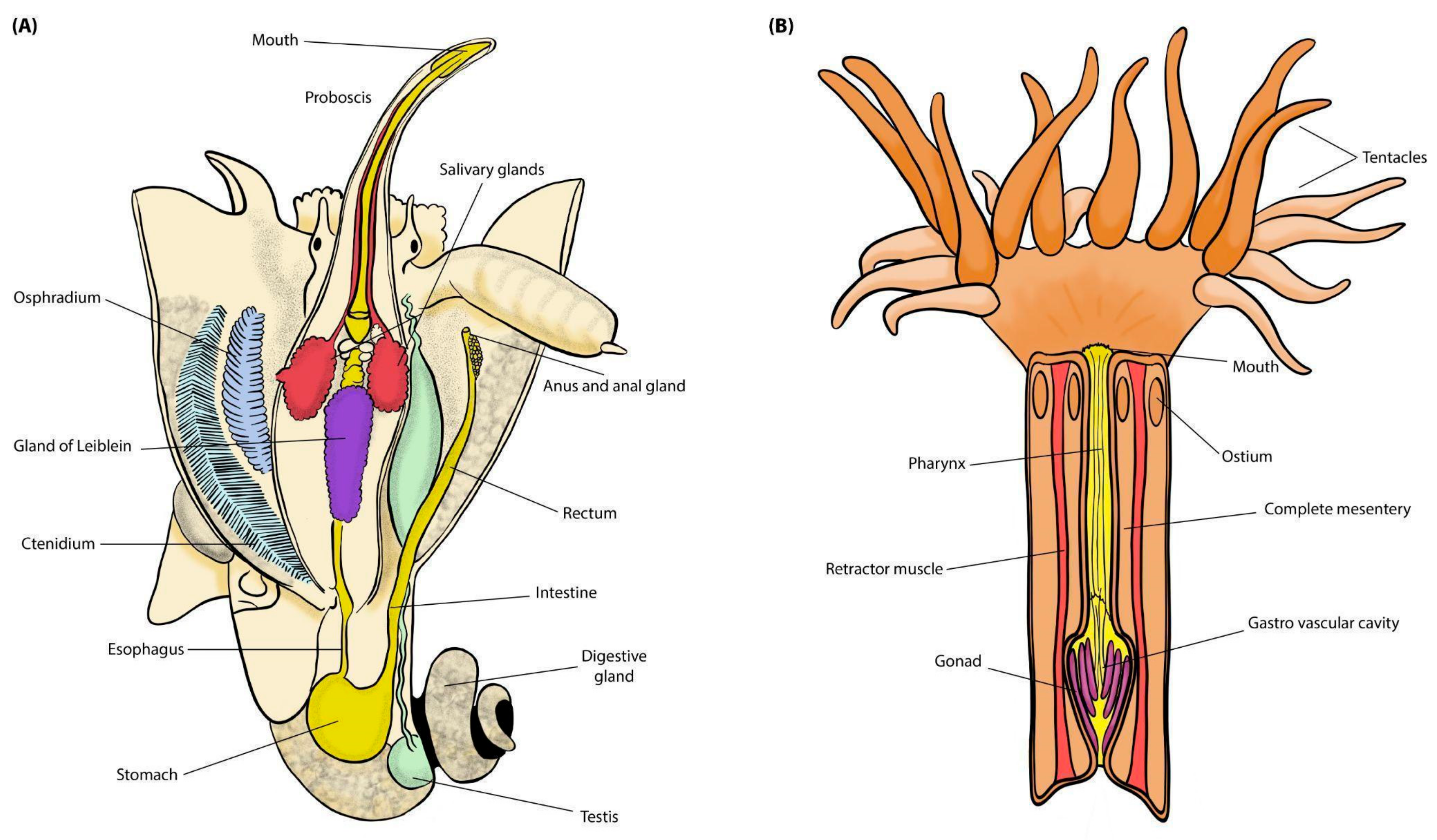
2. Material and Methods
2.1. Samples and Collecting Data
2.2. DNA Extraction and Molecular Analyses
2.3. Metabarcoding Analysis
3. Results
3.1. Overview of Sequencing Information
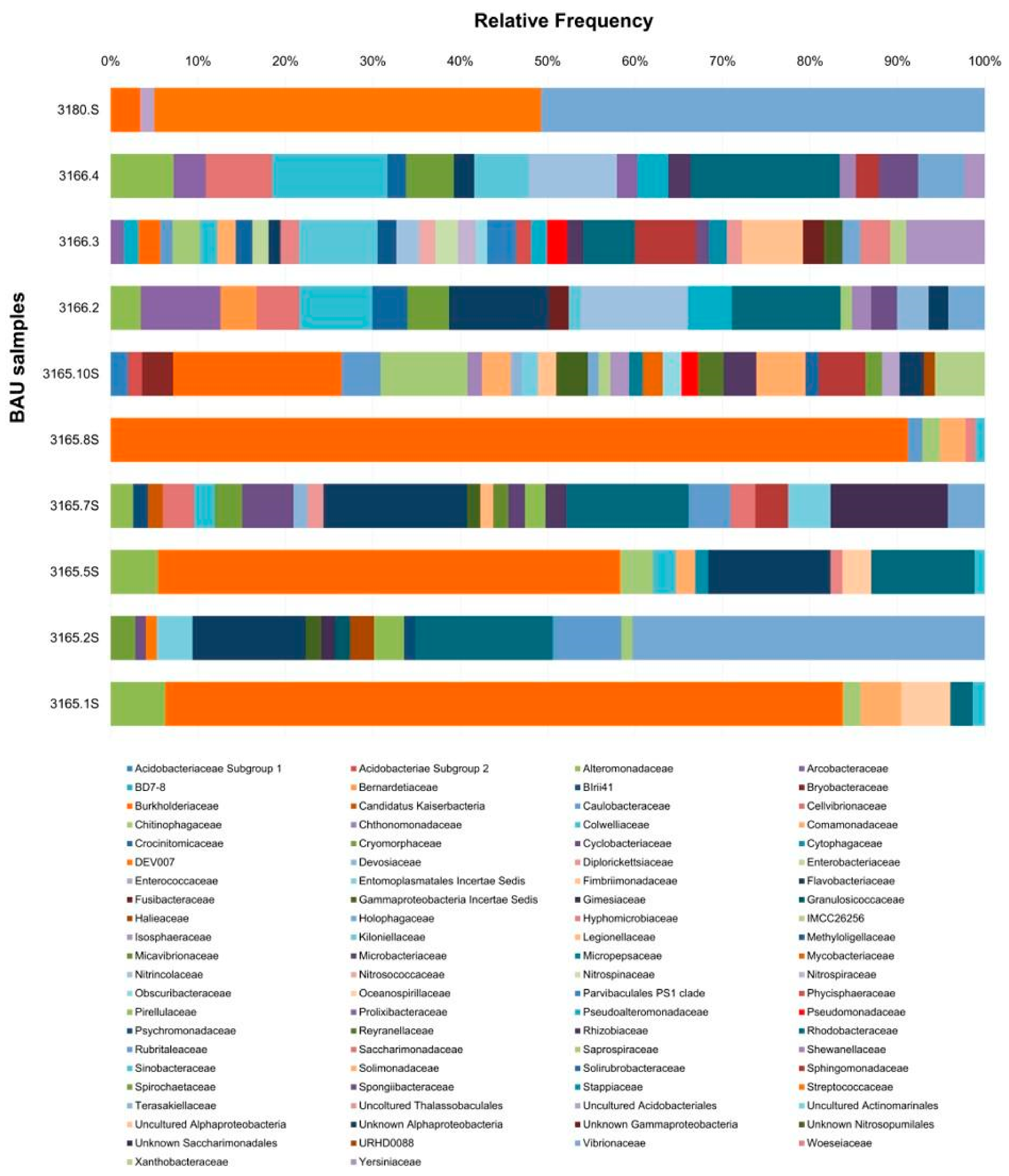
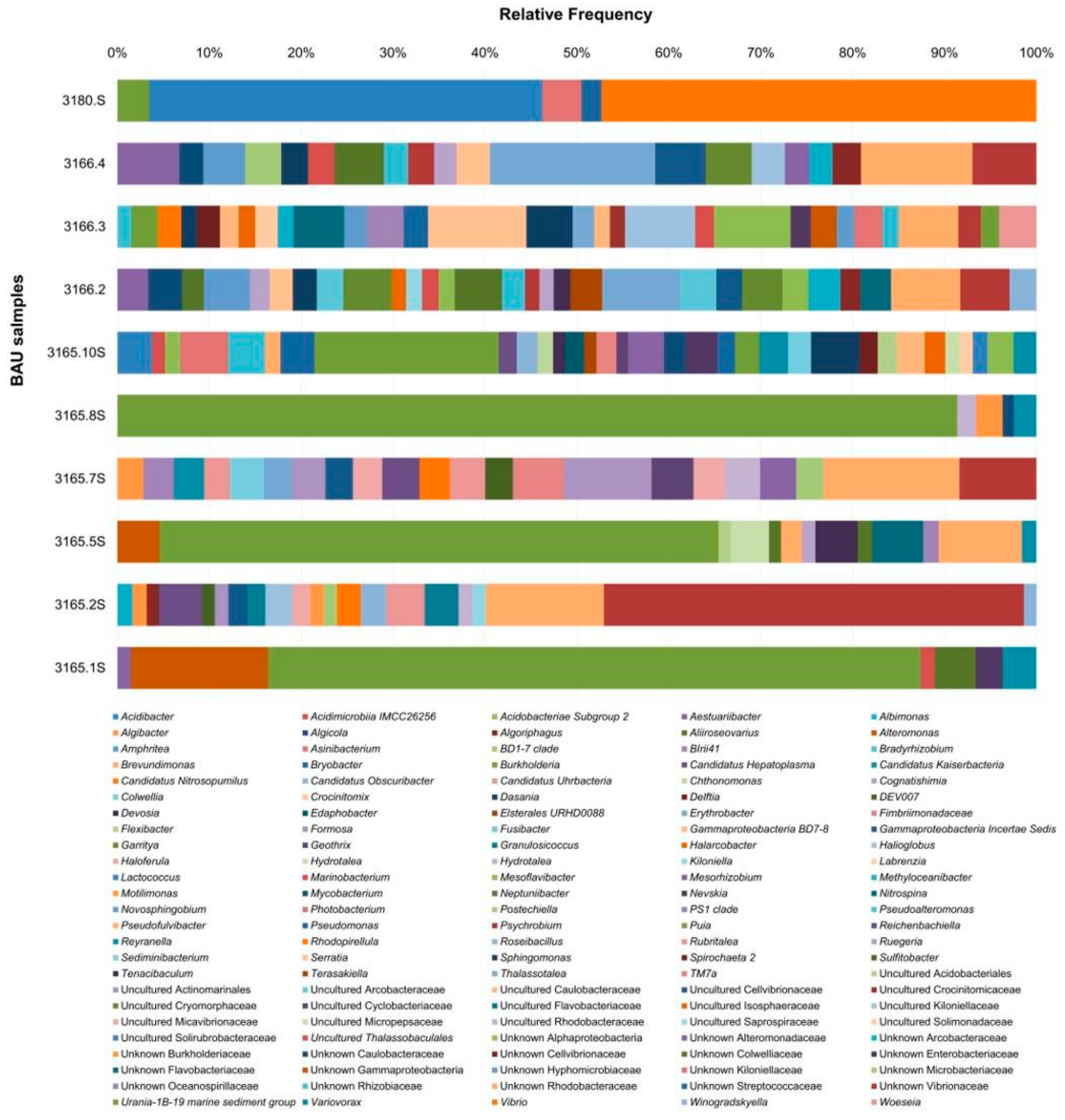
3.2. Bacterial Communities
3.2.1. Family Level
3.2.2. Genus Level
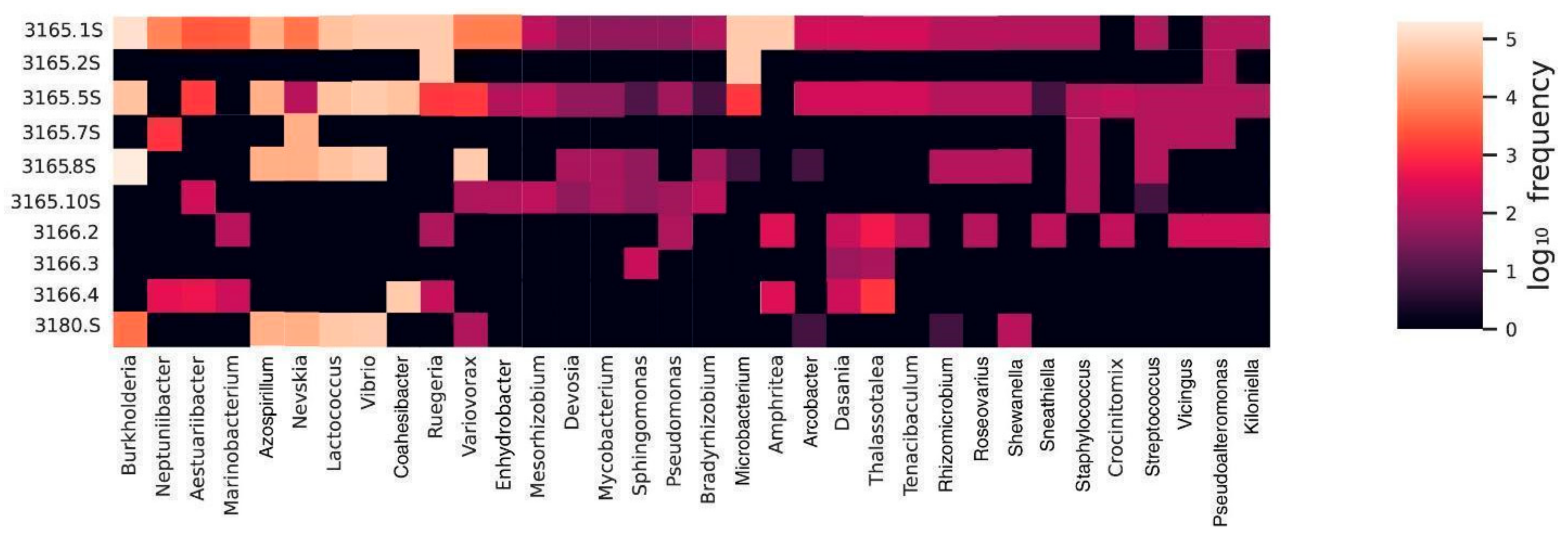
3.3. Microbiome Similarity of Snails and Corals
3.4. Alpha and Beta Diversity Analysis
4. Discussion
4.1. Structure of Bacterial Communities
4.2. Microbial Diversity
5. Conclusions
- -
- The bacterial taxa found exclusively or predominantly in the P. axinellae samples suggest the presence of holobiont components within the microbial community of this coral, as has been observed in other coral species.
- -
- Our findings indicate that the stomach microbiome of C. meyendorffii shows similarities to that of its coral host, P. axinellae, suggesting a possible influence of the coral’s microbiome on the gastropod’s gut. This is further supported by the observation that Babelomurex cariniferus and Parazoanthus axinellae, despite living in similar environments, exhibit distinct microbiome compositions. These differences highlight those specific ecological interactions, including potential horizontal transfer of bacterial taxa between organisms, significantly shape the microbiomes of these coral-associated species.
- -
- The microbial community identified in the stomachs of C. meyendorffii does not provide evidence for a primary role in parasitism, as we did not detect any bacteria known to be directly associated with corallivory, particularly those involved in neutralising or digesting the venomous substances of P. axinellae. This observation suggests that the microbiota responsible for toxin degradation may not be present in the digestive system but rather within the salivary glands that the Coralliophilinae have evolved over time. Future research should focus on investigating these glands to elucidate their potential role in toxin processing.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Stella, J.; Pratchett, M.; Hutchings, P.; Jones, G. Coral-associated invertebrates: Diversity, ecological importance and vulnerability to disturbance. Oceanogr. Mar. Biol. Annu. Rev. 2011, 49, 43–104. [Google Scholar] [CrossRef]
- Pita, L.; Rix, L.; Slaby, B.M.; Franke, A.; Hentschel, U. The sponge holobiont in a changing ocean: From microbes to ecosystems. Microbiome 2018, 6, 46. [Google Scholar] [CrossRef] [PubMed]
- Plaisance, L.; Matterson, K.; Fabricius, K.; Drovetski, S.; Meyer, C.; Knowlton, N. Effects of low pH on the coral reef cryptic invertebrate communities near CO2 vents in Papua New Guinea. PLoS ONE 2021, 16, e0258725. [Google Scholar] [CrossRef] [PubMed]
- Ashwood, L.M.; Norton, R.S.; Undheim, E.A.B.; Hurwood, D.A.; Prentis, P.J. Characterising functional venom profiles of anthozoans and medusozoans within their ecological context. Mar. Drugs 2020, 18, 202. [Google Scholar] [CrossRef]
- Robertson, R. Review of the predators and parasites of stony corals, with special reference to symbiotic prosobranch gastropods. Pac. Sci. 1970, 24, 43–54. [Google Scholar]
- Russini, V.; Fassio, G.; Nocella, E.; Houart, R.; Barco, A.; Puillandre, N.; Lozouet, P.; Modica, M.V.; Oliverio, M. Whelks, rock-snails, and allied: A new phylogenetic framework for the family Muricidae (Mollusca: Gastropoda). Eur. Zool. J. 2023, 90, 856–868. [Google Scholar] [CrossRef]
- Ponte, G.; Modica, M.V. Salivary glands in predatory mollusks: Evolutionary considerations. Front. Physiol. 2017, 8, 266744. [Google Scholar] [CrossRef]
- Oliverio, M.; Mariottini, P. A molecular framework for the phylogeny of Coralliophila and related muricoids. J. Molluscan Stud. 2001, 67, 215–224. [Google Scholar] [CrossRef]
- Oliverio, M.; Mariottini, P. Contrasting morphological and molecular variation in Coralliophila meyendorffii (Muricidae, Coralliophilinae). J. Molluscan Stud. 2001, 67, 243–246. [Google Scholar] [CrossRef][Green Version]
- Nocella, E.; Fassio, G.; Zuccon, D.; Puillandre, N.; Modica, M.V.; Oliverio, M. From coral reefs into the abyss: The evolution of corallivory in the Coralliophilinae (Neogastropoda, Muricidae). Coral Reefs 2024, 43, 1285–1302. [Google Scholar] [CrossRef]
- Ritcher, A.; Amor, M.J.; Dufort, M. The anatomy and ultrastructure of the gland of Leiblein of Bolinus brandaris and Coral-liophila meyendorffii, two neogastropod species with different ecology and feeding strategies. In Proceedings of the SEB Annual Main Meeting, Prague, Czech Republic, 30 June–3 July 2010. [Google Scholar]
- Guillen, P.O.; Jaramillo, K.B.; Genta-Jouve, G.; Thomas, O.P. Marine natural products from zoantharians: Bioactivity, biosynthesis, systematics, and ecological roles. Nat. Prod. Rep. 2020, 37, 515–540. [Google Scholar] [CrossRef] [PubMed]
- Nocella, E.; Zvonareva, S.S.; Fassio, G.; Pica, D.; Buge, B.; Villa, R.; Puillandre, N.; Modica, M.V.; Oliverio, M. Spicy food for the egg-cowries: The evolution of corallivory in the Ovulidae (Gastropoda: Cypraeoidea). Front. Mar. Sci. 2024, 10, 1323156. [Google Scholar] [CrossRef]
- Simone, L.R.L. A new species of Thaisella (Neogastropoda: Muricidae) from Caribbean Guatemala, with accounts on the anatomy and taxonomy of the genus in the western Atlantic. Arch. Molluskenkd. 2017, 146, 111–120. [Google Scholar] [CrossRef]
- Andrews, E.B.; Thorogood, K.E. An Ultrastructural Study of the gland of Leiblein of muricid and nassariid neogastropods in relation to function, with a discussion on its homologies in other caenogastropods. J. Molluscan Stud. 2005, 71, 269–300. [Google Scholar] [CrossRef]
- Mansour-Bek, J.J. Über die proteolytischen Enzyme von Murex anguliferus Lamk. Z. Vgl. Physiol. 1934, 20, 343–369. [Google Scholar] [CrossRef]
- Lau, D.C.; Leung, K.M. Feeding physiology of the carnivorous gastropod Thais clavigera (Kuster): Do they eat “soup”? J. Exp. Mar. Biol. Ecol. 2004, 312, 43–66. [Google Scholar] [CrossRef]
- Jourdan, M. On the Zoantharia Malacodermata of the shores of Marseilles. Ann. Mag. Nat. Hist. 1879, 4, 325–326. [Google Scholar] [CrossRef][Green Version]
- Galasso, C.; Corinaldesi, C.; Sansone, C. Carotenoids from marine organisms: Biological functions and industrial applications. Antioxidants 2017, 6, 96. [Google Scholar] [CrossRef]
- Glynn, P.W. Coral reef bleaching: Ecological perspectives. Coral Reefs 1993, 12, 1–17. [Google Scholar] [CrossRef]
- Modica, M.V.; Holford, M. The Neogastropoda: Evolutionary Innovations of Predatory Marine Snails with Remarkable Pharmacological Potential. In Evolutionary Biology—Concepts, Molecular and Morphological Evolution; Pontarotti, P., Ed.; Springer: Berlin/Heidelberg, Germany, 2010. [Google Scholar] [CrossRef]
- Gorson, J.; Fassio, G.; Lau, E.S.; Holford, M. Diet diversity in carnivorous terebrid snails is tied to the presence and absence of a venom gland. Toxins 2021, 13, 108. [Google Scholar] [CrossRef]
- Fassio, G.; Russo, P.; Bonomolo, G.; Fedosov, A.E.; Modica, M.V.; Nocella, E.; Oliverio, M. A molecular framework for the systematics of the Mediterranean spindle-shells (Gastropoda, Neogastropoda, Fasciolariidae, Fusininae). Mediterr. Mar. Sci. 2022, 23, 623–636. [Google Scholar] [CrossRef]
- Veron, J.E.N. Corals of the World; AIMS: Townsville, Australia, 2000; Volumes 1–3, p. 1410. [Google Scholar]
- Russini, V.; Fassio, G.; Chimenti, C.; Davolos, D. Discovering symbiosis in the supralittoral: Bacterial metabarcoding analysis from the hepatopancreas of Orchestia and Tylos (Crustacea). Symbiosis 2021, 83, 225–236. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef] [PubMed]
- Kruskal, W.H.; Wallis, W.A. Use of Ranks in One-Criterion Variance Analysis. J. Am. Stat. Assoc. 1952, 47, 583–621. [Google Scholar] [CrossRef]
- Anderson, M.J. Permutational Multivariate Analysis of Variance (PERMANOVA). In Wiley StatsRef: Statistics Reference Online; Wiley: Hoboken, NJ, USA, 2017; pp. 1–15. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Yang, F.; Xiao, Z.; Wei, Z.; Long, L. Bacterial Communities Associated with Healthy and Bleached Crustose Coralline Alga Porolithon onkodes. Front. Microbiol. 2021, 12, 646143. [Google Scholar] [CrossRef]
- Zhou, Z.; Wu, H.; Li, D.; Zeng, W.; Huang, J.; Wu, Z. Comparison of gut microbiome in the Chinese mud snail (Cipangopaludina chinensis) and the invasive golden apple snail (Pomacea canaliculata). PeerJ 2022, 10, e13245. [Google Scholar] [CrossRef]
- Quintanilla, E.; Rodrigues, C.F.; Henriques, I.; Hilário, A. Microbial associations of abyssal gorgonians and anemones (>4,000 m depth) at the Clarion-Clipperton Fracture Zone. Front. Microbiol. 2022, 13, 828469. [Google Scholar] [CrossRef]
- Bernasconi, R.; Stat, M.; Koenders, A.; Paparini, A.; Bunce, M.; Huggett, M.J. Establishment of coral-bacteria symbioses reveal changes in the core bacterial community with host ontogeny. Front. Microbiol. 2019, 10, 1529. [Google Scholar] [CrossRef]
- Lyu, T.; Zhu, J.; Yang, X.; Yang, W.; Zheng, Z. Responses of Gut Microbial Community Composition and Function of the Freshwater Gastropod Bellamya aeruginosa to Cyanobacterial Bloom. Front. Microbiol. 2022, 13, 906278. [Google Scholar] [CrossRef]
- van de Water, J.A.J.M.; Allemand, D.; Ferrier-Pagès, C. Bacterial symbionts of the precious coral Corallium rubrum are differentially distributed across colony-specific compartments and differ among colormorphs. Environ. Microbiol. Rep. 2024, 16, e13236. [Google Scholar] [CrossRef] [PubMed]
- Arboleda-Baena, C.; Pareja, C.B.; Pla, I.; Logares, R.; De la Iglesia, R.; Navarrete, S.A. Hidden interactions in the intertidal rocky shore: Variation in pedal mucus microbiota among marine grazers that feed on epilithic biofilm communities. PeerJ 2022, 10, e13642. [Google Scholar] [CrossRef] [PubMed]
- Wuerz, M.; Lawson, C.A.; Oakley, C.A.; Possell, M.; Wilkinson, S.P.; Grossman, A.R.; Weis, V.M.; Suggett, D.J.; Davy, S.K. Symbiont identity impacts the microbiome and volatilome of a model cnidarian-dinoflagellate symbiosis. Biology 2023, 12, 1014. [Google Scholar] [CrossRef] [PubMed]
- Osório, J.B.; Pereira, L.d.M.; Giongo, A.; Marconatto, L.; Potriquet, J.; Candido, R.R.F.; Mulvenna, J.; Jones, M.; Graeff-Teixeira, C.; Morassutti, A.L. Mollusk microbiota shift during Angiostrongylus cantonensis infection in the freshwater snail Biomphalaria glabrata and the terrestrial slug Phillocaulis soleiformis. Parasitol. Res. 2020, 119, 2495–2503. [Google Scholar] [CrossRef] [PubMed]
- Becker, C.C.; Brandt, M.; Miller, C.A.; Apprill, A. Microbial bioindicators of stony coral tissue loss disease identified in corals and overlying waters using a rapid field-based sequencing approach. Environ. Microbiol. 2022, 24, 1166–1182. [Google Scholar] [CrossRef]
- Messer, L.F.; Bourne, D.G.; Robbins, S.J.; Clay, M.; Bell, S.C.; McIlroy, S.J.; Tyson, G.W. A genome-centric view of the role of the Acropora kenti microbiome in coral health and resilience. Nat. Commun. 2024, 15, 2902. [Google Scholar] [CrossRef]
- Pollock, F.J.; McMinds, R.; Smith, S.; Bourne, D.G.; Willis, B.L.; Medina, M.; Thurber, R.V.; Zaneveld, J.R. Coral-associated bacteria demonstrate phylosymbiosis and cophylogeny. Nat. Commun. 2018, 9, 4921. [Google Scholar] [CrossRef]
- Hu, Z.; Chen, X.; Chang, J.; Yu, J.; Tong, Q.; Li, S.; Niu, H. Compositional and predicted functional analysis of the gut microbiota of Radix auricularia (Linnaeus) via high-throughput Illumina sequencing. PeerJ 2018, 6, e5537. [Google Scholar] [CrossRef]
- Yin, Q.; Liang, J.; Zheng, X.; Wang, Y.; Song, Z.-M.; Zhang, Y.; Xu, Y. Algibacter onchidii sp. nov., a symbiotic bacterium isolated from a marine invertebrate. Int. J. Syst. Evol. Microbiol. 2021, 71, 005102. [Google Scholar] [CrossRef]
- Schneider, Y.K.-H.; Hansen, K.; Isaksson, J.; Ullsten, S.; Hansen, E.H.; Andersen, J.H.; Schneider, Y.K.-H.; Hansen, K.; Isaksson, J.; Ullsten, S.; et al. Anti-bacterial effect and cytotoxicity assessment of lipid 430 Isolated from Algibacter sp. Molecules 2019, 24, 3991. [Google Scholar] [CrossRef]
- Gai, Y.; Huang, Z.; Lai, Q.; Shao, Z. Shewanella intestini sp. nov., isolated from the intestine of abalone, Haliotis diversicolor. Int. J. Syst. Evol. Microbiol. 2017, 67, 1901–1905. [Google Scholar] [CrossRef] [PubMed]
- Mizutani, Y.; Iehata, S.; Mori, T.; Oh, R.; Fukuzaki, S.; Tanaka, R. Diversity, enumeration, and isolation of Arcobacter spp. in the giant abalone, Haliotis gigantea. Microbiologyopen 2019, 8, e890. [Google Scholar] [CrossRef] [PubMed]
- Tanaka, R.; Cleenwerck, I.; Mizutani, Y.; Iehata, S.; Bossier, P.; Vandamme, P. Arcobacter haliotis sp. nov., isolated from abalone species Haliotis gigantea. Int. J. Syst. Evol. Microbiol. 2017, 67, 3050–3056. [Google Scholar] [CrossRef] [PubMed]
- Yang, A.; Li, W.; Tao, Z.; Ye, H.; Xu, Z.; Li, Y.; Gao, Y.; Yan, X. Vibrio harveyi isolated from marine aquaculture species in eastern China and virulence to the large yellow croaker (Larimichthys crocea). J. Appl. Microbiol. 2021, 131, 1710–1721. [Google Scholar] [CrossRef]
- Labella, A.M.; Arahal, D.R.; Castro, D.; Lemos, M.L.; Borrego, J.J. Revisiting the genus Photobacterium: Taxonomy, ecology and pathogenesis. Int. Microbiol. 2017, 20, 1–10. [Google Scholar] [CrossRef]
- Kaeding, A.J.; Ast, J.C.; Pearce, M.M.; Urbanczyk, H.; Kimura, S.; Endo, H.; Nakamura, M.; Dunlap, P.V. Phylogenetic Diversity and Cosymbiosis in the Bioluminescent Symbioses of “Photobacterium mandapamensis”. Appl. Environ. Microbiol. 2007, 73, 3173–3182. [Google Scholar] [CrossRef]
- Wang, Y.; Wang, H.; Liu, J.; Lai, Q.; Shao, Z.; Austin, B.; Zhang, X.-H. Aestuariibacter aggregatus sp. nov., a moderately halophilic bacterium isolated from seawater of the Yellow Sea. FEMS Microbiol. Lett. 2010, 309, 48–54. [Google Scholar] [CrossRef][Green Version]
- Hwang, C.Y.; Cho, B.C. Cohaesibacter gelatinilyticus gen. nov., sp. nov., a marine bacterium that forms a distinct branch in the order Rhizobiales, and proposal of Cohaesibacteraceae fam. nov. Int. J. Syst. Evol. Microbiol. 2008, 58, 267–277. [Google Scholar] [CrossRef]
- Lee, Y.K.; Hong, S.G.; Cho, H.H.; Cho, K.H.; Lee, H.K. Dasania marina gen. nov., sp. nov., of the order pseudomonadales, isolated from arctic marine sediment. J. Microbiol. 2007, 45, 505–509. [Google Scholar]
- Kim, M.; Hwang, J.; Kim, G.; Na, T.; Kim, T.-H.; Hyun, J.-H. Carbon cycling in the East Sea (Japan Sea): A review. Front. Mar. Sci. 2022, 9, 938935. [Google Scholar] [CrossRef]
- Wiese, J.; Saha, M.; Wenzel-Storjohann, A.; Weinberger, F.; Schmaljohann, R.; Imhoff, J.F. Vicingus serpentipes gen. nov., sp. nov., a new member of the Flavobacteriales from the North Sea. Int. J. Syst. Evol. Microbiol. 2018, 68, 333–340. [Google Scholar] [CrossRef]
- Holmström, C.; James, S.; Franks, A.; McCloy, S.; Kjelleberg, S. Antifouling activities expressed by marine surface associated Pseudoalteromonas species. FEMS Microbiol. Ecol. 2002, 41, 47–58. [Google Scholar] [CrossRef]
- Romanenko, L.A.; Tanaka, N.; Svetashev, V.I.; Mikhailov, V.V. Pseudomonas glareae sp. nov., a marine sediment-derived bacterium with antagonistic activity. Arch. Microbiol. 2015, 197, 693–699. [Google Scholar] [CrossRef] [PubMed]
- Nowlan, J.P.; Lumsden, J.S.; Russell, S. Advancements in characterizing Tenacibaculum infections in Canada. Pathogens 2020, 9, 1029. [Google Scholar] [CrossRef]
- Gerpe, D.; Lasa, A.; Lema, A.; Romalde, J.L. Metataxonomic analysis of tissue-associated microbiota in grooved carpet-shell (Ruditapes decussatus) and Manila (Ruditapes philippinarum) clams. Int. Microbiol. 2021, 24, 607–618. [Google Scholar] [CrossRef] [PubMed]
- Yu, J.; Sawabe, T.; Yamano, R.; Koike, S.; Sakai, Y.; Mino, S. Inferring potential causative microbial factors of intestinal atrophic disease in the sea cucumber Apostichopus japonicus. Front. Mar. Sci. 2023, 10, 1225318. [Google Scholar] [CrossRef]
- Shi, R.; Xu, S.; Qi, Z.; Zhu, Q.; Huang, H.; Weber, F. Influence of suspended mariculture on vertical distribution profiles of bacteria in sediment from Daya Bay, Southern China. Mar. Pollut. Bull. 2019, 146, 816–826. [Google Scholar] [CrossRef] [PubMed]
- Jang, H.; Yang, S.-H.; Seo, H.-S.; Lee, J.-H.; Kim, S.-J.; Kwon, K.K. Amphritea spongicola sp. nov., isolated from a marine sponge, and emended description of the genus Amphritea. Int. J. Syst. Evol. Microbiol. 2015, 65, 1866–1870. [Google Scholar] [CrossRef]
- Yamano, R.; Yu, J.; Jiang, C.; Harjuno Condro Haditomo, A.; Mino, S.; Sakai, Y.; Sawabe, T. Taxonomic revision of the genus Amphritea supported by genomic and in silico chemo-taxonomic analyses, and the proposal of Aliamphritea gen. nov. PLoS ONE 2022, 17, e0271174. [Google Scholar] [CrossRef]
- Yang, S.-H.; Seo, H.-S.; Lee, J.-H.; Kim, S.-J.; Kwon, K.K. Kiloniella spongiae sp. nov., isolated from a marine sponge and emended description of the genus Kiloniella Wiese et al. 2009 and Kiloniella laminariae. Int. J. Syst. Evol. Microbiol. 2015, 65, 230–234. [Google Scholar] [CrossRef]
- Karim, R.U.; Fukaya, K.; In, Y.; Sharma, A.R.; Harunari, E.; Oku, N.; Urabe, D.; Trianto, A.; Igarashi, Y. Marinoquinolones and marinobactoic acid: Antimicrobial and cytotoxic ortho-Dialkylbenzene-class metabolites produced by a marine obligate Gammaproteobacterium of the genus Marinobacterium. J. Nat. Prod. 2022, 85, 1763–1770. [Google Scholar] [CrossRef] [PubMed]
- Kitamura, R.; Miura, N.; Ito, M.; Takagi, T.; Yamashiro, H.; Nishikawa, Y.; Nishimura, Y.; Kobayashi, K.; Kataoka, M. Specific Detection of Coral-Associated Ruegeria, a Potential Probiotic Bacterium, in Corals and Subtropical Seawater. Mar. Biotechnol. 2021, 23, 576–589. [Google Scholar] [CrossRef] [PubMed]
- Durham, B.P.; Dearth, S.P.; Sharma, S.; Amin, S.A.; Smith, C.B.; Campagna, S.R.; Armbrust, E.V.; Moran, M.A. Recognition cascade and metabolite transfer in a marine bacteria-phytoplankton model system. Environ. Microbiol. 2017, 19, 3500–3513. [Google Scholar] [CrossRef] [PubMed]
- Doering, T.; Tandon, K.; Topa, S.H.; Pidot, S.J.; Blackall, L.L.; van Oppen, M.J.H. Genomic exploration of coral-associated bacteria: Identifying probiotic candidates to increase coral bleaching resilience in Galaxea fascicularis. Microbiome 2023, 11, 185. [Google Scholar] [CrossRef]
- Vancanneyt, M.; Schut, F.; Snauwaert, C.; Goris, J.; Swings, J.; Gottschal, J.C. Sphingomonas alaskensis sp. nov., a dominant bacterium from a marine oligotrophic environment. Int. J. Syst. Evol. Microbiol. 2001, 51, 73–79. [Google Scholar] [CrossRef]



| FAMILY | GENUS | |||
|---|---|---|---|---|
| Test | p-Value | Test | p-Value | |
| PERMANOVA | PSEUDO-F = 1.527733 | 0.038 | PSEUDO-F = 1.520639 | 0.039 |
| ANOSIM | R = 0.014403 | 0.416 | R = 0.024691 | 0.403 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Benvenuti, C.; Fassio, G.; Russini, V.; Modica, M.V.; Oliverio, M.; Davolos, D.; Nocella, E. A Metabarcoding Approach for Investigating the Stomach Microbiota of the Corallivorous Snail Coralliophila meyendorffii (Muricidae, Coralliophilinae) and Its Venomous Host, the Sea-Anemone Parazoanthus axinellae (Zoanthidea, Parazoanthidae). Microbiol. Res. 2024, 15, 2341-2357. https://doi.org/10.3390/microbiolres15040157
Benvenuti C, Fassio G, Russini V, Modica MV, Oliverio M, Davolos D, Nocella E. A Metabarcoding Approach for Investigating the Stomach Microbiota of the Corallivorous Snail Coralliophila meyendorffii (Muricidae, Coralliophilinae) and Its Venomous Host, the Sea-Anemone Parazoanthus axinellae (Zoanthidea, Parazoanthidae). Microbiology Research. 2024; 15(4):2341-2357. https://doi.org/10.3390/microbiolres15040157
Chicago/Turabian StyleBenvenuti, Chiara, Giulia Fassio, Valeria Russini, Maria Vittoria Modica, Marco Oliverio, Domenico Davolos, and Elisa Nocella. 2024. "A Metabarcoding Approach for Investigating the Stomach Microbiota of the Corallivorous Snail Coralliophila meyendorffii (Muricidae, Coralliophilinae) and Its Venomous Host, the Sea-Anemone Parazoanthus axinellae (Zoanthidea, Parazoanthidae)" Microbiology Research 15, no. 4: 2341-2357. https://doi.org/10.3390/microbiolres15040157
APA StyleBenvenuti, C., Fassio, G., Russini, V., Modica, M. V., Oliverio, M., Davolos, D., & Nocella, E. (2024). A Metabarcoding Approach for Investigating the Stomach Microbiota of the Corallivorous Snail Coralliophila meyendorffii (Muricidae, Coralliophilinae) and Its Venomous Host, the Sea-Anemone Parazoanthus axinellae (Zoanthidea, Parazoanthidae). Microbiology Research, 15(4), 2341-2357. https://doi.org/10.3390/microbiolres15040157






