The Genetic Homogeneity of Uganda’s East African Highland Bananas (Mutika/Lujugira) Does Not Match the Extensive Morphological Variation Identified in this Subgroup
Abstract
:1. Introduction
2. Materials and Methods
2.1. DNA Extraction
2.2. DNA Quantification
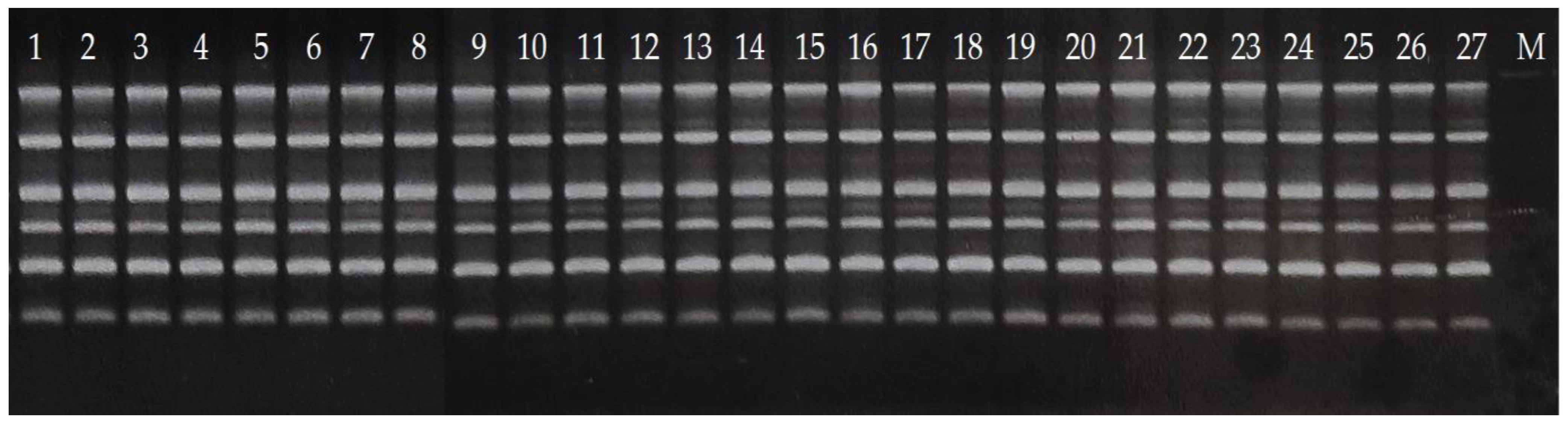
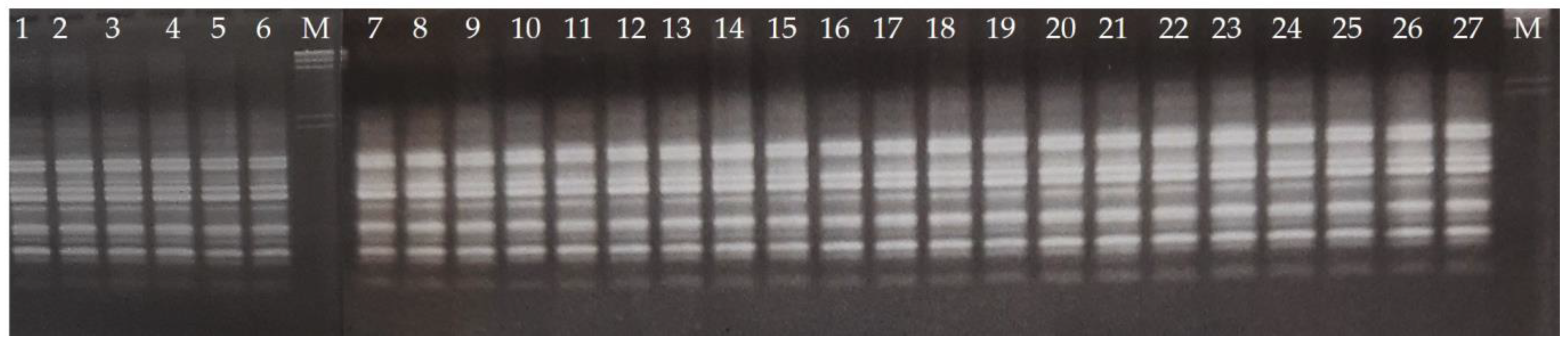
2.3. RAPD Analysis
2.4. Gel Electrophoresis
3. Results and Discussion
3.1. Reduced Diversity of Bananas via Domestication
3.2. Probable Mechanisms for Increased Phenotypic Diversity in the EAHB
3.3. Somatic Mutations
3.4. Transposable Elements as Agents of Diversity
3.5. Epigenetic Variations Contribute to Plant Evolution
3.6. Polyploidy
3.7. Chimerism
3.8. Genomic and Phenotypic Plasticity
4. Conclusions
Funding
Data Availability Statement
Conflicts of Interest
References
- Simmonds, N.W. The Evolution of the Bananas; Longmans: London, UK, 1962. [Google Scholar]
- Perrier, X.; De Langhe, E.; Donohue, M.; Lentfer, C.; Vrydaghs, L.; Bakry, F.; Carreel, F.; Hippolyte, I.; Horry, J.-P.; Jenny, C.; et al. Multidisciplinary Perspectives on Banana (Musa spp.) Domestication. Proc. Nat. Acad. Sci. USA 2011, 108, 11311–11318. [Google Scholar] [CrossRef] [PubMed]
- Němečková, A.; Christelová, P.; Čížková, J.; Nyine, M.; Van den houwe, I.; Svačina, R.; Uwimana, B.; Swennen, R.; Doležel, J.; Hřibová, E. Molecular and Cytogenetic Study of East African Highland Banana. Front. Plant Sci. 2018, 9, 339101. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Cottin, A.; Baurens, F.-C.; Labadie, K.; Hervouet, C.; Salmon, F.; Paulo-de-la-Reberdiere, N.; Van den Houwe, I.; Sardos, J.; Aury, J.-M.; et al. Interspecific Introgression Patterns Reveal the Origins of Worldwide Cultivated Bananas in New Guinea. Plant J. 2023, 113, 802–818. [Google Scholar] [CrossRef] [PubMed]
- Pillay, M.; Ogundiwin, E.; Nwakanma, D.C.; Ude, G.; Tenkouano, A. Analysis of Genetic Diversity and Relationships in East African Banana Germplasm. Theor. Appl. Genet. 2001, 102, 965–970. [Google Scholar] [CrossRef]
- Hinge, V.R.; Shaikh, I.M.; Chavhan, R.L.; Deshmukh, A.S.; Shelake, R.M.; Ghuge, S.A.; Dethe, A.M.; Suprasanna, P.; Kadam, U.S. Assessment of Genetic Diversity and Volatile Content of Commercially Grown Banana (Musa spp.) Cultivars. Sci. Rep. 2022, 12, 7979. [Google Scholar] [CrossRef] [PubMed]
- Simbare, A.; Sane, C.A.B.; Nduwimana, I.; Niyongere, C.; Omondi, B.A. Diminishing Farm Diversity of East African Highland Bananas in Banana Bunchy Top Disease Outbreak Areas of Burundi—The Effect of Both Disease and Control Approaches. Sustainability 2020, 12, 7467. [Google Scholar] [CrossRef]
- Van den houwe, I.; Chase, R.; Sardos, J.; Ruas, M.; Kempenaers, E.; Guignon, V.; Massart, S.; Carpentier, S.; Panis, B.; Rouard, M.; et al. Safeguarding and Using Global Banana Diversity: A Holistic Approach. CABI Agric. Biosci. 2020, 1, 15. [Google Scholar] [CrossRef]
- Swarup, S.; Cargill, E.J.; Crosby, K.; Flagel, L.; Kniskern, J.; Glenn, K.C. Genetic Diversity Is Indispensable for Plant Breeding to Improve Crops. Crop Sci. 2021, 61, 839–852. [Google Scholar] [CrossRef]
- Wahyudi, D.; Nursita, D.C.; Hapsari, L. Genetic Diversity among and within Genome Groups of Banana Cultivars Based on ISSR Markers. Int. J. Agric. Biol. 2022, 28, 366–374. [Google Scholar]
- Boonsrangsom, T.; Fuenghoi, C.; Premjet, D.; Suvittawat, K.; Ratanasut, K.; Sujipuli, K. Genetic Relationships and Genome Verification of Thai Banana Cultivars Using Random Amplification of Polymorphic DNA (RAPD) Markers. Biodiversitas 2023, 24, 3758–3765. [Google Scholar] [CrossRef]
- Slameto, S. Genetic Diversity and Molecular Analysis Using RAPD Markers of Banana Cultivars in the Five Regions of East Java, Indonesia. Biodiversitas 2023, 24, 5035–5043. [Google Scholar] [CrossRef]
- Bhalang, D.; Prabhuling, G.; Hipparagi, K.; Raghavendra, S.; Prakash, D.P.; Babu, A.G. Analysis of the Genetic Stability of Banana Tissue Culture Propagated Plantlets Cv. Ney Poovan (AB) Using Morphological and Molecular Markers. Int. J. Curr. Microbiol. App. Sci. 2018, 7, 1007–1018. [Google Scholar] [CrossRef]
- Fauré, S.; Noyer, J.L.; Horry, J.P.; Bakry, F.; Lanaud, C.; Gońzalez De León, D. A Molecular Marker-Based Linkage Map of Diploid Bananas (Musa acuminata). Theor. Appl. Genet. 1993, 87, 517–526. [Google Scholar] [CrossRef] [PubMed]
- Karamura, D.A.; International Network for the Improvement of Banana and Plantain. Numerical Taxonomic Studies of the East African Highland Bananas (Musa AAA—East Africa) in Uganda; INIBAP: Montpellier, France, 1999. [Google Scholar]
- Kitavi, M.; Downing, T.; Lorenzen, J.; Karamura, D.; Onyango, M.; Nyine, M.; Ferguson, M.; Spillane, C. The Triploid East African Highland Banana (EAHB) Genepool Is Genetically Uniform Arising from a Single Ancestral Clone That Underwent Population Expansion by Vegetative Propagation. Theor. Appl. Genet. 2016, 129, 547–561. [Google Scholar] [CrossRef] [PubMed]
- Christelová, P.; De Langhe, E.; Hřibová, E.; Čížková, J.; Sardos, J.; Hušáková, M.; Van den houwe, I.; Sutanto, A.; Kepler, A.K.; Swennen, R.; et al. Molecular and Cytological Characterization of the Global Musa Germplasm Collection Provides Insights into the Treasure of Banana Diversity. Biodivers. Conserv. 2017, 26, 801–824. [Google Scholar] [CrossRef]
- Rauf, S.; Silva, J.; Khan, A.; Naveed, A. Consequences of Plant Breeding on Genetic Diversity. Int. J. Plant Breed. 2010, 4, 1–21. [Google Scholar]
- Garcia, A.A.F.; Benchimol, L.L.; Barbosa, A.M.M.; Geraldi, I.O.; Souza, C.L., Jr.; de Souza, A.P. Comparison of RAPD, RFLP, AFLP and SSR Markers for Diversity Studies in Tropical Maize Inbred Lines. Genet. Mol. Biol. 2004, 27, 579–588. [Google Scholar] [CrossRef]
- Velasco-Ramírez, A.P.; Torres-Morán, M.I.; Molina-Moret, S.; Sánchez-González, J.d.J.; Santacruz-Ruvalcaba, F. Efficiency of RAPD, ISSR, AFLP and ISTR Markers for the Detection of Polymorphisms and Genetic Relationships in Camote de Cerro (Dioscorea spp.). Electron. J. Biotech. 2014, 17, 65–71. [Google Scholar] [CrossRef]
- Nyine, M.; Pillay, M. The Effect of Banana Breeding on the Diversity of East African Highland Banana (Musa), AAA). Acta Hortic. 2011, 897, 225–229. [Google Scholar] [CrossRef]
- Noyer, J.L.; Causse, S.; Tomekpe, K.; Bouet, A.; Baurens, F.C. A New Image of Plantain Diversity Assessed by SSR, AFLP and MSAP Markers. Genetica 2005, 124, 61–69. [Google Scholar] [CrossRef]
- Ssebuliba, R.; Talengera, D.; Makumbi, D.; Namanya, P.; Tenkouano, A.; Tushemereirwe, W.; Pillay, M. Reproductive Efficiency and Breeding Potential of East African Highland (Musa AAA-EA) Bananas. Field Crops Res. 2006, 95, 250–255. [Google Scholar] [CrossRef]
- Fungo, R.; Pillay, M. β-Carotene Content of Selected Banana Genotypes from Uganda. Afr. J. Biotech. 2011, 10, 5423–5430. [Google Scholar]
- Pillay, M.; Fungo, R. Diversity of Iron and Zinc Content in Bananas from East and Central Africa. HortScience 2016, 51, 320–324. [Google Scholar] [CrossRef]
- Ben-Ari, G.; Lavi, U. 11—Marker-Assisted Selection in Plant Breeding. In Plant Biotechnology and Agriculture; Altman, A., Hasegawa, P.M., Eds.; Academic Press: San Diego, CA, USA, 2012; pp. 163–184. [Google Scholar] [CrossRef]
- Vieira, M.L.C.; Santini, L.; Diniz, A.L.; Munhoz, C.d.F. Microsatellite Markers: What They Mean and Why They Are so Useful. Genet. Mol. Biol. 2016, 39, 312–328. [Google Scholar] [CrossRef] [PubMed]
- Dela Cruz, F.M.; Laude, R.; Sandoval, C.M.; Gueco, L.; Huelgas, V.; Garcia, R.; Mendoza, E.M. Genetic Characterization of Philippine Saba Germplasm Collection Using Microsatellite Markers. Philipp. J. Sci. 2020, 149, 169–197. [Google Scholar] [CrossRef]
- Workneh, S.T.; Alemu, S.K.; Olani, G.; Debebe, A.; Berhanu, B.; Dagnew, A.; Assefa, W. Molecular Characterization of Banana Genotypes by SSR Markers. Afr. J. Plant Sci. 2022, 16, 258–269. [Google Scholar] [CrossRef]
- Biswas, M.K.; Bagchi, M.; Biswas, D.; Harikrishna, J.A.; Liu, Y.; Li, C.; Sheng, O.; Mayer, C.; Yi, G.; Deng, G. Genome-Wide Novel Genic Microsatellite Marker Resource Development and Validation for Genetic Diversity and Population Structure Analysis of Banana. Genes 2020, 11, 1479. [Google Scholar] [CrossRef] [PubMed]
- Nzawele, D.; Kanyenga, A.; Kusolwa, P.; Rweyemamu, C.; Maerere, A. Genetic Diversity in Banana and Plantains Cultivars from Eastern DRC and Tanzania Using SSR and Morphological Markers, Their Phylogenetic Classification and Principal Components Analyses; IntechOpen: Rijeka, Croatia, 2018. [Google Scholar] [CrossRef]
- Tugume, A.K.; Lubega, G.W.; Rubaihayo, P.R.; Tugume, A.K.; Lubega, G.W.; Rubaihayo, P.R. Genetic Diversity of East African Highland Bananas Using AFLP. Infomusa 2002, 11, 28–32. [Google Scholar]
- Ude, G.; Pillay, M.; Ogundiwin, E.; Tenkouano, A. Genetic Diversity in an African Plantain Core Collection Using AFLP and RAPD Markers. Theor. Appl. Genet. 2003, 107, 248–255. [Google Scholar] [CrossRef]
- Ermini, J.L.; Tenaglia, G.C.; Pratta, G.R. Molecular Diversity in Selected Banana Clones (Musa AAA) “Cavendish”) Adapted to the Subtropical Environment of Formosa Province (Argentina). Am. J. Plant Sci. 2018, 9, 2504–2513. [Google Scholar] [CrossRef]
- Dhivya, S.; Ashutosh, S.; Gowtham, I.; Baskar, V.; Harini, A.B.; Mukunthakumar, S.; Sathishkumar, R. Molecular Identification and Evolutionary Relationships between the Subspecies of Musa by DNA Barcodes. BMC Genom. 2020, 21, 659. [Google Scholar] [CrossRef] [PubMed]
- Borborah, K.; Saikia, D.; Rehman, M.; Islam, A.; Mahanta, S.; Chutia, J.; Borthakur, S.K.; Tanti, B. Comparative Analysis of Genetic Diversity in Some Non-Commercial Cultivars of Musa L. from Assam, India, Using Morphometric and ISSR Markers. Int. J. Fruit Sci. 2020, 20 (Suppl. S2), 1814–1828. [Google Scholar] [CrossRef]
- Noor, S.; Muhammad, A.; Shahzad, A.; Hussain, I.; Zeshan, M.; Ali, K.; Begum, S.; Aqeel, M.; Numan, M.; Muazzam Naz, R.M.; et al. Inter Simple Sequence Repeat-Based Genetic Divergence and Varietal Identification of Banana in Pakistan. Agronomy 2022, 12, 2932. [Google Scholar] [CrossRef]
- Beaton, K.; Mazadza, A.; Chikwambi, Z. Identification of Zimbabwe’s Locally Grown Banana (Musa spp.) Cultivars Using Morphology and Genome-Targeted Sequencing. J. Gen. Eng. Biotech. 2023, 21, 118. [Google Scholar] [CrossRef]
- Safhi, F.A.; Alshamrani, S.M.; Alshaya, D.S.; Hussein, M.A.A.; Abd El-Moneim, D. Genetic Diversity Analysis of Banana Cultivars (Musa sp.) in Saudi Arabia Based on AFLP Marker. Curr. Issues Mol. Biol. 2023, 45, 1810–1819. [Google Scholar] [CrossRef]
- Mbo Nkoulou, L.F.; Tchinda Ninla, L.A.; Cros, D.; Martin, G.; Ndiang, Z.; Houegban, J.; Ngalle, H.B.; Bell, J.M.; Achigan-Dako, E.G. Analysis of Genetic Diversity and Agronomic Variation in Banana Sub-Populations for Genomic Selection under Drought Stress in Southern Benin. Gene 2023, 859, 147210. [Google Scholar] [CrossRef]
- Premjet, D.; Boonsrangsom, T.; Sujipuli, K.; Rattanasut, K.; Kongbungkerd, A.; Premjet, S. Morphological and Molecular Characterizations of Musa (ABB) “Mali-Ong” in Thailand. Biology 2022, 11, 1429. [Google Scholar] [CrossRef] [PubMed]
- Flint-Garcia, S.A. Genetics and Consequences of Crop Domestication. J. Agric. Food Chem. 2013, 61, 8267–8276. [Google Scholar] [CrossRef]
- Comai, L. The Advantages and Disadvantages of Being Polyploid. Nat. Rev. Genet. 2005, 6, 836–846. [Google Scholar] [CrossRef]
- Budar, F.; Roux, F. The Role of Organelle Genomes in Plant Adaptation: Time to Get to Work! Plant Signal. Behav. 2011, 6, 635–639. [Google Scholar] [CrossRef]
- Forneck, A. Plant Breeding: Clonality—A Concept for Stability and Variability During Vegetative Propagation. In Progress in Botany; Esser, K., Lüttge, U., Beyschlag, W., Murata, J., Eds.; Progress in Botany; Springer: Berlin/Heidelberg, Germany, 2005; Volume 66, pp. 164–183. [Google Scholar] [CrossRef]
- Infante, D.; González, G.; Peraza-Echeverría, L.; Keb-Llanes, M. Asexual Genetic Variability in Agave fourcroydes. Plant Sci. 2003, 164, 223–230. [Google Scholar] [CrossRef]
- Riaz, S.; De Lorenzis, G.; Velasco, D.; Koehmstedt, A.; Maghradze, D.; Bobokashvili, Z.; Musayev, M.; Zdunic, G.; Laucou, V.; Andrew Walker, M.; et al. Genetic Diversity Analysis of Cultivated and Wild Grapevine (Vitis vinifera L.) Accessions around the Mediterranean Basin and Central Asia. BMC Plant Biol. 2018, 18, 137. [Google Scholar] [CrossRef]
- Bakry, F.; Carreel, F.; Jenny, C.; Horry, J.-P. Genetic Improvement of Banana. In Breeding Plantation Tree Crops: Tropical Species; Jain, S.M., Priyadarshan, P.M., Eds.; Springer: New York, NY, USA, 2009; pp. 3–50. [Google Scholar] [CrossRef]
- Jeridi, M.; Bakry, F.; Escoute, J.; Fondi, E.; Carreel, F.; Ferchichi, A.; D’Hont, A.; Rodier-Goud, M. Homoeologous Chromosome Pairing between the A and B Genomes of Musa spp. Revealed by Genomic in Situ Hybridization. Ann. Bot. 2011, 108, 975–981. [Google Scholar] [CrossRef] [PubMed]
- Baurens, F.-C.; Martin, G.; Hervouet, C.; Salmon, F.; Yohomé, D.; Ricci, S.; Rouard, M.; Habas, R.; Lemainque, A.; Yahiaoui, N.; et al. Recombination and Large Structural Variations Shape Interspecific Edible Bananas Genomes. Mol. Biol. Evol. 2019, 36, 97–111. [Google Scholar] [CrossRef] [PubMed]
- Martin, G.; Baurens, F.; Hervouet, C.; Salmon, F.; Delos, J.; Labadie, K.; Perdereau, A.; Mournet, P.; Blois, L.; Dupouy, M.; et al. Chromosome Reciprocal Translocations Have Accompanied Subspecies Evolution in Bananas. Plant J. 2020, 104, 1698–1711. [Google Scholar] [CrossRef]
- Meyer, R.S.; Purugganan, M.D. Evolution of Crop Species: Genetics of Domestication and Diversification. Nat. Rev. Genet. 2013, 14, 840–852. [Google Scholar] [CrossRef]
- Shi, J.; Lai, J. Patterns of Genomic Changes with Crop Domestication and Breeding. Curr. Opin. Plant Biol. 2015, 24, 47–53. [Google Scholar] [CrossRef] [PubMed]
- Kantar, M.B.; Nashoba, A.R.; Anderson, J.E.; Blackman, B.K.; Rieseberg, L.H. The Genetics and Genomics of Plant Domestication. BioScience 2017, 67, 971–982. [Google Scholar] [CrossRef]
- Gepts, P. Crop Domestication as a Long-Term Selection Experiment. In Plant Breeding Reviews; John Wiley & Sons, Ltd.: Hoboken, NJ, USA, 2003; pp. 1–44. [Google Scholar] [CrossRef]
- Denham, T.; Barton, H.; Castillo, C.; Crowther, A.; Dotte-Sarout, E.; Florin, S.A.; Pritchard, J.; Barron, A.; Zhang, Y.; Fuller, D.Q. The Domestication Syndrome in Vegetatively Propagated Field Crops. Ann. Bot. 2020, 125, 581–597. [Google Scholar] [CrossRef]
- Perrier, X.; Jenny, C.; Bakry, F.; Karamura, D.; Kitavi, M.; Dubois, C.; Hervouet, C.; Philippson, G.; De Langhe, E. East African Diploid and Triploid Bananas: A Genetic Complex Transported from South-East Asia. Ann. Bot. 2019, 123, 19–36. [Google Scholar] [CrossRef]
- Raboin, L.-M.; Carreel, F.; Noyer, J.-L.; Baurens, F.-C.; Horry, J.-P.; Bakry, F.; Montcel, H.T.D.; Ganry, J.; Lanaud, C.; Lagoda, P.J.L. Diploid Ancestors of Triploid Export Banana Cultivars: Molecular Identification of 2n Restitution Gamete Donors and n Gamete Donors. Mol. Breed. 2005, 16, 333–341. [Google Scholar] [CrossRef]
- McKey, D.; Elias, M.; Pujol, B.; Duputié, A. The Evolutionary Ecology of Clonally Propagated Domesticated Plants. New Phytol. 2010, 186, 318–332. [Google Scholar] [CrossRef] [PubMed]
- Carreel, F.; Gonzalez de Leon, D.; Lagoda, P.; Lanaud, C.; Jenny, C.; Horry, J.P.; Tezenas du Montcel, H. Ascertaining Maternal and Paternal Lineage within Musa by Chloroplast and Mitochondrial DNA RFLP Analyses. Genome 2002, 45, 679–692. [Google Scholar] [CrossRef]
- De Langhe, E.; Hřibová, E.; Carpentier, S.; Doležel, J.; Swennen, R. Did Backcrossing Contribute to the Origin of Hybrid Edible Bananas? Ann. Bot. 2010, 106, 849–857. [Google Scholar] [CrossRef] [PubMed]
- Smýkal, P.; Nelson, M.N.; Berger, J.D.; Von Wettberg, E.J.B. The Impact of Genetic Changes during Crop Domestication. Agronomy 2018, 8, 119. [Google Scholar] [CrossRef]
- Nankar, A.N.; Tringovska, I.; Grozeva, S.; Ganeva, D.; Kostova, D. Tomato Phenotypic Diversity Determined by Combined Approaches of Conventional and High-Throughput Tomato Analyzer Phenotyping. Plants 2020, 9, 197. [Google Scholar] [CrossRef]
- Troyer, A.F. Background of U.S. Hybrid Corn. Crop Sci. 1999, 39, 601–626. [Google Scholar] [CrossRef]
- Duncan, E.J.; Gluckman, P.D.; Dearden, P.K. Epigenetics, Plasticity, and Evolution: How Do We Link Epigenetic Change to Phenotype? J. Exp. Zool. B Mol. Dev. Evol. 2014, 322, 208–220. [Google Scholar] [CrossRef] [PubMed]
- Šimoníková, D.; Němečková, A.; Čížková, J.; Brown, A.; Swennen, R.; Doležel, J.; Hřibová, E. Chromosome Painting in Cultivated Bananas and Their Wild Relatives (Musa spp.) Reveals Differences in Chromosome Structure. Int. J. Mol. Sci. 2020, 21, 7915. [Google Scholar] [CrossRef]
- Karamura, D.; Karamura, E.; Tushemereirwe, W.; Rubaihayo, P.R.; Markham, R. Somatic Mutations and Their Implications to the Conservation Strategies of the East African Highland Bananas (Musa spp.). Acta Hortic. 2010, 879, 615–622. [Google Scholar] [CrossRef]
- Whitham, T.G.; Slobodchikoff, C.N. Evolution by Individuals, Plant-Herbivore Interactions, and Mosaics of Genetic Variability: The Adaptive Significance of Somatic Mutations in Plants. Oecologia 1981, 49, 287–292. [Google Scholar] [CrossRef]
- Holsinger, K.E. Reproductive Systems and Evolution in Vascular Plants. Proc. Natl. Acad. Sci. USA 2000, 97, 7037–7042. [Google Scholar] [CrossRef]
- Caetano-anollés, G. High Genome-Wide Mutation Rates in Vegetatively Propagated Bermudagrass. Mol. Ecol. 1999, 8, 1211–1221. [Google Scholar] [CrossRef]
- Karamura, D.; Karamura, E.; Blomme, G. General Plant Morphology of Musa. In Banana Breeding: Progress and Challenges; Pillay, M., Tenkouano, A., Eds.; CRC Press: Boca Raton, FL, USA, 2011; pp. 1–20. [Google Scholar] [CrossRef]
- Hou, B.-H.; Tsai, Y.-H.; Chiang, M.-H.; Tsao, S.-M.; Huang, S.-H.; Chao, C.-P.; Chen, H.-M. Cultivar-Specific Markers, Mutations, and Chimerism of Cavendish Banana Somaclonal Variants Resistant to Fusarium oxysporum f. sp. cubense Tropical Race 4. BMC Genom. 2022, 23, 470. [Google Scholar] [CrossRef]
- Yu, L.; Nie, Y.; Jiao, J.; Jian, L.; Zhao, J. The Sequencing-Based Mapping Method for Effectively Cloning Plant Mutated Genes. Int. J. Mol. Sci. 2021, 22, 6224. [Google Scholar] [CrossRef] [PubMed]
- Desjardins, Y.; Dubuc, J.-F.; Badr, A. In Vitro Culture of Plants: A Stressful Activity! Acta Hortic. 2009, 812, 29–50. [Google Scholar] [CrossRef]
- Şen, A. Oxidative Stress Studies in Plant Tissue Culture. In Antioxidant Enzyme; IntechOpen: Rijeka, Croatia, 2012. [Google Scholar] [CrossRef]
- Niu, X.-M.; Xu, Y.-C.; Li, Z.-W.; Bian, Y.-T.; Hou, X.-H.; Chen, J.-F.; Zou, Y.-P.; Jiang, J.; Wu, Q.; Ge, S.; et al. Transposable Elements Drive Rapid Phenotypic Variation in Capsella rubella. Proc. Natl. Acad. Sci. USA 2019, 116, 6908–6913. [Google Scholar] [CrossRef] [PubMed]
- Colonna Romano, N.; Fanti, L. Transposable Elements: Major Players in Shaping Genomic and Evolutionary Patterns. Cells 2022, 11, 1048. [Google Scholar] [CrossRef] [PubMed]
- Lisch, D. How Important Are Transposons for Plant Evolution? Nat. Rev. Genet. 2013, 14, 49–61. [Google Scholar] [CrossRef]
- Vitte, C.; Fustier, M.-A.; Alix, K.; Tenaillon, M.I. The Bright Side of Transposons in Crop Evolution. Brief. Funct. Genom. 2014, 13, 276–295. [Google Scholar] [CrossRef]
- Razali, N.M.; Cheah, B.H.; Nadarajah, K. Transposable Elements Adaptive Role in Genome Plasticity, Pathogenicity and Evolution in Fungal Phytopathogens. Int. J. Mol. Sci. 2019, 20, 3597. [Google Scholar] [CrossRef]
- Wei, L.; Cao, X. The Effect of Transposable Elements on Phenotypic Variation: Insights from Plants to Humans. Sci. China Life Sci. 2016, 59, 24–37. [Google Scholar] [CrossRef] [PubMed]
- Horváth, V.; Merenciano, M.; González, J. Revisiting the Relationship between Transposable Elements and the Eukaryotic Stress Response. Trends Genet. 2017, 33, 832–841. [Google Scholar] [CrossRef] [PubMed]
- Wang, M.-Z.; Li, H.-L.; Li, J.-M.; Yu, F.-H. Correlations between Genetic, Epigenetic and Phenotypic Variation of an Introduced Clonal Herb. Heredity 2020, 124, 146–155. [Google Scholar] [CrossRef] [PubMed]
- Balint-Kurti, P.J.; Clendennen, S.K.; Doleželová, M.; Valárik, M.; Doležel, J.; Beetham, P.R.; May, G.D. Identification and Chromosomal Localization of the Monkey Retrotransposon in Musa sp. Mol. Gen. Genet. 2000, 263, 908–915. [Google Scholar] [CrossRef] [PubMed]
- Pratama, S.N.; Dwivany, F.M.; Nugrahapraja, H. Comparative Genomics of Copia and Gypsy Retroelements in Three Banana Genomes: A, B, and S Genomes. Pertanika J. Trop. Agric. Sci. 2021, 44, 697–711. [Google Scholar] [CrossRef]
- Nouroz, F.; Noreen, S.; Ahmad, H.; Heslop-Harrison, J.S.P. The Landscape and Structural Diversity of LTR Retrotransposons in Musa Genome. Mol. Genet. Genom. 2017, 292, 1051–1067. [Google Scholar] [CrossRef] [PubMed]
- Kapazoglou, A.; Ganopoulos, I.; Tani, E.; Tsaftaris, A. Chapter Nine—Epigenetics, Epigenomics and Crop Improvement. In Advances in Botanical Research; Kuntz, M., Ed.; Transgenic Plants; Academic Press: Cambridge, MA, USA, 2018; Volume 86, pp. 287–324. [Google Scholar] [CrossRef]
- Gupta, C.; Salgotra, R.K. Epigenetics and Its Role in Effecting Agronomical Traits. Front. Plant Sci. 2022, 13, 925688. [Google Scholar] [CrossRef]
- Kitavi, M.; Cashell, R.; Ferguson, M.; Lorenzen, J.; Nyine, M.; McKeown, P.C.; Spillane, C. Heritable Epigenetic Diversity for Conservation and Utilization of Epigenetic Germplasm Resources of Clonal East African Highland Banana (EAHB) Accessions. Theor. Appl. Genet. 2020, 133, 2605–2625. [Google Scholar] [CrossRef] [PubMed]
- Ranganath, G. Genome-Wide Characterization of Small RNA, Gene Expression and DNA Methylation Changes in Response to Salt Stress in Musa acuminata. Ph.D. Thesis, University of Malaya, Kuala Lumpur, Malaysia, 2018. [Google Scholar]
- Flagel, L.E.; Wendel, J.F. Gene Duplication and Evolutionary Novelty in Plants. New Phytol. 2009, 183, 557–564. [Google Scholar] [CrossRef]
- Qiao, X.; Li, Q.; Yin, H.; Qi, K.; Li, L.; Wang, R.; Zhang, S.; Paterson, A.H. Gene Duplication and Evolution in Recurring Polyploidization–Diploidization Cycles in Plants. Genome Biol. 2019, 20, 38. [Google Scholar] [CrossRef] [PubMed]
- Madlung, A. Polyploidy and Its Effect on Evolutionary Success: Old Questions Revisited with New Tools. Heredity 2013, 110, 99–104. [Google Scholar] [CrossRef] [PubMed]
- Cheng, F.; Wu, J.; Cai, X.; Liang, J.; Freeling, M.; Wang, X. Gene Retention, Fractionation and Subgenome Differences in Polyploid Plants. Nat. Plants 2018, 4, 258–268. [Google Scholar] [CrossRef] [PubMed]
- Weiss-Schneeweiss, H.; Emadzade, K.; Jang, T.-S.; Schneeweiss, G.M. Evolutionary Consequences, Constraints and Potential of Polyploidy in Plants. Cytogenet. Genome Res. 2013, 140, 137–150. [Google Scholar] [CrossRef]
- Wang, Z.; Fan, G.; Dong, Y.; Zhai, X.; Deng, M.; Zhao, Z.; Liu, W.; Cao, Y. Implications of Polyploidy Events on the Phenotype, Microstructure, and Proteome of Paulownia Australis. PLoS ONE 2017, 12, e0172633. [Google Scholar] [CrossRef] [PubMed]
- Sattler, M.C.; Carvalho, C.R.; Clarindo, W.R. The Polyploidy and Its Key Role in Plant Breeding. Planta 2016, 243, 281–296. [Google Scholar] [CrossRef] [PubMed]
- Simmonds, N.W. The Strength of Banana Petioles in Relation to Ploidy. Ann. Bot. 1952, 16, 341–347. [Google Scholar] [CrossRef]
- Vandenhout, H.; Ortiz, R.; Vuylsteke, D.R.; Swennen, R.L.; Bai, K. Effect of Ploidy on Stomatal and Other Quantitative Traits in Plantain and Banana Hybrids. Euphytica 1995, 83, 117–122. [Google Scholar] [CrossRef]
- Kanchanapoom, K.; Koarapatchaikul, K. In Vitro Induction of Tetraploid Plants from Callus Cultures of Diploid Bananas (Musa acuminata, AA Group) ‘Kluai Leb Mu Nang’ and ‘Kluai Sa’. Euphytica 2012, 183, 111–117. [Google Scholar] [CrossRef]
- Do Amaral, C.M.; De Almeida Dos Santos-Serejo, J.; De Oliveira E Silva, S.; Da Silva Ledo, C.A.; Amorim, E.P. Agronomic Characterization of Autotetraploid Banana Plants Derived from ‘Pisang Lilin’ (AA) Obtained through Chromosome Doubling. Euphytica 2015, 202, 435–443. [Google Scholar] [CrossRef]
- Amah, D.; van Biljon, A.; Maziya-Dixon, B.; Labuschagne, M.; Swennen, R. Effects of In Vitro Polyploidization on Agronomic Characteristics and Fruit Carotenoid Content; Implications for Banana Genetic Improvement. Front. Plant Sci. 2019, 10, 480198. [Google Scholar] [CrossRef] [PubMed]
- Viehmannová, I.; Trávníčková, M.; Špatenková, E.; Černá, M.; Trávníček, P. Induced Polyploidization and Its Influence on Yield, Morphological, and Qualitative Characteristics of Microtubers in Ullucus tuberosus. Plant Cell Tissue Organ Cult. 2012, 109, 83–90. [Google Scholar] [CrossRef]
- Tavan, M.; Mirjalili, M.H.; Karimzadeh, G. In Vitro Polyploidy Induction: Changes in Morphological, Anatomical and Phytochemical Characteristics of Thymus persicus (Lamiaceae). Plant Cell Tissue Organ Cult. 2015, 122, 573–583. [Google Scholar] [CrossRef]
- Cenci, A.; Hueber, Y.; Zorrilla-Fontanesi, Y.; van Wesemael, J.; Kissel, E.; Gislard, M.; Sardos, J.; Swennen, R.; Roux, N.; Carpentier, S.C.; et al. Effect of Paleopolyploidy and Allopolyploidy on Gene Expression in Banana. BMC Genom. 2019, 20, 244. [Google Scholar] [CrossRef] [PubMed]
- Tilney-Bassett, R.A.E. Plant Chimeras; Cambridge University Press: Cambridge, UK, 1991. [Google Scholar]
- Israeli, Y.; Lahav, E.; Reuveni, O. In Vitro Culture of Bananas. In Bananas and Plantains; Gowen, S., Ed.; Springer: Dordrecht, The Netherlands, 1995; pp. 147–178. [Google Scholar] [CrossRef]
- Franks, T.; Botta, R.; Thomas, M.R.; Franks, J. Chimerism in Grapevines: Implications for Cultivar Identity, Ancestry and Genetic Improvement. Theor. Appl. Genet. 2002, 104, 192–199. [Google Scholar] [CrossRef] [PubMed]
- Hocquigny, S.; Merdinoglu, D.; Heloir, M.-C.; Pelsy, F. Chimerism and genetic diversity within the cultivar group of Pinots. Acta Hortic. 2003, 603, 535–544. [Google Scholar] [CrossRef]
- Blomme, G.; Swennen, R.L.; Tenkouano, A. Environmental Influences on Shoot and Root Growth in Banana and Plantain. Afr. Crop Sci. Conf. Proc. 2005, 7, 1163–1167. [Google Scholar]
- Taulya, G.; van Asten, P.J.A.; Leffelaar, P.A.; Giller, K.E. Phenological Development of East African Highland Banana Involves Trade-Offs between Physiological Age and Chronological Age. Eur. J. Agron. 2014, 60, 41–53. [Google Scholar] [CrossRef]
- Zhang, K.; Wang, X.; Cheng, F. Plant Polyploidy: Origin, Evolution, and Its Influence on Crop Domestication. Hortic. Plant J. 2019, 5, 231–239. [Google Scholar] [CrossRef]
- Leitch, A.R.; Leitch, I.J. Genomic Plasticity and the Diversity of Polyploid Plants. Science 2008, 320, 481–483. [Google Scholar] [CrossRef]
- Slotkin, R.K.; Martienssen, R. Transposable Elements and the Epigenetic Regulation of the Genome. Nat. Rev. Genet. 2007, 8, 272–285. [Google Scholar] [CrossRef] [PubMed]
- Ding, M.; Chen, Z.J. Epigenetic Perspectives on the Evolution and Domestication of Polyploid Plant and Crops. Curr. Opin. Plant Biol. 2018, 42, 37–48. [Google Scholar] [CrossRef] [PubMed]
- Sardos, J.; Breton, C.; Perrier, X.; Van Den Houwe, I.; Carpentier, S.; Paofa, J.; Rouard, M.; Roux, N. Hybridization, Missing Wild Ancestors and the Domestication of Cultivated Diploid Bananas. Front. Plant Sci. 2022, 13, 969220. [Google Scholar] [CrossRef] [PubMed]

| Serial No. | Genotypes | Genome Composition | Clone Set | Use |
|---|---|---|---|---|
| 1 | Entukura | AAA | Nfuuka | Cooking |
| 2 | Enzirabahima | AAA | Nfuuka | Cooking |
| 3 | Nabusa | AAA | Nfuuka | Cooking |
| 4 | Namwezi | AAA | Nfuuka | Cooking |
| 5 | Nante | AAA | Nfuuka | Cooking |
| 6 | Ndyabalangira | AAA | Nfuuka | Cooking |
| 7 | Nfuuka | AAA | Nfuuka | Cooking |
| 8 | Tereza | AAA | Nfuuka | Cooking |
| 9 | Kabucuragye | AAA | Musakala | Cooking |
| 10 | Mayovu(e) | AAA | Musakala | Cooking |
| 11 | Mukazialanda | AAA | Musakala | Cooking |
| 12 | Nakibizzi | AAA | Musakala | Cooking |
| 13 | Namunwe | AAA | Musakala | Cooking |
| 14 | Siira | AAA | Musakala | Cooking |
| 15 | Kazirakwe | AAA | Nakabulu | Cooking |
| 16 | Kibuzi | AAA | Nakitembe | Cooking |
| 17 | Mbwazirume | AAA | Nakitembe | Cooking |
| 18 | Nakasabira | AAA | Nakitembe | Cooking |
| 19 | Nakawere | AAA | Nakitembe | Cooking |
| 20 | Nakyetengu | AAA | Nakitembe | Cooking |
| 21 | Nandigobe | AAA | Nakitembe | Cooking |
| 22 | Salalugazi | AAA | Nakitembe | Cooking |
| 23 | Enkara | AAA | Mbidde | Beer |
| 24 | Kabula | AAA | Mbidde | Beer |
| 25 | Nalukira | AAA | Mbidde | Beer |
| 26 | Murure | AAA | unknown | Unknown |
| 27 | Nsowe | AAA | unknown | Unknown |
| Primer No. | Primer | Primer Sequence |
|---|---|---|
| 1 | A17 | 5′-GACCGCTTGT-3′ |
| 2 | A18 | 5′-AGGTGACCGT-3′ |
| 3 | B5 | 5′-TGCGCCCTTC-3′ |
| 4 | B10 | 5′-CTGCTGGGAC-3′ |
| 5 | B17 | 5′-AGGGAACGAG-3′ |
| 6 | B19 | 5′-ACCCCCGAAG-3′ |
| 7 | C8 | 5′-TGGACCGGTG-3′ |
| 8 | C12 | 5′-TGTCATCCCC-3′ |
| 9 | C15 | 5′-GACGGATCAG-3′ |
| 10 | C16 | 5′-CACACTCCAG-3′ |
| 11 | C18 | 5′-TGAGTGGGTG-3′ |
| 12 | C20 | 5′-ACTTCGCCAC-3′ |
| 13 | D2 | 5′-GGACCCAACC-3′ |
| 14 | D4 | 5′-TCTGGTGAGG-3′ |
| 15 | D8 | 5′-GTGTGCCCCA-3′ |
| 16 | D10 | 5′-GGTCTACACC-3′ |
| 17 | D11 | 5′-AGCGCCATTG-3′ |
| 18 | D13 | 5′-GGGGTGACGA-3′ |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the author. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pillay, M. The Genetic Homogeneity of Uganda’s East African Highland Bananas (Mutika/Lujugira) Does Not Match the Extensive Morphological Variation Identified in this Subgroup. Int. J. Plant Biol. 2024, 15, 267-280. https://doi.org/10.3390/ijpb15020023
Pillay M. The Genetic Homogeneity of Uganda’s East African Highland Bananas (Mutika/Lujugira) Does Not Match the Extensive Morphological Variation Identified in this Subgroup. International Journal of Plant Biology. 2024; 15(2):267-280. https://doi.org/10.3390/ijpb15020023
Chicago/Turabian StylePillay, Michael. 2024. "The Genetic Homogeneity of Uganda’s East African Highland Bananas (Mutika/Lujugira) Does Not Match the Extensive Morphological Variation Identified in this Subgroup" International Journal of Plant Biology 15, no. 2: 267-280. https://doi.org/10.3390/ijpb15020023
APA StylePillay, M. (2024). The Genetic Homogeneity of Uganda’s East African Highland Bananas (Mutika/Lujugira) Does Not Match the Extensive Morphological Variation Identified in this Subgroup. International Journal of Plant Biology, 15(2), 267-280. https://doi.org/10.3390/ijpb15020023






