Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation
Abstract
:1. Introduction
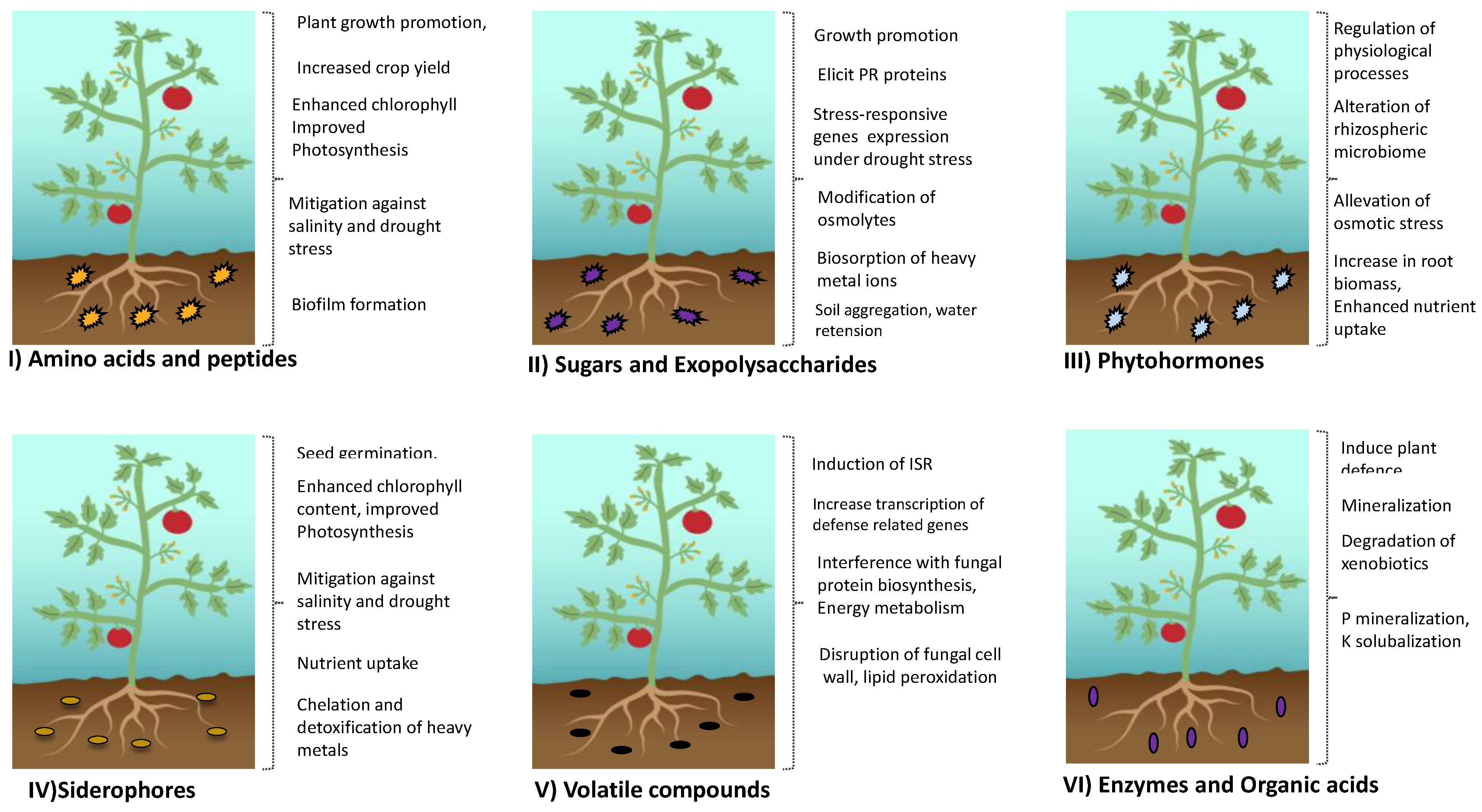
2. Microbial Exudates and Their Composition
2.1. Siderophores
2.1.1. Catecholate Siderophores
2.1.2. Hydroxamate Siderophores
2.1.3. Carboxylate and Mixed-Type Siderophores
2.2. Exopolysaccharides (EPSs)
2.3. Phytohormones
2.4. Volatile Organic Compounds (VOCs)
2.5. Organic Acids and Amino Acids
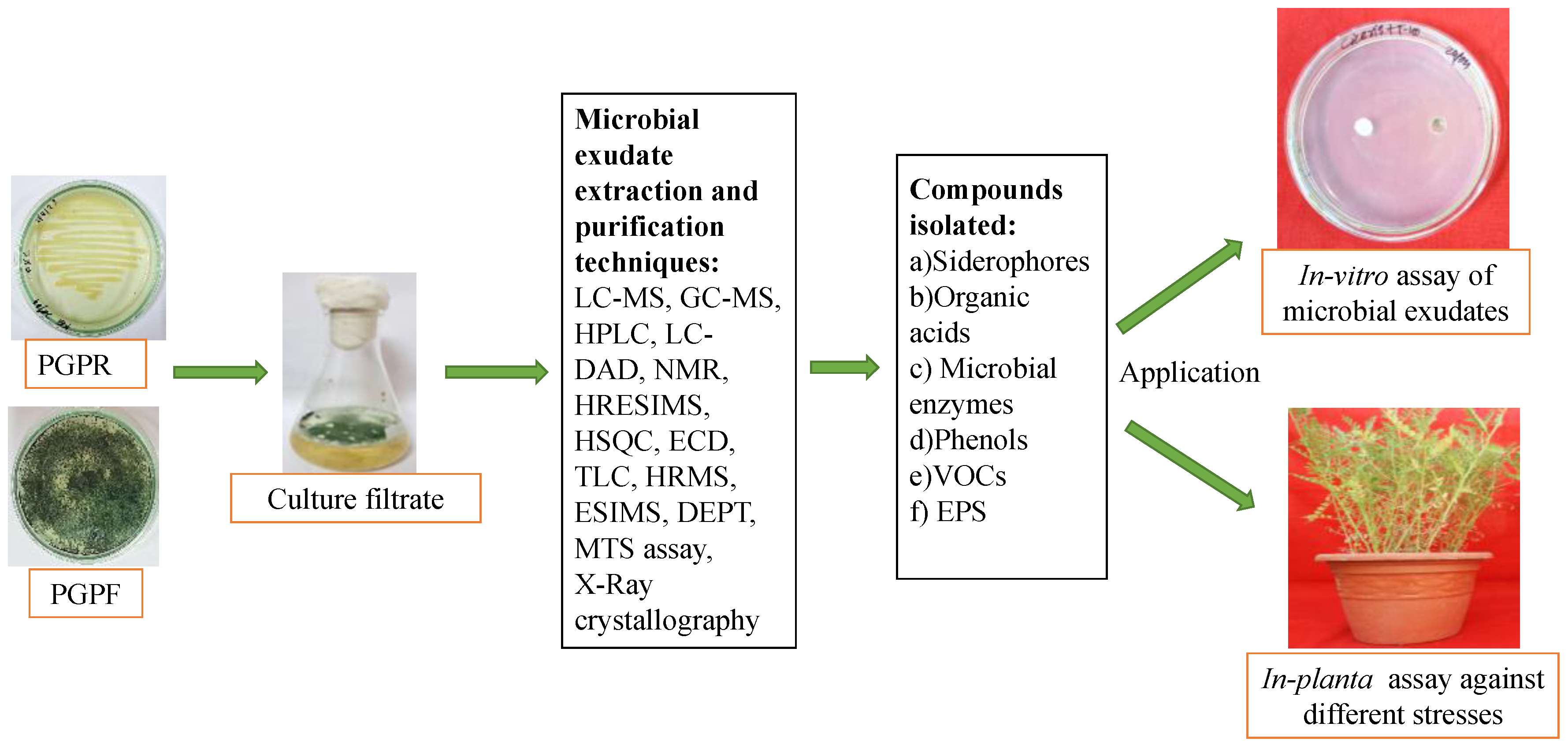
3. Identification and Characterization of Microbial Biostimulants
3.1. Biological Assays
3.1.1. In Vitro Study
3.1.2. In-Pot Assay
3.1.3. On-Field and Hydroponics Study
3.2. Biochemical Assay
3.3. Molecular Identification
| Microbe | Detection Techniques | Compounds Detected | Property of Compound | Reference |
|---|---|---|---|---|
| Trichoderma harzianum | OSMAC, extraction with ethyl acetate, LC-MS, GC-MS, X-ray analysis, plant growth, antifungal assay, cytotoxicity assay. | Siderophores, (ferricrocin and coprogen B), harzianic acid (HA) and its derivatives, butenolides and a novel metabolite, 5-hydroxy-2, 3-dimethyl-7-methoxychromone | Antifungal, anticancerous, no cytotoxic effect | [92] |
| Alcaligenes faecalis | Co-cultivation with fragments of Sclerotium rolfsii, extraction by ethyl acetate, HPLC, poisoned food technique, in-plant assay of defence and growth promotion. | Higher concentration of shikimic acid and gallic acid in CFS during co-cultivation. Higher concentration of defence enzymes in plants challenged and sprayed with CFS of co-cultivated A. faecalis. | Antifungal, plant growth promoter, and plant defense promoter | [72] |
| Actinomycetes (Micromonospora sp. UR56 and Actinokinespora sp. EG49) | Co-cultivation with Actinomycetes or other non-actinomycete bacteria, fungi, cell-derived components, and/or algae.OSMAC. | 1,6-Dicarboxylate | Antibacterial | [93] |
| Carbazoquinocin G | Antimicrobial | [94] | ||
| Malformin C | Increase in cytotoxic activity | [95] | ||
| Trichoderma spp. | α,α-diphenyl-β- picrylhydrazyl (DPPH) free radical assay for total phenolic, ascorbic acid, total antioxidant capacity, anthocyanin characterization, fruit protein analysis by bioinformatics and Nano LC-ESI-Q-Orbitrap MS/MS. | 6-pentyl-α-pyrone (6PP), harzianic acid (HA), and hydrophobin 1 (HYTLO1) | Growth promotion of strawberry, more synthesis of proteins, activated defense response in plants after treatment with specified compounds | [96] |
| Trichoderma brevicompactum | Preparative TLC, NMR, HR-ESI-MS, X-ray crystallography | Trichodermarins G–N, trichodermol, trichodermin, trichoderminol, trichodermarins A and B, 2,4,12-trihydroxy apotrichothecene | Antifungal and antimicroalgal activities | [97] |
| T. brevicompactum TPU199 | Fermentation with sodium halides, LC-MS, NMR | Trichobreols A–C | Antifungal activity | [98] |
| T. longibrachiatum | Extraction with ethyl acetate, silica gel vaccum liquid chromatography, HPLC, HR-ESI-MS, NMR, HSQC, ECD spectra, microdilution. | Trichothecinol A, 8-deoxy-trichothecin, trichothecinol B, Trichodermene A | Antifungal activity | [99] |
| T. atroviride B7 | Extraction with ethyl acetate, TLC, HPLC, CC, preparative TLC, semi-preparative HPLC, NMR. HRMS, COSY, key HMBC and key ROESY correlation of compounds, MTS assay for cytotoxicity | Harzianols F–J, 3S-hydroxyharzianone, harziandione, harzianol A | Potent antibacterial activity and moderate cytotoxicity | [100] |
| T. virens FKI-7573 | Molecular identification, MS, NMR, ECD, and chemical degradation and comparison with DNPD. | Trichothioneic acid | Potent antioxidant activity | [101] |
| T. afroharzianum Fes1712 | Overexpression of talae1, insertion of transformant plasmids (nested PCR and vector-based strategy) of E.coli into T. afroharzianum Fes1712 for secondary metabolite production. Ethly acetate extraction, CC, semi preparative HPLC, HRMS, NMR, ECD, bioactivity (96-well titer plate microdilution). | (R,3E,5E)-1-(3,5-dihydroxy-2,4- dimethylphenyl)-1-hydroxyhepta-3,5-dien-2-one, (R,3E,5E)-1-(3,5-dihydroxy-2,4- dimethylphenyl)-1-methoxyhepta-3,5-dien-2-one | Moderate antifungal activity | [91] |
| T. harzianum QTYC77 | Ethyl acetate extraction, NMR, HRMS, COSY spectra, HMBC spectra, HMQC spectra, DEPT spectra, UV spectra, CD spectra, IR spectra, UHPLC-QTOF-MS | Azaphilones D and E | Moderate antibacterial activity | [102] |
| T. harzianum D13 | Ethyl acetate filtrate, ECDspectra, spectrophotometer, The 1D (1H, 13C, and NOE) and 2D NMR spectra [HMQC, (COSY), (HMBC), and (NOESY)], ECD spectra, ESI-MS, and HRESIMS, HPLC, CC, 96-well microtitre plate assay for antifungal activity. | Nafuredin C, nafuredin A | Moderate antifungal activity | [103] |
| T. asperellum IRAN 3062C and T. longibrachiatum IRAN 3067C w | Co-cultivation, methanol/ethanol extraction, reverse-phase HPLC, ESI-MS, RNA-extraction-based expression of tex1 peptaibol synthetase gene. | Increased expression of tex1 peptaibol synthetase gene and increased synthesis of Peptiabol when co-cultivated with plant pathogens | Antifungal activity | [73] |
4. Microbial Exudates as Biostimulants
4.1. Microbial Exudates in Promoting Plant Growth and Health
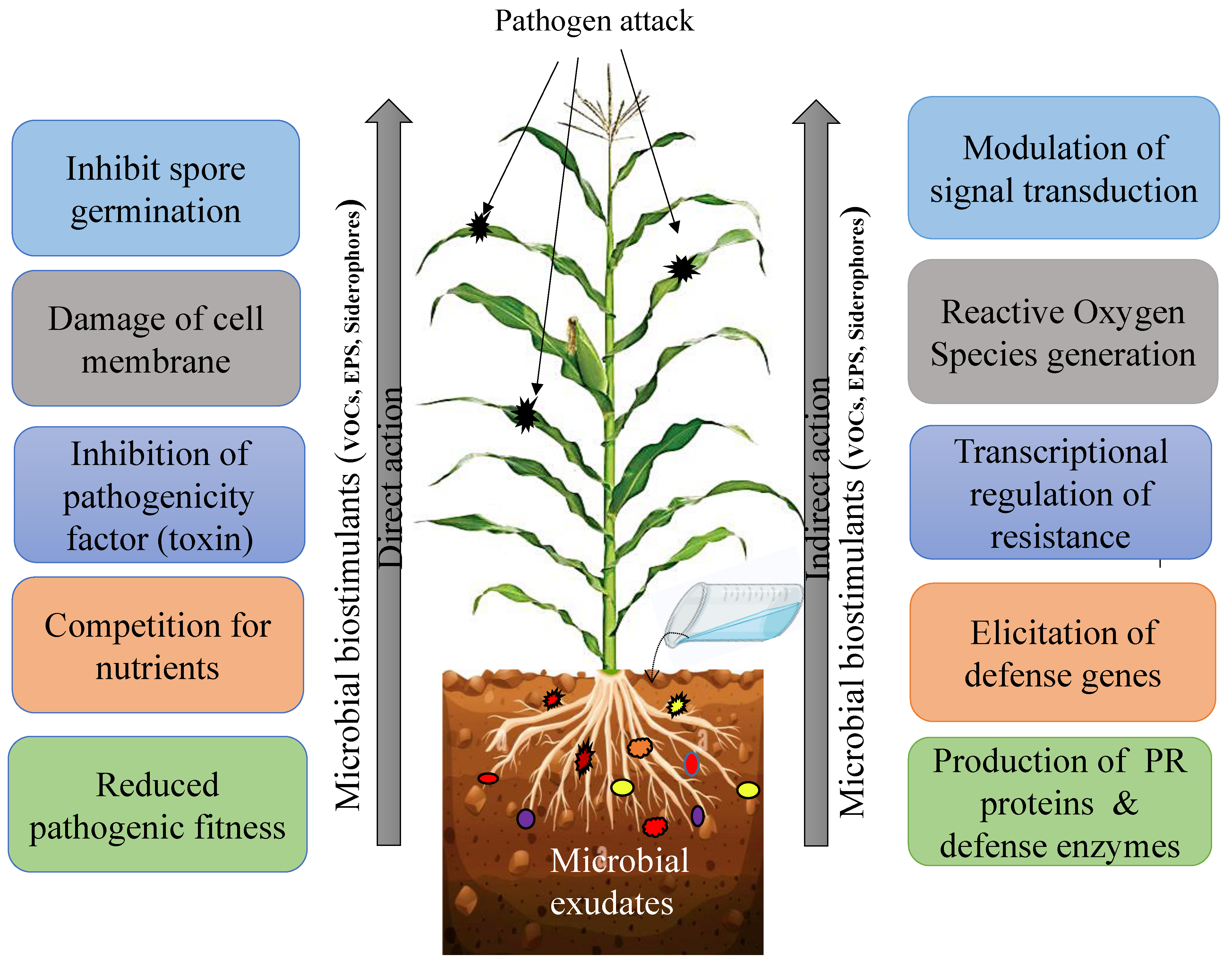
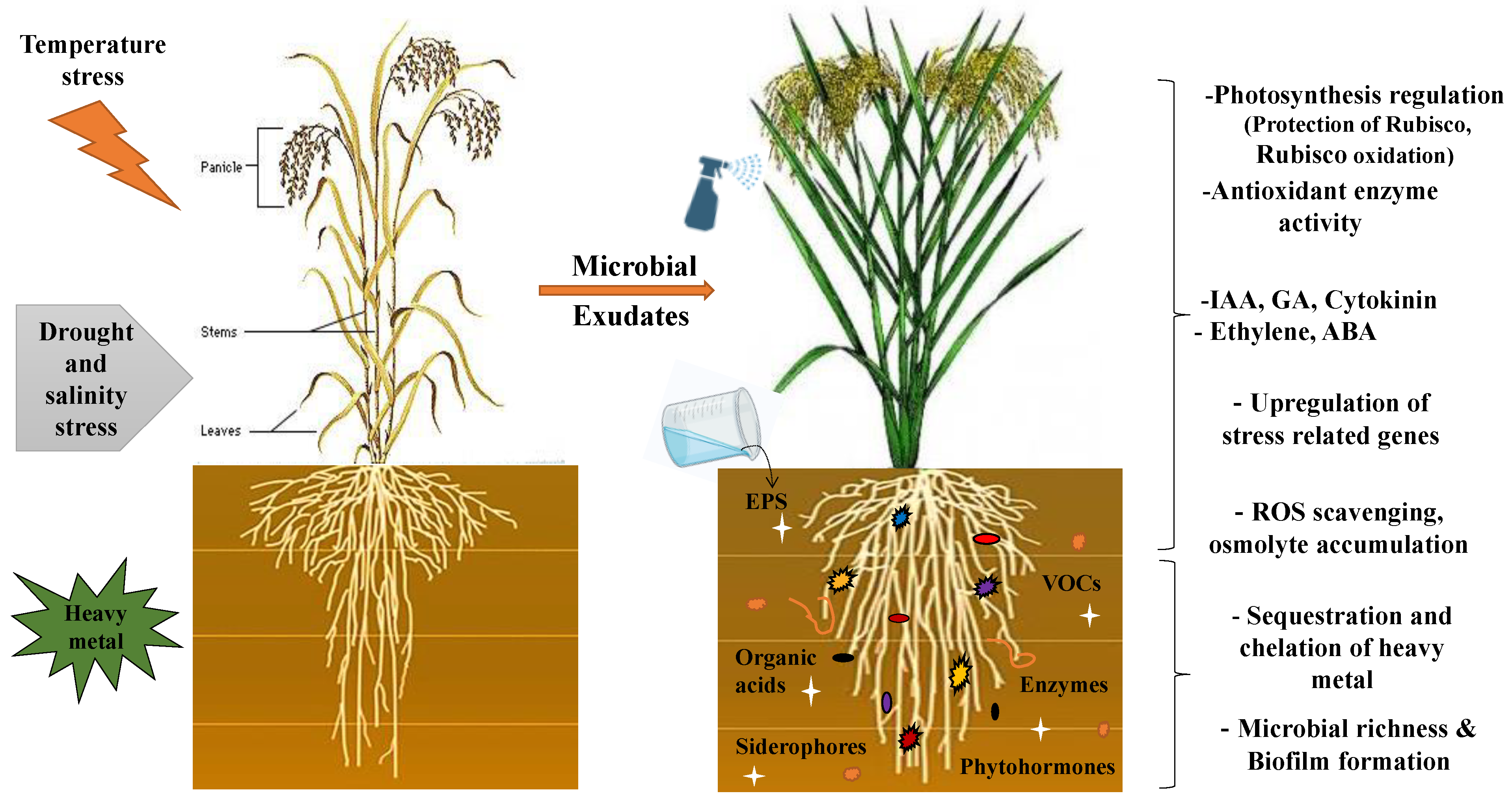
4.2. Microbial Exudates in Alleviating Biotic and Abiotic Stress
4.2.1. Microbial Exudate as Plant Protectants
4.2.2. Alleviation of Abiotic Stress
5. Microbial Exudates as Environmental Protectors
6. Impact of Microbial Exudates on the Plant Microbiome
6.1. Microbial Exudates as Food for Other Microbes
6.2. Microbial Exudates as Signaling Molecules for Other Microbes
6.3. Microbial Exudates Promote Niche Adaptation
7. Limitations and Constraints
8. Conclusions and Prospects
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Abdel Latef, A.A.H.; Abu Alhmad, M.F.; Kordrostami, M.; Abo-Baker, A.-B.A.-E.; Zakir, A. Inoculation with Azospirillumlipoferum or Azotobacterchroococcum reinforces maize growth byimproving physiological activities under saline conditions. J. Plant Growth Regul. 2020, 39, 1293–1306. [Google Scholar] [CrossRef]
- Mayak, S.; Tirosh, T.; Glick, B.R. Plant growth promoting bacteria that confer resistance towater stress in tomato and pepper. Plant Sci. 2004, 166, 525–530. [Google Scholar] [CrossRef]
- Hui, L.J.; Kim, S.D. Induction of drought stress resistance by multi-functional PGPR Bacillus licheniformis K11 in pepper. Plant Pathol. J. 2013, 29, 201–208. [Google Scholar] [CrossRef]
- Zhang, H.; Kim, M.S.; Sun, Y.; Dowd, S.E.; Shi, H.; Paré, P.W. Soil bacteria confer plant salttolerance by tissue-specific regulation of the sodium transporter HKT1. Mol. Plant Microbe Interact. 2008, 21, 737–744. [Google Scholar] [CrossRef] [PubMed]
- Atouei, M.T.; Pourbabaee, A.A.; Shorafa, M. Alleviation of salinity stress on some growthparameters of wheat by exopolysaccharide-producing bacteria. Iran. J. Sci. Technol. Trans. Sci. 2019, 43, 2725–2733. [Google Scholar] [CrossRef]
- Alami, Y.; Achouak, W.; Marol, C.; Heulin, T. Rhizosphere soil aggregation and plant growthpromotion of sunflowers by an exopolysaccharide-producing Rhizobium sp. strain isolated from sunflower roots. Appl. Environ. Microbiol. 2000, 66, 3393–3398. [Google Scholar] [CrossRef]
- Lu, X.; Liu, S.F.; Yue, L.; Zhao, X.; Zhang, Y.B.; Xie, Z.K.; Wang, R.Y. Epsc involved in the encoding of exopolysaccharides produced by Bacillus amyloliquefaciens FZB42 act to boost the drought tolerance of Arabidopsis thaliana. Int. J. Mol. Sci. 2018, 219, 3795. [Google Scholar] [CrossRef]
- Cohen, A.C.; Travaglia, C.N.; Bottini, R.; Piccoli, P.N. Participation of abscisic acid andgibberellins produced by endophytic Azospirillum in the alleviation of drought effects in maize. Botanique 2009, 87, 455–462. [Google Scholar] [CrossRef]
- Liu, W.; Mu, W.; Zhu, B.Y.; Du, Y.C.; Liu, F. Antagonistic activities of volatiles from four strains of Bacillus spp. and Paeni bacillus spp. against soil-borne plant pathogens. Agric. Sci. China 2008, 7, 1104–1114. [Google Scholar] [CrossRef]
- Kang, S.M.; Radhakrishnan, R.; Khan, A.L.; Kim, M.J.; Park, J.M.; Kim, B.R.; Shin, D.H.; Lee, I.J. Gibberellin secreting rhizobacterium, Pseudomonas putida H-2-3 modulates the hormonal andstress physiology of soybean to improve the plant growth under saline and drought conditions. Plant Physiol. Biochem. 2014, 84, 115–124. [Google Scholar] [CrossRef]
- Thakur, M.; Medintz, I.L.; Walper, S.A. Enzymatic bioremediation of organophosphatecompounds—Progress and remaining challenges. Front. Bioeng. Biotechnol. 2019, 7, 289. [Google Scholar] [CrossRef] [PubMed]
- European Commission. The European Parliament and the Council of the European Union Regulation (EU) 2019/1009 of the European Parliament and of the Council of 5 June 2019 laying down rules on the making available on the market of EU fertilising products and amending Regulation (EC) No. 1069/2009 and (EC) No. 1107/2009 and repealing Regulation. Off. J. Eur. Union 2019, 2019, 1–114. Available online: https://eur-lex.europa.eu/legal-content/EN/TXT/PDF/?uri=CELEX:32019R1009&from=EN (accessed on 26 July 2021).
- Castiglione, A.M.; Mannino, G.; Contartese, V.; Bertea, C.M.; Ertani, A. Microbial biostimulants as response to modern agriculture needs: Composition, role and application of these innovative products. Plants 2021, 10, 1533. [Google Scholar] [CrossRef] [PubMed]
- Veliz-Vallejos, D.F.; van Noorden, G.E.; Yuan, M.; Mathesius, U. A SinorhizobiumMeliloti-Specific N-Acyl Homoserine Lactone Quorum-Sensing Signal Increases Nodule Numbers in Medicago Truncatula Independent of Autoregulation. Front. Plant Sci. 2014, 5, 551. [Google Scholar] [CrossRef] [PubMed]
- Banchio, E.; Xie, X.; Zhang, H.; Paré, P.W. Soil Bacteria Elevate Essential Oil Accumulation and Emissions in Sweet Basil. J. Agric. Food Chem. 2009, 57, 653–657. [Google Scholar] [CrossRef]
- Hanif, M.K.; Malik, K.A.; Hameed, S.; Saddique, M.J.; Fatima, K.; Naqqash, T.; Majeed, A.; Iqbal, M.J.; Imran, A. Growth Stimulatory Effect of AHL Producing Serratia spp. from Potato on Homologous and Non-Homologous Host Plants. Microbiol. Res. 2020, 238, 126506. [Google Scholar] [CrossRef]
- Shrestha, A.; Grimm, M.; Ojiro, I.; Krumwiede, J.; Schikora, A. Impact of Quorum Sensing Molecules on Plant Growth and Immune System. Front. Microbiol. 2020, 11, 1545. [Google Scholar] [CrossRef]
- Kumar, M.; Mishra, S.; Dixit, V.; Kumar, M.; Agarwal, L.; Chauhan, P.S.; Nautiyal, C.S. Synergistic Effect of Pseudomonas putida and Bacillus amyloliquefaciens Ameliorates Drought Stress in Chickpea (Cicer arietinum L.). Plant SignalBehav. 2016, 11, e1071004. [Google Scholar] [CrossRef]
- Sultana, S.; Alam, S.; Karim, M.M. Screening of Siderophore-Producing Salt-Tolerant Rhizobacteria Suitable for Supporting Plant Growth in Saline Soils with Iron Limitation. J. Agri. Food Res. 2021, 4, 100150. [Google Scholar] [CrossRef]
- Hofmann, M.; Heine, T.; Malik, L.; Hofmann, S.; Joffroy, K.; Senges, C.H.R.; Bandow, J.E.; Tischler, D. Screening for Microbial Metal-Chelating Siderophores for the Removal of Metal Ions from Solutions. Microorganisms 2021, 9, 111. [Google Scholar] [CrossRef]
- Ahmed, E.; Holmstrom, S.J.M. Siderophores in Environmental Research: Roles and Applications. Microb. Biotechnol. 2014, 7, 196–208. [Google Scholar] [CrossRef] [PubMed]
- Schalk, I.J.; Hannauer, M.; Braud, A. Mini Review New Roles for Bacterial. Environ. Microbiol. 2011, 13, 2844–2854. [Google Scholar] [CrossRef] [PubMed]
- Rajkumar, M.; Ae, N.; Prasad, M.N.V.; Freitas, H. Potential of Siderophore-Producing Bacteria for Improving Heavy Metal Phytoextraction. Trends Biotechnol. 2010, 28, 142–149. [Google Scholar] [CrossRef] [PubMed]
- Ito, T. Enzymatic Determination of Itoic Acid, a Bacillus subtilis Siderophore, and 2, 3-Dihydroxybenzoic Acid. Appl. Environ. Microbiol. 1993, 59, 2343–2345. [Google Scholar] [CrossRef]
- Shah, S.; Karkhanis, V.; Desai, A. Isolation and Characterization of Siderophore, with Antimicrobial Activity, from Azospirillumlipoferum. M. Curr. Microbiol. 1992, 25, 347–351. [Google Scholar] [CrossRef]
- Tindale, A.E.; Mehrotra, M.; Ottem, D.; Page, W.J. Dual Regulation of Catecholate Siderophore Biosynthesis in Azotobactervinelandii by Iron and Oxidative Stress. Microbiology 2000, 146, 1617–1626. [Google Scholar] [CrossRef]
- Eng-Wilmot, D.L.; Van der Helm, D. Molecular and Crystal Structure of the Linear Tricatechol Siderophore, Agrobactin. J. Am. Chem. Soc. 1980, 102, 7719–7725. [Google Scholar] [CrossRef]
- Storey, E.P.; Boghozian, R.; Little, J.L.; Lowman, D.W.; Chakraborty, R. Characterization of ‘Schizokinen’; a Dihydroxamate-Type Siderophore Produced by Rhizobium leguminosarum IARI 917. BioMetals 2006, 19, 637–649. [Google Scholar] [CrossRef]
- Kumari, S.; Kiran, S.; Kumari, S.; Kumar, P.; Singh, A. Optimization of Siderophore Production by Bacillus subtilis DR2 and Its Effect on Growth of Coriandrum Sativum. Russ. Agricult. Sci. 2022, 48, 467–475. [Google Scholar] [CrossRef]
- Igiehon, N.O.; Babalola, O.O.; Aremu, B.R. Genomic Insights into Plant Growth Promoting Rhizobia Capable of Enhancing Soybean Germination under Drought Stress. BMC Microbiol. 2019, 19, 159. [Google Scholar] [CrossRef]
- Ghazy, N.; El-Nahrawy, S. Siderophore Production by Bacillus subtilis MF497446 and Pseudomonas koreensis MG209738 and Their Efficacy in Controlling Cephalosporium maydis in Maize Plant. Arch. Microbiol. 2021, 203, 1195–1209. [Google Scholar] [CrossRef] [PubMed]
- Lynch, D.; O’Brien, J.; Welch, T.; Clarke, P.; Cuív, P.O.; Crosa, J.H.; O’Connell, M. Genetic Organization of the Region Encoding Regulation, Biosynthesis, and Transport of Rhizobactin 1021, a Siderophore Produced by Sinorhizobium meliloti. J.Bacteriol. 2001, 183, 2576–2585. [Google Scholar] [CrossRef]
- Drechsel, H.; Tschierske, M.; Thieken, A.; Jung, G.; Zähner, H.; Winkelmann, G. The Carboxylate Type Siderophore rhizoferrin and Its Analogs Produced by Directed Fermentation. J. Indust. Microbiol. 1995, 14, 105–112. [Google Scholar] [CrossRef]
- Campestre, M.P.; Castagno, L.N.; Estrella, M.J.; Ruiz, O.A. Lotus japonicus plants of the Gifu B-129 ecotype subjected to alkaline stress improve their Fe2+ bio-availability through inoculation with Pantoea eucalypti M91. J. Plant Physiol. 2016, 192, 47–55. [Google Scholar] [CrossRef] [PubMed]
- Mishra, A.; Jha, B. MicrobialExopolysaccharides. InThe Prokaryotes: Applied Bacteriology and Biotechnology, 4th ed.; Rosenberg, E., DeLong, E.F., Thompson, F., Lory, S., Stackebrandt, E., Eds.; Springer: Berlin/Heidelberg, Germany, 2013; pp. 179–192. [Google Scholar]
- Schulz-Bohm, K.; Martín-Sánchez, L.; Garbeva, P. Microbial Volatiles: Small Molecules with an Important Role in Intra- and Inter-Kingdom Interactions. Front. Microbiol. 2017, 8, 2484. [Google Scholar] [CrossRef]
- Margaritis, A.; Pace, G.W. Microbial Polysaccharides. In Comprehensive Biotechnology; Moo-Young, M., Ed.; Pergamon Press: Oxford, UK, 1985; Volume 3, pp. 1005–1044. [Google Scholar]
- Nandal, K.; Sehrawat, A.R.; Yadav, A.S.; Vashishat, R.K.; Boora, K.S. High Temperature-Induced Changes in Exopolysaccharides, Lipopolysaccharides, and Protein Profile of Heat-Resistant Mutants of Rhizobium sp. (Cajanus). Microbiol. Res. 2005, 160, 367–373. [Google Scholar] [CrossRef]
- Upadhyay, S.K.; Singh, J.S.; Singh, D.P. Exopolysaccharide-Producing Plant Growth-Promoting Rhizobacteria under Salinity Condition. Pedosphere 2011, 21, 214–222. [Google Scholar] [CrossRef]
- Pulsawat, W.; Leksawasdi, N.; Rogers, P.L.; Foster, L.J.R. Anions Effects on Biosorption of Mn(II) by Extracellular Polymeric Substance (EPS) from Rhizobium etli. Biotechnol. Lett. 2003, 25, 1267–1270. [Google Scholar] [CrossRef]
- Morcillo, R.J.L.; Manzanera, M. The Effects of Plant-Associated Bacterial Exopolysaccharides on Plant Abiotic Stress Tolerance. Metabolites 2021, 11, 337. [Google Scholar] [CrossRef]
- Carminati, A.; Schneider, C.L.; Moradi, A.B.; Zarebanadkouki, M.; Vetterlein, D.; Vogel, H.J.; Hildebrandt, A.; Weller, U.; Schüler, L.; Oswald, S.E. How the Rhizosphere May Favor Water Availability to Roots. Vadose Zone J. 2011, 10, 988–998. [Google Scholar] [CrossRef]
- Naseem, H.; Bano, A. Role of Plant Growth-Promoting Rhizobacteria and Their Exopolysaccharide in Drought Tolerance of Maize. J. Plant Interact. 2014, 9, 689–701. [Google Scholar] [CrossRef]
- Dar, A.; Zahir, Z.A.; Iqbal, M.; Mehmood, A.; Javed, A.; Hussain, A.; Ahmad, M. Efficacy of Rhizobacterial Exopolysaccharides in Improving Plant Growth, Physiology, and Soil Properties. Environ. Monit. Assess. 2021, 193, 1–15. [Google Scholar] [CrossRef] [PubMed]
- Ishii, S.; Koki, J.; Unno, H.; Hori, K. Two Morphological Types of Cell Appendages on a Strongly Adhesive Bacterium, Acinetobacter sp. Strain Tol 5. Appl. Environ. Microbiol. 2004, 70, 5026–5029. [Google Scholar] [CrossRef]
- Fujishige, N.A.; Kapadia, N.N.; Hirsch, A.M. A Feeling for the Micro-Organism: Structure on a Small Scale. Biofilms on Plant Roots. Bot. J. Linn. Soc. 2006, 150, 79–88. [Google Scholar] [CrossRef]
- Hori, K.; Matsumoto, S. Bacterial Adhesion: From Mechanism to Control. Biochem. Eng. J. 2010, 48, 424–434. [Google Scholar] [CrossRef]
- Fazli, M.; Almblad, H.; Rybtke, M.L.; Givskov, M.; Eberl, L.; Tolker-Nielsen, T. Regulation of Biofilm Formation in Pseudomonas and Burkholderia Species. Environ. Microbiol. 2014, 16, 1961–1981. [Google Scholar] [CrossRef] [PubMed]
- Cheng, C.; Shang-Guan, W.; He, L.; Sheng, X. Effect of Exopolysaccharide-Producing Bacteria on Water-Stable Macro-Aggregate Formation in Soil. Geomicrobiol. J. 2020, 37, 738–745. [Google Scholar] [CrossRef]
- Zheng, W.; Zeng, S.; Bais, H.; LaManna, J.M.; Hussey, D.S.; Jacobson, D.L.; Jin, Y. Plant Growth-Promoting Rhizobacteria (PGPR) Reduce Evaporation and Increase Soil Water Retention. Water Resour. Res. 2018, 54, 3673–3687. [Google Scholar] [CrossRef]
- Benard, P.; Bickel, S.; Kaestner, A.; Lehmann, P.; Carminati, A. Extracellular Polymeric Substances from Soil-Grown Bacteria Delay Evaporative Drying. Adv. Water Resour. 2023, 172, 104364. [Google Scholar] [CrossRef]
- Kudoyarova, G.; Arkhipova, T.; Korshunova, T.; Bakaeva, M.; Loginov, O.; Dodd, I.C. Phytohormone Mediation of Interactions between Plants and Non-Symbiotic Growth-Promoting Bacteria under Edaphic Stresses. Front. Plant Sci. 2019, 10, 1368. [Google Scholar] [CrossRef]
- Kudoyarova, G.R.; Melentiev, A.I.; Martynenko, E.V.; Timergalina, L.N.; Arkhipova, T.N.; Shendel, G.V.; Kuz’mina, L.Y.; Dodd, I.C.; Veselov, S.Y. Cytokinin Producing Bacteria Stimulate Amino Acid Deposition by Wheat Roots. Plant Physiol. Biochem. 2014, 83, 285–291. [Google Scholar] [CrossRef] [PubMed]
- Wheatley, R.; Hackett, C.; Bruce, A.; Kundzewicz, A. Effect of Substrate Composition on Production of Volatile Organic Compounds from Trichoderma spp. Inhibitory to Wood Decay Fungi. Int. Biodeterior. Biodegrad. 1997, 39, 199–205. [Google Scholar] [CrossRef]
- Chiron, N.; Micherlot, D. Odeurs des Champignons: Chimie et Rôle dans les Interactions Biotiques-une Revue. Cryptogam.-Mycol. 2005, 26, 299–364. [Google Scholar]
- Morath, S.U.; Hung, R.; Bennett, J.W. Fungal Volatile Organic Compounds: A Review with Emphasis on Their Biotechnological Potential. Fungal Biol. Rev. 2012, 26, 73–83. [Google Scholar] [CrossRef]
- Schulz, S.; Dickschat, J.S. Bacterial Volatiles: The Smell of Small Organisms. Nat. Prod. Rep. 2007, 24, 814–842. [Google Scholar] [CrossRef]
- Blom, D.; Fabbri, C.; Connor, E.C.; Schiestl, F.P.; Klauser, D.R.; Boller, T.; Eberl, L.; Weisskopf, L. Production of Plant Growth Modulating Volatiles Is Widespread among Rhizosphere Bacteria and Strongly Depends on Culture Conditions. Environ. Microbiol. 2011, 13, 3047–3058. [Google Scholar] [CrossRef]
- Peñuelas, J.; Asensio, D.; Tholl, D.; Wenke, K.; Rosenkranz, M.; Piechulla, B.; Schnitzler, J. Biogenic Volatile Emissions from the Soil. Plant Cell Environ. 2014, 37, 1866–1891. [Google Scholar] [CrossRef]
- Schenkel, D.; Lemfack, M.C.; Piechulla, B.; Splivallo, R. A Metaanalysis Approach for Assessing the Diversity and Specificity of Belowground Root and Microbial Volatiles. Front. Plant Sci. 2015, 6, 707. [Google Scholar] [CrossRef]
- Bennett, J.W.; Hung, R.; Lee, S.; Padhi, S. Fungal and Bacterial Volatile Organic Compounds: An Overview and Their Role as Ecological Signaling Agents. In Fungal Associations; Hock, B., Ed.; The Mycota; Springer: Berlin/Heidelberg, Germany, 2012; Volume 9, pp. 373–393. [Google Scholar] [CrossRef]
- Ryu, C.M.; Farag, M.A.; Hu, C.H.; Reddy, M.S.; Wei, H.X.; Paré, P.W.; Kloepper, J.W. Bacterial Volatiles Promote Growth in Arabidopsis. Proc. Natl. Acad. Sci. USA 2003, 100, 4927–4932. [Google Scholar] [CrossRef]
- Zhao, X.; Zhou, J.; Tian, R.; Liu, Y. Microbial Volatile Organic Compounds: Antifungal Mechanisms, Applications, and Challenges. Front. Microbiol. 2022, 13, 922450. [Google Scholar] [CrossRef]
- Li, H.; Bölscher, T.; Winnick, M.; Tfaily, M.M.; Cardon, Z.G.; Keiluweit, M. Simple Plant and Microbial Exudates Destabilize Mineral-Associated Organic Matter via Multiple Pathways. Environ. Sci. Technol. 2021, 55, 3389–3398. [Google Scholar] [CrossRef] [PubMed]
- Chari, N.R.; Taylor, B.N. Soil Organic Matter Formation and Loss Are Mediated by Root Exudates in a Temperate Forest. Nat. Geosci. 2022, 15, 1011–1016. [Google Scholar] [CrossRef]
- Nwachukwu, B.C.; Ayangbenro, A.S.; Babalola, O.O. Elucidating the Rhizosphere Associated Bacteria for Environmental Sustainability. Agriculture 2021, 11, 75. [Google Scholar] [CrossRef]
- Jilling, A.; Keiluweit, M.; Contosta, A.R.; Frey, S.; Schimel, J.; Schnecker, J.; Smith, R.G.; Tiemann, L.; Grandy, A.S. Minerals in the Rhizosphere: Overlooked Mediators of Soil Nitrogen Availability to Plants and Microbes. Biogeochemistry 2018, 139, 103–122. [Google Scholar] [CrossRef]
- Walton, C.L.; Khalid, M.; Bible, A.N.; Kertesz, V.; Retterer, S.T.; Morrell-Falvey, J.; Cahill, J.F. In Situ Detection of Amino Acids from Bacterial Biofilms and Plant Root Exudates by Liquid Microjunction Surface-Sampling Probe Mass Spectrometry. J. Am. Soc. Mass. Spectrom. 2022, 33, 1615–1625. [Google Scholar] [CrossRef]
- Wang, Y.; Luo, D.; Xiong, Z.; Wang, Z.; Gao, M. Changes in rhizosphere phosphorus fractions and phosphate-mineralizing microbial populations in acid soil as influenced by organic acid exudation. Soil. Tillage Res. 2023, 225, 105543. [Google Scholar] [CrossRef]
- Romera, F.J.; García, M.J.; Lucena, C.; Martínez-Medina, A.; Aparicio, M.A.; Ramos, J.; Alcántara, E.; Angulo, M.; Pérez-Vicente, R. Induced systemic resistance (ISR) and Fe deficiency responses in dicot plants. Front. Plant Sci. 2019, 10, 287. [Google Scholar] [CrossRef]
- Zhang, J.L.; Tang, W.L.; Huang, Q.R.; Li, Y.Z.; Wei, M.L.; Jiang, L.L.; Liu, C.; Yu, X.; Zhu, H.W.; Chen, G.Z.; et al. Trichoderma: A treasure house of structurally diverse secondary metabolites with medicinal importance. Front. Microb. 2021, 12, 723828. [Google Scholar] [CrossRef]
- Ray, S.; Singh, P.; Singh, J.; Singh, S.; Sarma, B.K.; Singh, H.B. Killed fungal pathogen triggers antifungal metabolites in Alcaligenes faecalis for plant defense. Physiol. Mol. Plant Pathol. 2023, 125, 101996. [Google Scholar] [CrossRef]
- Tamandegani, P.R.; Marik, T.; Zafari, D.; Balázs, D.; Vágvölgyi, C.; Szekeres, A.; Kredics, L. Changes in Peptaibol Production of Trichoderma Species during In Vitro Antagonistic Interactions with Fungal Plant Pathogens. Biomolecules 2020, 10, 730. [Google Scholar] [CrossRef]
- Shamikh, Y.I.; El Shamy, A.A.; Gaber, Y.; Abdelmohsen, U.R.; Madkour, H.A.; Horn, H.; Hassan, H.M.; Elmaidomy, A.H.; Alkhalifah, D.H.M.; Hozzein, W.N. Actinomycetes from the Red Sea sponge Coscinodermamathewsi: Isolation, diversity, and potential for bioactive compounds discovery. Microorganisms 2020, 8, 783. [Google Scholar] [CrossRef]
- Ugena, L.; Hýlová, A.; Podlešáková, K.; Humplík, J.F.; Doležal, K.; De Diego, N.; Spíchal, L. Characterization of biostimulant mode of action using novel multi-trait high-throughput screening of Arabidopsis germination and rosette growth. Front. Plant Sci. 2018, 9, 1327. [Google Scholar] [CrossRef]
- Ogunsanya, H.Y.; Motti, P.; Li, J.; Trinh, H.K.; Xu, L.; Bernaert, N.; Van Droogenbroeck, B.; Murvanidze, N.; Werbrouck, S.P.; Mangelinckx, S.; et al. Belgian endive-derived biostimulants promote shoot and root growth in vitro. Sci. Rep. 2022, 12, 8792. [Google Scholar] [CrossRef]
- Saporta, R.; Bou, C.; Frías, V.; Mulet, J.M. A method for a fast evaluation of the biostimulant potential of different natural extracts for promoting growth or tolerance against abiotic stress. Agronomy 2019, 9, 143. [Google Scholar] [CrossRef]
- Jiménez-Arias, D.; Morales-Sierra, S.; Borges, A.A.; Herrera, A.J.; Luis, J.C. New biostimulants screening method for crop seedlings under water deficit stress. Agronomy 2022, 12, 728. [Google Scholar] [CrossRef]
- Hines, S.; van der Zwan, T.; Shiell, K.; Shotton, K.; Prithiviraj, B. Alkaline extract of the seaweed Ascophyllum nodosum stimulates arbuscular mycorrhizal fungi and their endomycorrhization of plant roots. Sci. Rep. 2021, 11, 13491. [Google Scholar] [CrossRef] [PubMed]
- Meher, J.; Lenka, S.; Sarkar, A.; Sarma, B.K. Transcriptional regulation of OsWRKY genes in response to individual and overlapped challenges of Magnaportheoryzae and drought in indica genotypes of rice. Environ. Exp. Bot. 2023, 207, 105221. [Google Scholar] [CrossRef]
- Kumar, S.; Mohapatra, T. Dynamics of DNA Methylation and Its Functions in Plant Growth and Development. Front. Plant Sci. 2021, 12, 596236. [Google Scholar] [CrossRef] [PubMed]
- Quievreux, M.; Falesse, W.; Lengrand, S.; Delaplace, P.; Dieryck, B.; Dumont de Chassart, S.; Dumont de Chassart, T.; Legrève, A.; Jardin, P. Testing biostimulants for validating the claims: A multi-level analysis. In Proceedings of the 5th Biostimulants World Congress, Miami, FL, USA, 29 November–2 December 2021; Available online: https://hdl.handle.net/2268/266111 (accessed on 7 April 2022).
- De Diego, N.; Spíchal, L. Presence and future of plant phenotyping approaches in biostimulant research and development. J. Exp. Bot. 2022, 73, 5199–5212. [Google Scholar] [CrossRef]
- Khiralla, A.; Spina, R.; Varbanov, M.; Philippot, S.; Lemiere, P.; Slezack-Deschaumes, S.; André, P.; Mohamed, I.; Yagi, S.M.; Laurain-Mattar, D. Evaluation of antiviral, antibacterial and antiproliferative activities of the endophytic fungus Curvulariapapendorfii, and isolation of a new polyhydroxyacid. Microorganisms 2020, 8, 1353. [Google Scholar] [CrossRef]
- Papaianni, M.; Ricciardelli, A.; Fulgione, A.; d’Errico, G.; Zoina, A.; Lorito, M.; Woo, S.L.; Vinale, F.; Capparelli, R. Antibiofilm Activity of a Trichoderma Metabolite against Xanthomonas campestris pv. campestris, Alone and in Association with a Phage. Microorganisms 2020, 8, 620. [Google Scholar] [CrossRef] [PubMed]
- Voitsekhovskaia, I.; Paulus, C.; Dahlem, C.; Rebets, Y.; Nadmid, S.; Zapp, J.; Axenov-Gribanov, D.; Rückert, C.; Timofeyev, M.; Kalinowski, J.; et al. Baikalomycins AC, New Aquayamycin-type angucyclines isolated from Lake Baikal derived Streptomyces sp. IB201691-2A. Microorganisms 2020, 8, 680. [Google Scholar] [CrossRef] [PubMed]
- Hamed, A.A.; Soldatou, S.; Qader, M.M.; Arjunan, S.; Miranda, K.J.; Casolari, F.; Pavesi, C.; Diyaolu, O.A.; Thissera, B.; Eshelli, M.; et al. Screening fungal endophytes derived from under-explored Egyptian marine habitats for antimicrobial and antioxidant properties in factionalised textiles. Microorganisms 2020, 8, 1617. [Google Scholar] [CrossRef] [PubMed]
- Hwang, S.; Le, L.T.H.L.; Jo, S.I.; Shin, J.; Lee, M.J.; Oh, D.C. Pentaminomycins C–E: Cyclic pentapeptides as autophagy inducers from a mealworm beetle gut bacterium. Microorganisms 2020, 8, 1390. [Google Scholar] [CrossRef] [PubMed]
- Liu, L.; Hu, Z.; Li, S.; Yang, H.; Li, S.; Lv, C.; Zaynab, M.; Cheng, C.H.; Chen, H.; Yang, X. Comparative transcriptomic analysis uncovers genes responsible for the DHA enhancement in the mutant Aurantiochytrium sp. Microorganisms 2020, 8, 529. [Google Scholar] [CrossRef] [PubMed]
- Wolff, P.B. Genomics-Driven Discovery, Characterization, and Engineering of Fungal Secondary Metabolites. Ph.D. Thesis, Technical University of Denmark, Kongens Lyngby, Denmark, 2020; p. 220. [Google Scholar]
- Ding, Z.; Wang, X.; Kong, F.D.; Huang, H.M.; Zhao, Y.N.; Liu, M.; Wang, Z.P.; Han, J. Overexpression of global regulator talae1 leads to the discovery of new antifungal polyketides from endophytic fungus Trichoderma afroharzianum. Front. Microbiol. 2020, 11, 622785. [Google Scholar] [CrossRef] [PubMed]
- Staropoli, A.; Iacomino, G.; De Cicco, P.; Woo, S.L.; Di Costanzo, L.; Vinale, F. Induced secondary metabolites of the beneficial fungus Trichoderma harzianum M10 through OSMAC approach. Chem. Biol. Technol. Agric. 2023, 10, 1–11. [Google Scholar] [CrossRef]
- Hifnawy, S.; Hassan, M.; Mohammed, H.M.; Fouda, R.M.; Sayed, M.; Hamed, A.M.A.; AbouZid, A.F.; Rateb, S.; Alhadrami, M.E.; Abdelmohsen, U.R. Induction of antibacterial metabolites by co-cultivation of two red-sea-sponge-associated actinomycetes Micromonospora sp. UR56 and Actinokinespora sp. EG49. Mar. Drugs 2020, 18, 243. [Google Scholar] [CrossRef]
- Liang, L.; Wang, G.; Haltli, B.; Marchbank, D.H.; Stryhn, H.; Correa, H.; Kerr, R.G. Metabolomic comparison and assessment of co-cultivation and a heat-killed inducer strategy in activation of cryptic biosynthetic pathways. J. Nat. Prod. 2020, 83, 2696–2705. [Google Scholar] [CrossRef]
- Jomori, T.; Hara, Y.; Sasaoka, M.; Harada, K.; Setiawan, A.; Hirata, K.; Kimishima, A.; Arai, M. Mycobacterium smegmatis alters the production of secondary metabolites by marine-derived Aspergillus niger. J. Nat. Med. 2020, 74, 76–82. [Google Scholar] [CrossRef]
- Lombardi, N.; Salzano, A.M.; Troise, A.D.; Scaloni, A.; Vitaglione, P.; Vinale, F.; Marra, R.; Caira, S.; Lorito, M.; d’Errico, G.; et al. Effect of Trichoderma bioactive metabolite treatments on the production, quality, and protein profile of strawberry fruits. J. Agric. Food Chem. 2020, 68, 7246–7258. [Google Scholar] [CrossRef] [PubMed]
- Shi, Z.Z.; Liu, X.H.; Li, X.N.; Ji, N.Y. Antifungal and antimicroalgal trichothecene sesquiterpenes from the marine algicolous fungus Trichoderma brevicompactum A-DL-9-2. J. Agric. Food Chem. 2020, 68, 15440–15448. [Google Scholar] [CrossRef] [PubMed]
- Yamazaki, H.; Takahashi, O.; Kirikoshi, R.; Yagi, A.; Ogasawara, T.; Bunya, Y.; Rotinsulu, H.; Uchida, R.; Namikoshi, M. Epipolythiodiketopiperazine and trichothecene derivatives from the NaI-containing fermentation of marine-derived Trichoderma cf. brevicompactum. J. Antibiot. 2020, 73, 559–567. [Google Scholar] [CrossRef] [PubMed]
- Du, F.Y.; Ju, G.L.; Xiao, L.; Zhou, Y.M.; Wu, X. Sesquiterpenes and cyclodepsipeptides from marine-derived fungus Trichoderma longibrachiatum and their antagonistic activities against soil-borne pathogens. Mar. Drugs 2020, 18, 165. [Google Scholar] [CrossRef]
- Li, W.Y.; Liu, Y.; Lin, Y.T.; Liu, Y.C.; Guo, K.; Li, X.N.; Luo, S.H.; Li, S.H. Antibacterial harziane diterpenoids from a fungal symbiont Trichoderma atroviride isolated from Colquhounia coccinea var. mollis. Phytochemistry 2020, 170, 112198. [Google Scholar] [CrossRef]
- Miyano, R.; Matsuo, H.; Mokudai, T.; Noguchi, Y.; Higo, M.; Nonaka, K.; Niwano, Y.; Sunazuka, T.; Shiomi, K.; Takahashi, Y.; et al. Trichothioneic acid, a new antioxidant compound produced by the fungal strain Trichoderma virens FKI-7573. J. Biosci. Bioeng. 2020, 129, 508–513. [Google Scholar] [CrossRef]
- Zhang, S.; Sun, F.; Liu, L.; Bao, L.; Fang, W.; Yin, C.; Zhang, Y. Dragonfly associated Trichoderma harzianum QTYC77 is not only a potential biological control agent of Fusarium oxysporum f. sp. cucumerinum but also a source of new antibacterial agents. J. Agric. Food Chem. 2020, 68, 14161–14167. [Google Scholar] [CrossRef]
- Zhao, D.L.; Zhang, X.F.; Huang, R.H.; Wang, D.; Wang, X.Q.; Li, Y.Q.; Zheng, C.J.; Zhang, P.; Zhang, C.S. Antifungal nafuredin and epithiodiketopiperazine derivatives from the mangrove-derived fungus Trichoderma harzianum D13. Front. Microbiol. 2020, 11, 1495. [Google Scholar] [CrossRef]
- Duzan, H.M.; Mabood, F.; Zhou, X.; Souleimanov, A.; Smith, D.L. Nod factor induces soybean resistance to powdery mildew. Plant Physiol. Biochem. 2005, 43, 1022–1030. [Google Scholar] [CrossRef]
- Choi, H.K.; Song, G.C.; Yi, H.S.; Ryu, C.M. Field evaluation of the bacterial volatile derivative 3-pentanol in priming for induced resistance in pepper. J. Chem. Ecol. 2014, 40, 882–892. [Google Scholar] [CrossRef]
- Prudent, M.; Salon, C.; Smith, D.L.; Emery, R.J.N. Nod factor supply under water stress conditions modulates cytokinin biosynthesis and enhances nodule formation and N nutrition in soybean. Plant Signal. Behav. 2016, 11, e1212799. [Google Scholar] [CrossRef] [PubMed]
- Piechulla, B.; Lemfack, M.C.; Kai, M. Effects of discrete bioactive microbial volatiles on plants and fungi. Plant Cell Environ. 2017, 40, 2042–2067. [Google Scholar] [CrossRef] [PubMed]
- Zhou, D.; Huang, X.F.; Chaparro, J.M.; Badri, D.V.; Manter, D.K.; Vivanco, J.M.; Guo, J. Root and bacterial secretions regulate the interaction between plants and PGPR leading to distinct plant growth promotion effects. Plant Soil. 2016, 401, 259–272. [Google Scholar] [CrossRef]
- Zhao, Q.; Yang, X.Y.; Li, Y.; Liu, F.; Cao, X.Y.; Jia, Z.H.; Song, S.S. N-3-oxo-hexanoyl-homoserine lactone, a bacterial quorum sensing signal, enhances salt tolerance in Arabidopsis and wheat. Bot. Stud. 2020, 61, 8. [Google Scholar] [CrossRef] [PubMed]
- Shrestha, A.; Elhady, A.; Adss, S.; Wehner, G.; Böttcher, C.; Heuer, H.; Ordon, F.; Schikora, A. Genetic differences in barley govern the responsiveness to N-Acyl homoserine lactone. Phytobiomes J. 2019, 3, 191–202. [Google Scholar] [CrossRef]
- Sayyed, R.Z.; Reddy, M.S. Bacterial Plant Biostimulants: A sustainable way toward improving growth, productivity and health of crops. Sustainability 2021, 13, 2856. [Google Scholar] [CrossRef]
- Kumar, D.H.; Aloke, P. Role of biostimulants in crop production: An overview. Int. J. Agric. Sci. Vet. Med. 2020, 8, 2. [Google Scholar] [CrossRef]
- Mine, A.; Sato, M.; Tsuda, K. Toward a systems understanding of plant–microbe interactions. Front. Plant Sci. 2014, 5, 42. [Google Scholar] [CrossRef]
- Ruzzi, M.; Aroca, R. Plant growth-promoting rhizobacteria act as biostimulants in horticulture. Sci. Hortic. 2015, 196, 124–134. [Google Scholar] [CrossRef]
- Paradikovic, N.; Teklic, T.; Zeljkovic, S.; Lisjak, M.; Spoljarevic, M. Biostimulants research in some horticultural plant species—A review. Food Energy Secur. 2019, 8, e00162. [Google Scholar] [CrossRef]
- Kour, D.; Rana, K.L.; Yadav, A.N.; Yadav, N.; Kumar, M.; Kumar, V.; Vyas, P.; Dhaliwal, H.S.; Saxena, A.K. Microbial biofertilizers: Bioresources and eco-friendly technologies for agricultural and environmental sustainability. Biocatal. Agric. Biotechnol. 2020, 23, 101487. [Google Scholar] [CrossRef]
- Santos-Torres, M.; Romero-Perdomo, F.; Mendoza-Labrador, J.; Gutiérrez, A.Y.; Vargas, C.; Castro-Rincon, E.; Caro-Quintero, A.; Uribe-Velez, D.; Estrada-Bonilla, G.A. Genomic and phenotypic analysis of rock phosphate-solubilizing rhizobacteria. Rhizosphere 2021, 17, 100290. [Google Scholar] [CrossRef]
- Hii, Y.S.; Yen San, C.; Lau, S.W.; Danquah, M.K. Isolation and characterisation of phosphate solubilizing microorganisms from peat. Biocatal. Agric. Biotechnol. 2020, 26, 101643. [Google Scholar] [CrossRef]
- Santos, M.S.; Nogueira, M.A.; Hungria, M. Outstanding impact of Azospirillumbrasilense strains ab-v5 and ab-v6 on Brazilian agriculture: Lessons that farmers are receptive to adopt new microbial inoculants. Rev. Bras. Cienc. Solo. 2021, 45. [Google Scholar] [CrossRef]
- Radhakrishnan, R.; Hashem, A.; Abd Allah, E.F. Bacillus: A biological tool for crop improvement through bio-molecular changes in adverse environments. Front. Physiol. 2017, 8, 667. [Google Scholar] [CrossRef] [PubMed]
- Kang, S.M.; Khan, A.L.; Waqas, M.; You, Y.H.; Kim, J.H.; Kim, J.G.; Hamayun, M.; Lee, I.J. Plant growth-promoting rhizobacteria reduce adverse effects of salinity and osmotic stress by regulating phytohormones and antioxidants in Cucumis sativus. J. Plant Interact. 2014, 9, 673–682. [Google Scholar] [CrossRef]
- Okada, K.; Abe, H.; Arimura, G.I. Jasmonates induce both defense responses and communication in monocotyledonous and dicotyledonous plants. Plant Cell Physiol. 2015, 56, 16–27. [Google Scholar] [CrossRef]
- Rijavec, T.; Lapanje, A. Hydrogen cyanide in the rhizosphere: Not suppressing plant pathogens, but rather regulating availability of phosphate. Front. Microbiol. 2016, 7, 1785. [Google Scholar] [CrossRef]
- Llorente, B.E.; Alasia, M.A.; Larraburu, E.E. Biofertilization with Azospirillumbrasilense improves in vitro culture of Handroanthusochraceus, a forestry, ornamental and medicinal plant. New Biotechnol. 2016, 33, 32–40. [Google Scholar] [CrossRef]
- Lim, J.H.; Kim, S.D. Synergistic plant growth promotion by the indigenous auxins producing PGPR Bacillus subtilis AH18 and Bacillus licheniformis K11. J. Korean Soc. Appl. Biol. Chem. 2009, 52, 531–538. [Google Scholar] [CrossRef]
- Ortiz-Castro, R.; Campos-Garcıa, J.; Lopez-Bucio, J. Pseudomonas putida and Pseudomonas fluorescens influence Arabidopsis root system architecture through an auxin response mediated by bioactive cyclodipeptides. J. Plant Growth Regul. 2020, 39, 254–265. [Google Scholar] [CrossRef]
- Yadav, S.; Kaushik, R.; Saxena, A.K.; Arora, D.K. Diversity and phylogeny of plant growth-promoting bacilli from moderately acidic soil. J. Basic. Microbiol. 2011, 51, 98–106. [Google Scholar] [CrossRef] [PubMed]
- Hellequin, E.; Monard, C.; Chorin, M.; Le Bris, N.; Daburon, V.; Klarzynski, O.; Binet, F. Responses of active soil microorganisms facing a soil biostimulant input compared to plant legacy effects. Sci. Rep. 2020, 10, 13727. [Google Scholar] [CrossRef]
- Jha, Y.; Subramanian, R.B. PGPR regulate the caspase-like activity, programmed cell death, and antioxidant enzyme activity in paddy under salinity. Physiol. Mol. Biol. Plants 2014, 20, 201–207. [Google Scholar] [CrossRef] [PubMed]
- El-Ghany, M.F.A.; Attia, M. Effect of exopolysaccharide-producing bacteria and melatonin on faba bean production in saline and non-saline soil. Agronomy 2020, 10, 316. [Google Scholar] [CrossRef]
- Zainab, N.; Din, B.U.; Javed, M.T.; Afridi, M.S.; Mukhtar, T.; Kamran, M.A.; Khan, A.A.; Ali, J.; Jatoi, W.N.; Munis, M.F.H.; et al. Deciphering metal toxicity responses of flax (Linumusitatissimum L.) with exopolysaccharide and ACC-deaminase producing bacteria in industrially contaminated soils. Plant Physiol. Biochem. 2020, 152, 90–99. [Google Scholar] [CrossRef] [PubMed]
- Kour, D.; Rana, K.L.; Yadav, A.N.; Sheikh, I.; Kumar, V.; Dhaliwal, H.S.; Saxena, A.K. Amelioration of drought stress in Foxtail millet (Setariaitalica L.) by P-solubilizing drought-tolerant microbes with multifarious plant growth promoting attributes. Environ. Sustain. 2020, 3, 23–34. [Google Scholar] [CrossRef]
- Manganiello, G.; Sacco, A.; Ercolano, M.R.; Vinale, F.; Lanzuise, S.; Pascale, A.; Napolitano, M.; Lombardi, N.; Lorito, M.; Woo, S.L. Modulation of tomato response to Rhizoctonia solani by Trichoderma harzianum and its secondary metabolite harzianic acid. Front. Microbiol. 2018, 9, 1966. [Google Scholar] [CrossRef]
- Gu, Q.; Yang, Y.; Yuan, Q.; Shi, G.; Wu, L.; Lou, Z.; Huo, R.; Wu, H.; Borriss, R.; Gao, X. Bacillomycin D produced by Bacillus amyloliquefaciens is involved in the antagonistic interaction with the plant-pathogenic fungus Fusarium graminearum. Appl. Environ. Microbiol. 2017, 83, e01075-17. [Google Scholar] [CrossRef]
- Don, S.M.Y.; Schmidtke, L.M.; Gambetta, J.M.; Steel, C.C.J.R.I.M. Volatile organic compounds produced by Aureobasidium pullulans induce electrolyte loss and oxidative stress in Botrytis cinerea and Alternaria alternata. Res. Microbiol. 2020, 172, 103788. [Google Scholar] [CrossRef]
- Meena, M.; Zehra, A. Tomato: A model plant to study plant-pathogen interactions. Food Sci. Nutr. Technol. 2019, 4, 000171. [Google Scholar] [CrossRef]
- Khaing, A.; Theint, W.T.; Thi, O.K.; Fu, P. Antagonistic activity of indigenous rhizobacteria through biosynthesis of indole-3-acetic acid (IAA), hydrogen cyanide (HCN), and siderophores. Aust. J. Biotechnol. Bioeng. 2021, 8, 1110. [Google Scholar] [CrossRef]
- Rachidi, F.; Benhima, R.; Sbabou, L.; El Arroussi, H. Microalgae polysaccharides bio-stimulating effect on tomato plants: Growth and metabolic distribution. Biotechnol. Rep. 2020, 25, e00426. [Google Scholar] [CrossRef] [PubMed]
- Jantaro, S.; Kanwal, S. Low-Molecular-Weight Nitrogenous Compounds (GABA and Polyamines) in Blue-Green Algae. In Algal Green Chemistry; Rastogi, R.P., Madamwar, D., Eds.; Elsevier: Gujarat, India, 2017; pp. 149–169. [Google Scholar]
- Sharma, C.K.; Vishnoi, V.K.; Dubey, R.C.; Maheshwari, D.K. A twin rhizospheric bacterial consortium induces systemic resistance to a phytopathogen Macrophominaphaseolina in mung bean. Rhizosphere 2018, 5, 71–75. [Google Scholar] [CrossRef]
- Ling, L.; Zhao, Y.; Tu, Y.; Yang, C.; Ma, W.; Feng, S.; Lu, L.; Zhang, J. The inhibitory effect of volatile organic compounds produced by Bacillus subtilis CL2 on pathogenic fungi of wolfberry. J. Basic. Microbiol. 2021, 61, 110–121. [Google Scholar] [CrossRef] [PubMed]
- Großkinsky, D.K.; Tafner, R.; Moreno, M.V.; Stenglein, S.A.; García de Salamone, I.E.; Nelson, L.M.; Novák, O.; Strnad, M.; Van der Graaff, E.; Roitsch, T. Cytokinin production by Pseudomonas fluorescens G20–18 determines biocontrol activity against Pseudomonas syringae in Arabidopsis. Sci. Rep. 2016, 6, 23310. [Google Scholar] [CrossRef] [PubMed]
- Akhtar, S.S.; Mekureyaw, M.F.; Pandey, C.; Roitsch, T. Role of cytokinins for interactions of plants with microbial pathogens and pest insects. Front. Plant Sci. 2020, 10, 1777. [Google Scholar] [CrossRef]
- Lurthy, T.; Cantat, C.; Jeudy, C.; Declerck, P.; Gallardo, K.; Barraud, C.; Leroy, F.; Ourry, A.; Lemanceau, P.; Salon, C.; et al. Impact of bacterial siderophores on iron status and ionome in pea. Front. Plant Sci. 2020, 11, 730. [Google Scholar] [CrossRef]
- Silpa, D.; Brahmaji, R.P.; Kranthi, K.G. Biocontrol activity of Siderophores producing Bacillus licheniformis DS3 against several pathogenic fungi in Black gram [Vigna mungo (L.) hepper]. Int. J. Cur. Res. 2018, 10, 71590–71594. [Google Scholar] [CrossRef]
- Gunupuru, L.R.; Patel, J.S.; Sumarah, M.W.; Renaud, J.B.; Mantin, E.G.; Prithiviraj, B. A plant biostimulant made from the marine brown algae Ascophyllum nodosum and chitosan reduce Fusarium head blight and mycotoxin contamination in wheat. PLoS ONE 2019, 14, 0220562. [Google Scholar] [CrossRef]
- Mohammed, A.F. Effectiveness of exopolysaccharides and biofilm forming plant growth promoting rhizobacteria on salinity tolerance of faba bean (Viciafaba L.). Afr. J. Microbiol. Res. 2018, 12, 399–404. [Google Scholar] [CrossRef]
- Sivapriya, S.L.; Priya, P.R. Selection of Hyper Exopolysaccharide Producing and Cyst Forming Azotobacter Isolates for Better Survival under Stress Conditions. Int. J. Curr. Microbiol. App. Sci. 2017, 6, 2310–2320. [Google Scholar] [CrossRef]
- Ghosh, D.; Gupta, A.; Mohapatra, S. A comparative analysis of exopolysaccharide and phytohormone secretions by four drought-tolerant rhizobacterial strains and their impact on osmotic-stress mitigation in Arabidopsis thaliana. World J. Microbiol. Biotechnol. 2019, 35, 90. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.A.; Asaf, S.; Khan, A.L.; Ullah, I.; Ali, S.; Kang, S.M.; Lee, I.J. Alleviation of salt stress response in soybean plants with the endophytic bacterial isolate Curtobacterium sp. SAK1. Ann. Microbiol. 2019, 69, 797–800. [Google Scholar] [CrossRef]
- Rangseekaew, P.; Barros-Rodriguez, A.; Pathom-aree, W.; Manzanera, M. Deep-Sea actinobacteria mitigate salinity stress in tomato seedlings and their biosafety testing. Plants 2021, 10, 1687. [Google Scholar] [CrossRef]
- Sathya, A.; Vijayabharathi, R.; Gopalakrishnan, S. Plant growth-promoting actinobacteria: A new strategy for enhancing sustainable production and protection of grain legumes. 3Biotech 2017, 7, 102. [Google Scholar] [CrossRef]
- Silaochkina, O.; Garshina, D.; Pusenkova, L. Effect of endophytic Bacillus subtilis on drought stress tolerance of Triticum aestivum plants of steppe volga and forest-steppe west siberian agroecological groups. In Proceedings of the 2nd International Conference Plants and Microbes: The Future of Biotechnology, Saratov, Russia, 5–9 September 2020. [Google Scholar] [CrossRef]
- Silambarasan, S.; Logeswari, P.; Valentine, A.; Cornejo, P.; Kannan, V.R. Pseudomonas citronellolis strain SLP6 enhances the phytoremediation efficiency of Helianthus annuus in copper contaminated soils under salinity stress. Plant Soil. 2020, 457, 241–253. [Google Scholar] [CrossRef]
- Mukherjee, P.; Mitra, A.; Roy, M. Halomonas rhizobacteria of Avicennia marina of Indian Sundarbans promote rice growth under saline and heavy metal stresses through exopolysaccharide production. Front. Microbiol. 2019, 10, 1207. [Google Scholar] [CrossRef]
- Yasmin, H.; Rashid, U.; Hassan, M.N.; Nosheen, A.; Naz, R.; Ilyas, N.; Sajjad, M.; Azmat, A.; Alyemeni, M.N. Volatile organic compounds produced by Pseudomonas pseudoalcaligenes alleviated drought stress by modulating defense system in maize (Zea mays L.). Physiol. Planta 2021, 172, 896–911. [Google Scholar] [CrossRef]
- Desale, P.; Patel, B.; Singh, S.; Malhotra, A.; Nawani, N. Plant growth promoting properties of Halobacillus sp. and Halomonas sp. in presence of salinity and heavy metals. J. Basic. Microbiol. 2014, 54, 781–791. [Google Scholar] [CrossRef]
- Lahsini, A.I.; Sallami, A.; Obtel, M.; Douira, A.; El Modafar, C.; Benkerroum, N.; Talbi, C.; Chakhchar, A.; Filali-Maltouf, A. Isolation and molecular identification of an indigenous abiotic stress-tolerant plant growth-promoting rhizobacteria from the rhizosphere of the olive tree in southern Morocco. Rhizosphere. 2022, 23, 100554. [Google Scholar] [CrossRef]
- Nadeem, S.M.; Ahmad, M.; Tufail, M.A.; Asghar, H.N.; Nazli, F.; Zahir, Z.A. Appraising the potential of EPS-producing rhizobacteria with ACC-deaminase activity to improve growth and physiology of maize under drought stress. Physiol. Plant 2021, 172, 463–476. [Google Scholar] [CrossRef] [PubMed]
- Mukhtar, T.; Rehman, S.U.; Smith, D.; Sultan, T.; Seleiman, M.F.; Alsadon, A.A.; Amna, A.S.; Chaudhary, H.J.; Solieman, T.H.; Ibrahim, A.A. Mitigation of heat stress in Solanum lycopersicum L. by ACC-deaminase and exopolysaccharide producing Bacillus cereus: Effects on biochemical profiling. Sustainability 2020, 12, 2159. [Google Scholar] [CrossRef]
- Chattopadhyay, A.; Purohit, J.; Tiwari, K.K.; Deshmukh, R. Targeting transcription factors for plant disease resistance: Shifting paradigm. Curr. Sci. 2019, 117, 1598–1607. [Google Scholar] [CrossRef]
- Yuan, J.; Raza, W.; Shen, Q.; Huang, Q. Antifungal activity of Bacillus amyloliquefaciens NJN-6 volatile compounds against Fusarium oxysporum f. sp. cubense. Appl. Environ. Microbiol. 2012, 78, 5942–5944. [Google Scholar] [CrossRef]
- Cordero, P.; Principe, A.; Jofre, E.; Mori, G.; Fischer, S. Inhibition of the phytopathogenic fungus Fusarium proliferatum by volatile compounds produced by Pseudomonas. Arch. Microbiol. 2014, 196, 803–809. [Google Scholar] [CrossRef]
- Teli, B.; Purohit, J.; Rashid, M.M.; Jailani, A.A.K.; Chattopadhyay, A. Omics insight on Fusarium head blight of wheat for translational research perspective. Curr. Genom. 2020, 21, 411–428. [Google Scholar] [CrossRef]
- Giorgio, A.; De Stradis, A.; Lo Cantore, P.; Iacobellis, N.S. Biocide effects of volatile organic compounds produced by potential biocontrol rhizobacteria on Sclerotinia sclerotiorum. Front. Microbiol. 2015, 6, 1056. [Google Scholar] [CrossRef]
- Groenhagen, U.; Baumgartner, R.; Bailly, A.; Gardiner, A.; Eberl, L.; Schulz, S.; Weisskopf, L. Production of bioactive volatiles by different Burkholderiaambifaria strains. J. Chem. Ecol. 2013, 39, 892–906. [Google Scholar] [CrossRef]
- Sanchez-Fernandez, R.E.; Diaz, D.; Duarte, G.; Lappe-Oliveras, P.; Sanchez, S.; Macias-Rubalcava, M.L. Antifungal volatile organic compounds from the endophyte Nodulisporium sp. strain GS4d2II1a: A qualitative change in the intraspecific and interspecific interactions with Pythium aphanidermatum. Microorganisms 2016, 8, 620. [Google Scholar] [CrossRef]
- Sharma, R.; Chauhan, A.; Shirkot, C.K. Characterization of plant growth-promoting Bacillus strains and their potential as crop protectants against Phytophthora capsici in tomato. Biol. Agric. Hortic. 2015, 31, 230–244. [Google Scholar] [CrossRef]
- Peleg, I.; Feldman, K. Minrav Industries Ltd.Bacillus firmus CNCM I-1582 or Bacillus cereus CNCM I-1562 for Controlling Nematodes. U.S. Patent 6: 406690, 2002. Application granted on 18 June 2002. [Google Scholar]
- Susic, N.; Janezic, S.; Rupnik, M.; Stare, B.G. Whole genome sequencing and comparative genomics of two nematicidal Bacillus strains reveals a wide range of possible virulence factors. G3 Genes Genomes Genet. 2020, 10, 881–890. [Google Scholar] [CrossRef] [PubMed]
- Ruiu, L.; Floris, I.; Satta, A.; Ellar, D.J. Toxicity of a Brevibacilluslaterosporus strain lacking parasporal crystals against Musca domestica and Aedes aegypti. Biol. Control 2007, 43, 136–143. [Google Scholar] [CrossRef]
- Ruiu, L. Brevibacilluslaterosporus, a pathogen of invertebrates and a broad-spectrum antimicrobial species. Insects 2013, 4, 476–492. [Google Scholar] [CrossRef] [PubMed]
- Marche, M.G.; Camiolo, S.; Porceddu, A.; Ruiu, L. Survey of Brevibacilluslaterosporus insecticidal protein genes and virulence factors. J. Invertebr. Pathol. 2018, 155, 38–43. [Google Scholar] [CrossRef]
- Shehata, M.G.; Badr, A.N.; El Sohaimy, S.A.; Asker, D.; Awad, T.S. Characterization of antifungal metabolites produced by novel lactic acid bacterium and their potential application as food biopreservatives. Ann. Agric. Sci. 2019, 64, 71–78. [Google Scholar] [CrossRef]
- Wang, Z.; Mei, X.; Du, M.; Chen, K.; Jiang, M.; Wang, K.; Zalán, Z.; Kan, J. Potential modes of action of Pseudomonas fluorescens ZX during biocontrol of blue mold decay on postharvest citrus. J. Sci. Food Agric. 2020, 100, 744–754. [Google Scholar] [CrossRef]
- Wang, Z.; Zhong, T.; Chen, K.; Du, M.; Chen, G.; Chen, X.; Wang, K.; Zalán, Z.; Takács, K.; Kan, J. Antifungal activity of volatile organic compounds produced by Pseudomonas fluorescens ZX and potential biocontrol of blue mold decay on postharvest citrus. Food Control 2021, 120, 107499. [Google Scholar] [CrossRef]
- Blainski, J.M.; da Rocha Neto, A.C.; Schimidt, E.C.; Voltolini, J.A.; Rossi, M.J.; Di Piero, R.M. Exopolysaccharides from Lactobacillus plantarum induce biochemical and physiological alterations in tomato plant against bacterial spot. Appl. Microbiol. Biotechnol. 2018, 102, 4741–4753. [Google Scholar] [CrossRef]
- Ramadan, E.M.; AbdelHafez, A.A.; Hassan, E.A.; Saber, F.M. Plant growth-promoting rhizobacteria and their potential for biocontrol of phytopathogens. Afr. J. Microbiol. Res. 2016, 10, 486–504. [Google Scholar] [CrossRef]
- Arseneault, T.; Goyer, C.; Filion, M. Pseudomonas fluorescens LBUM223 increases potato yield and reduces common scab symptoms in the field. Phytopathology 2015, 105, 1311–1317. [Google Scholar] [CrossRef] [PubMed]
- Fukami, J.; Ollero, F.J.; Megías, M.; Hungria, M. Phytohormones and induction of plant-stress tolerance and defense genes by seed and foliar inoculation with Azospirillumbrasilense cells and metabolites promote maize growth. AMB Express 2017, 7, 153. [Google Scholar] [CrossRef] [PubMed]
- Vacheron, J.; Renoud, S.; Muller, D.; Babalola, O.O.; Prigent-Combaret, C. Alleviation of abiotic and biotic stresses in plants by Azospirillum. In Handbook for Azospirillum, Technical Issues and Protocol; Cassán, F., Okon, Y., Creus, C., Eds.; Springer International Publishing: Cham, Switzerland, 2015; pp. 333–365. [Google Scholar]
- Luz, C.; D’Opazo, V.; Quiles, J.M.; Romano, R.; Mañes, J.; Meca, G. Biopreservation of tomatoes using fermented media by lactic acid bacteria. LWT 2020, 130, 109618. [Google Scholar] [CrossRef]
- Juodeikiene, G.; Bartkiene, E.; Cernauskas, D.; Cizeikiene, D.; Zadeike, D.; Lele, V.; Bartkevics, V. Antifungal activity of lactic acid bacteria and their application for Fusarium mycotoxin reduction in malting wheat grains. LWT 2018, 89, 307–314. [Google Scholar] [CrossRef]
- Cao, H.; Meng, D.; Zhang, W.; Ye, T.; Yuan, M.; Yu, J.; Wu, X.; Li, Y.; Yin, F.; Fu, C.; et al. Growth inhibition of Fusarium graminearum and deoxynivalenol detoxification by lactic acid bacteria and their application in sourdough bread. Int. J. Food Sci. Technol. 2021, 56, 2304–2314. [Google Scholar] [CrossRef]
- Enebe, M.C.; Babaloa, O.O. The impact of microbes in the orchestration of plants’ resistance to biotic stress: A disease management approach. Appl. Microbiol. Biotechnol. 2019, 103, 9–25. [Google Scholar] [CrossRef]
- Kantar, C.; Demiray, H.; Dogan, N.M. Role of microbial exopolymeric substances on chromium sorption and transport in heterogeneous subsurface soil. II. Binding of Cr(III) in EPS/soil system. Chemosphere 2010, 82, 1489–1495. [Google Scholar] [CrossRef]
- Priester, J.H.; Olson, S.G.; Webb, S.M.; Neu, M.P.; Hersman, L.E.; Holden, P.A. Enhanced exopolymer production and chromium stabilization in Pseudomonas putida unsaturated biofilms. Appl. Environ. Microbiol. 2006, 72, 1988–1996. [Google Scholar] [CrossRef]
- Kantar, C.; Cetin, Z.; Demiray, H. In situ stabilization of chromium (VI) in polluted soils using organic ligands: The role of galacturonic, glucuronic and alginic acids. J. Hazard. Mater. 2008, 159, 287–293. [Google Scholar] [CrossRef]
- Cetin, Z.; Kantar, C.; Alpaslan, M. Interactions between uronic acids and chromium (III). Environ. Toxicol. Chem. 2008, 28, 1599–1608. [Google Scholar] [CrossRef]
- Nair, A.; Juwarkar, A.A.; Singh, S.K. Production and characterization of siderophores and its application in arsenic removal from contaminated soil. Water Air Soil Pollut. 2007, 180, 199–212. [Google Scholar] [CrossRef]
- O’Brien, S.; Hodgson, D.J.; Buckling, A. Social evolution of toxic metal bioremediation in Pseudomonas aeruginosa. Proc. R. Soc. B Biol. Sci. 2014, 281, 20140858. [Google Scholar] [CrossRef] [PubMed]
- Rizvi, A.; Khan, M.S. Heavy metal-induced oxidative damage and root morphology alterations of maize (Zea mays L.) plants and stress mitigation by metal tolerant nitrogen-fixing Azotobacterchroococcum. Ecotoxicol. Environ. Saf. 2018, 157, 9–20. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Xiong, D.; Zhao, P.; Yu, X.; Tu, B.; Wang, G. Effect of applying an arsenic-resistant and plant growth-promoting Rhizobacterium to enhance soil arsenic phyto-remediation by Populusdeltoides LH05-17. J. Appl. Microbiol. 2011, 111, 1065–1074. [Google Scholar] [CrossRef] [PubMed]
- Gamalero, E.; Glick, B.R. Bacterial modulation of plant ethylene levels. Plant Physiol. 2015, 169, 13–22. [Google Scholar] [CrossRef]
- Vurukonda, S.S.K.P.; Vardharajula, S.; Shrivastava, M.; Ali, S.Z. Enhancement of drought stress tolerance in crops by plant growth-promoting rhizobacteria. Microbiol. Res. 2016, 184, 13–24. [Google Scholar] [CrossRef]
- Jochum, M.D.; McWilliams, K.L.; Borrego, E.J.; Kolomiets, M.V.; Niu, G.; Pierson, E.A.; Jo, Y.K. Bioprospecting plant growth promoting rhizobacteria that mitigate drought stress in grasses. Front. Microbiol. 2019, 10, 2106. [Google Scholar] [CrossRef]
- Khan, I.; Samrah, A.A.; Ikram, R.; Rizwan, M.; Akhtar, N.; Yasmin, H.; Sayyed, R.Z.; Ali, S.; Ilyas, N. Effects of 24-epibrassinolide on plant growth, antioxidants defense system, and endogenous hormones in two wheat varieties under drought stress. Physiol. Plant. 2020, 172, 696–706. [Google Scholar] [CrossRef]
- Singh, J.; Singh, P.; Ray, S.; Rajput, R.S.; Singh, H.B. Plant growth-promoting rhizobacteria: Benign and useful substitute for mitigation of biotic and abiotic stresses. In Plant Growth Promoting Rhizobacteria for Sustainable Stress Management; Springer: Singapore, 2019; pp. 81–101. [Google Scholar]
- Schobert, C.; Kockenberger, W.; Komor, E. Uptake of amino acids by plants from the soil: A comparative study with castor bean seedlings grown under natural and axenic soil conditions. Plant Soil. 1998, 109, 181. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of proline under changing environments. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Holtmann, G.; Bremer, E. Thermoprotection of Bacillus subtilis by exogenously provided Glycine Betaine and structurally related compatible solutes: Involvement of Opu transporters. J. Bacteriol. 2004, 186, 1683–1693. [Google Scholar] [CrossRef]
- Fariduddin, Q.; Varshney, P.; Yusuf, M.; Ali, A.; Ahmad, A. Dissecting the role of glycine betaine in plants under abiotic stress. Plant Stress 2013, 7, 8–18. [Google Scholar]
- Rahman, M.S.; Miyake, H.; Takeoka, Y. Effects of exogenous glycine-betaine on growth and ultra-structure of salt-stressed rice seedlings (Oryza sativa L.). Plant Prod. Sci. 2002, 5, 33–44. [Google Scholar] [CrossRef]
- Abdelrahman, M.; Jogaiah, S.; Burritt, D.J.; Tran, L.P. Legume genetic resources and transcriptome dynamics under abiotic stress conditions. Plant Cell Environ. 2018, 41, 1949–2225. [Google Scholar] [CrossRef] [PubMed]
- Gong, M.; Tang, M.; Chen, H.; Zhang, Q.; Feng, X. Effects of two Glomus species on the growth and physiological performance of Sophora davidii seedlings under water stress. New For. 2013, 44, 399–408. [Google Scholar] [CrossRef]
- Miglani, R.; Parveen, N.; Kumar, A.; Ansari, M.A.; Khanna, S.; Rawat, G.; Panda, A.K.; Bisht, S.S.; Upadhyay, J.; Ansari, M.N. Degradation of xenobiotic pollutants: An environmentally sustainable approach. Metabolites 2022, 12, 818. [Google Scholar] [CrossRef]
- Pande, V.; Pandey, S.C.; Sati, D.; Bhatt, P.; Samant, M. Microbial interventions in bioremediation of heavy metal contaminants in agroecosystem. Front. Microbiol. 2022, 13, 824084. [Google Scholar] [CrossRef] [PubMed]
- Datta, S.; Singh, S.; Kumar, V.; Dhanjal, D.S.; Sidhu, G.K.; Amin, D.S.; Kumar, S.; Singh, J.; Singh, J. Endophytic bacteria in xenobiotic degradation. In Microbial Endophytes; Woodhead Publishing: Cambridge, UK, 2020; pp. 125–156. [Google Scholar] [CrossRef]
- Bhandari, S.; Poudel, D.K.; Marahatha, R.; Dawadi, S.; Khadayat, K.; Phuyal, S.; Shrestha, S.; Gaire, S.; Basnet, K.; Khadka, U.; et al. Microbial enzymes used in bioremediation. J. Chem. 2021, 8849512. [Google Scholar] [CrossRef]
- Charles, S.; Ratier, A.; Baudrot, V.; Multari, G.; Siberchicot, A.; Wu, D.; Lopes, C. Taking full advantage of modelling to better assess environmental risk due to xenobiotics-the all-in-one facility MOSAIC. Environ. Sci. Pollut. Res. Int. 2022, 29, 29244–29257. [Google Scholar] [CrossRef]
- Gangola, S.; Joshi, S.; Kumar, S.; Pandey, S.C. Comparative analysis of fungal and bacterial enzymes in biodegradation of xenobiotic compounds. In Smart Bioremediation Technologies; Academic Press: Cambridge, MA, USA, 2019; pp. 169–189. [Google Scholar] [CrossRef]
- Sinha, S.; Chattopadhyay, P.; Pan, I.; Debashis, B.; Das, K.; Sen, S.K. Microbial transformation of xenobiotics for environmental bioremediation. Afr. J. Biotechnol. 2009, 8, 6016–6027. [Google Scholar] [CrossRef]
- Atashgahi, S.; Shetty, S.A.; Smidt, H.; De Vos, W.M. Flux, impact and fate of halogenated xenobiotic compounds in the gut. Front. Physiol. 2018, 9, 888. [Google Scholar] [CrossRef] [PubMed]
- Jeffries, T.C.; Rayu, S.; Nielsen, U.N.; Lai, K.; Ijaz, A.; Nazaries, L.; Singh, B.K. Metagenomic Functional Potential Predicts Degradation Rates of a Model Organophosphorus Xenobiotic in Pesticide Contaminated Soils. Front. Microbiol. 2018, 9, 147. [Google Scholar] [CrossRef] [PubMed]
- Davolos, D.; Russo, F.; Canfora, L.; Malusà, E.; Tartanus, M.; Furmanczyk, E.M.; Persiani, A.M. A Genomic and Transcriptomic Study on the DDT-Resistant Trichoderma hamatum FBL 587: First Genetic Data into Mycoremediation Strategies for DDT-Polluted Sites. Microorganisms 2021, 9, 1680. [Google Scholar] [CrossRef]
- Meng, D.; Zhang, L.; Meng, J.; Tian, Q.; Zhai, L.; Hao, Z.; Guan, Z.; Cai, Y.; Liao, X. Evaluation of the Strain Bacillus amyloliquefaciens YP6 in Phoxim degradation via transcriptomic data and product analysis. Molecules 2019, 24, 399. [Google Scholar] [CrossRef]
- Dhiman, N.; Jasrotia, T.; Sharma, P.; Negi, S.; Chaudhary, S.; Kumar, R.; Mahnashi, M.H.; Umar, A.; Kumar, R. Immobilization interaction between xenobiotic and Bjerkanderaadusta for the biodegradation of atrazine. Chemosphere 2020, 257, 127060. [Google Scholar] [CrossRef] [PubMed]
- AlMatar, M.; Makky, E.A. Cladosporium cladosporioides from the perspectives of medical and biotechnological approaches. 3Biotech 2016, 6, 4. [Google Scholar] [CrossRef]
- Esparza-Naranjo, S.B.; da Silva, G.F.; Duque-Castaño, D.C.; Araújo, W.L.; Peres, C.K.; Boroski, M.; Bonugli-Santos, R.C. Potential for the biodegradation of atrazine using leaf litter fungi from a subtropical protection area. Curr. Microbiol. 2019, 78, 358–368. [Google Scholar] [CrossRef]
- Al-Hawash, A.B.; Alkooranee, J.T.; Zhang, X.; Ma, F. Fungal Degradation of Polycyclic Aromatic Hydrocarbons. Int. J. Pure Appl. Biosci. 2018, 6, 8–24. [Google Scholar] [CrossRef]
- Ijoma, G.N.; Tekere, M. Potential microbial applications of co-cultures involving ligninolytic fungi in the Bioremediation of recalcitrant xenobiotic compounds. Int. J. Environ. Sci. Technol. 2017, 14, 1787–1806. [Google Scholar] [CrossRef]
- Kantharaj, P.; Boobalan, B.; Sooriamuthu, S.; Mani, R. Lignocellulose Degrading Enzymes from Fungi and Their Industrial Applications. Int. J. Curr. Res. Rev. 2017, 9, 1–12. [Google Scholar] [CrossRef]
- Dash, D.M.; Osborne, W.J. Rapid biodegradation and biofilm-mediated bioremoval of organophosphorus pesticides using an indigenous Kosakoniaoryzae strain-VITPSCQ3 in a vertical-flow packed bed biofilm bioreactor. Ecotox. Environ. Saf. 2020, 192, 110290. [Google Scholar] [CrossRef] [PubMed]
- Zhou, Z.; Wu, X.; Lin, Z.; Pang, S.; Mishra, S.; Chen, S. Biodegradation of fipronil: Current state of mechanisms of biodegradation and future perspectives. Appl. Microbiol. Biotechnol. 2021, 105, 7695–7708. [Google Scholar] [CrossRef] [PubMed]
- Cairns, T.C.; Zheng, X.; Zheng, P.; Sun, J.; Meyer, V. Turning inside out: Filamentous fungal secretion and its applications in biotechnology, agriculture, and the clinic. J.Fungi 2021, 7, 535. [Google Scholar] [CrossRef]
- Morcillo, R.J.L.; Baroja-Fernández, E.; López-Serrano, L.; Leal-López, J.; Muñoz, F.J.; Bahaji, A.; Férez-Gómez, A.; Pozueta-Romero, J. Cell-free microbial culture filtrates as candidate biostimulants to enhance plant growth and yield and activate soil- and plant-associated beneficial microbiota. Front. Plant Sci. 2022, 13, 1040515. [Google Scholar] [CrossRef] [PubMed]
- Feng, H.; Fu, R.; Hou, X.; Lv, Y.; Zhang, N.; Liu, Y.; Xu, Z.; Miao, Y.; Krell, T.; Shen, Q.; et al. Chemotaxis of beneficial Rhizobacteria to root exudates: The first step towards root-microbe rhizosphere interactions. Int. J. Mol. Sci. 2021, 22, 6655. [Google Scholar] [CrossRef]
- Goswami, D.; Thakker, J.N.; Dhandhukia, P.C. Portraying mechanics of plant growth-promoting rhizobacteria (PGPR): A review. Cogent Food Agric. 2016, 2, 1127500. [Google Scholar] [CrossRef]
- Cretoiu, M.S.; Korthals, G.W.; Visser, J.H.; van Elsas, J.D. Chitin amendment increases soil suppressiveness toward plant pathogens and modulates the actinobacterial and oxalobacteraceal communities in an experimental agricultural field. Appl. Environ. Microbiol. 2013, 79, 5291–5301. [Google Scholar] [CrossRef]
- De Tender, C.; Mesuere, B.; Van der Jeugt, F.; Haegeman, A.; Ruttink, T.; Vandecasteele, B.; Dawyndt, P.; Debode, J.; Kuramae, E.E. Peat substrate amended with chitin modulates the N-cycle, siderophore and chitinase responses in the lettuce rhizobiome. Sci. Rep. 2019, 9, 9890. [Google Scholar] [CrossRef]
- Sun, J.; Li, S.; Fan, C.; Cui, K.; Tan, H.; Qiao, L.; Lu, L. N-Acetylglucosamine Promotes Tomato Plant Growth by Shaping the Community Structure and Metabolism of the Rhizosphere Microbiome. Microbiol. Spectr. 2022, 10, e0035822. [Google Scholar] [CrossRef]
- Macias-Benitez, S.; Garcia-Martinez, A.M.; Caballero Jimenez, P.; Gonzalez, J.M.; Tejada Moral, M.; Parrado Rubio, J. Rhizospheric Organic Acids as Biostimulants: Monitoring Feedbacks on Soil Microorganisms and Biochemical Properties. Front. Plant Sci. 2020, 11, 633. [Google Scholar] [CrossRef]
- Acin-Albiac, M.; García-Jiménez, B.; Marín Garrido, C.; Borda Casas, E.; Velasco-Alvarez, J.; Serra, N.S.; Acedo, A. Lettuce Soil Microbiome Modulated by an L-α-Amino Acid-Based Biostimulant. Agronomy 2023, 13, 344. [Google Scholar] [CrossRef]
- Kim, D.R.; Jeon, C.W.; Cho, G.; Thomashow, L.S.; Weller, D.M.; Paik, M.J.; Lee, Y.B.; Kwak, Y.S. Glutamic Acid Reshapes the Plant Microbiota to Protect Plants against Pathogens. Microbiome 2021, 9, 244. [Google Scholar] [CrossRef] [PubMed]
- Colla, G.; Hoagland, L.; Ruzzi, M.; Cardarelli, M.; Bonini, P.; Canaguier, R.; Rouphael, Y. Biostimulant Action of Protein Hydrolysates: Unraveling Their Effects on Plant Physiology and Microbiome. Front. Plant Sci. 2017, 8, 2202. [Google Scholar] [CrossRef] [PubMed]
- Luziatelli, F.; Ficca, A.G.; Colla, G.; BaldassarreŠvecová, E.; Ruzzi, M. Foliar Application of Vegetal-Derived Bioactive Compounds Stimulates the Growth of Beneficial Bacteria and Enhances Microbiome Biodiversity in Lettuce. Front. Plant Sci. 2019, 10, 60. [Google Scholar] [CrossRef] [PubMed]
- Ibal, J.-C.; Park, M.-K.; Park, G.-S.; Jung, B.-K.; Park, T.-H.; Kim, M.-S.; Kang, G.-U.; Park, Y.-J.; Shin, J.-H. Use of Acyl-Homoserine Lactones Leads to Improved Growth of Ginseng Seedlings and Shifts in Soil Microbiome Structure. Agronomy 2021, 11, 2177. [Google Scholar] [CrossRef]
- Spaepen, S.; Vanderleyden, J.; Remans, R. Indole-3-Acetic Acid in Microbial and Microorganism-Plant Signaling. FEMS Microbiol. Rev. 2007, 31, 425–448. [Google Scholar] [CrossRef]
- Duca, D.R.; Glick, B.R. Indole-3-Acetic Acid Biosynthesis and Its Regulation in Plant-Associated Bacteria. Appl. Microbiol. Biotechnol. 2020, 104, 8607–8619. [Google Scholar] [CrossRef]
- Eichmann, R.; Richards, L.; Schäfer, P. Hormones as Go-Betweens in Plant Microbiome Assembly. Plant J. 2021, 105, 518–541. [Google Scholar] [CrossRef]
- Spaepen, S.; Das, F.; Luyten, E.; Michiels, J.; Vanderleyden, J. Research Letter: Indole-3-Acetic Acid-Regulated Genes in Rhizobium etli CNPAF512. FEMS Microbiol. Lett. 2009, 291, 195–200. [Google Scholar] [CrossRef]
- Van Puyvelde, S.; Cloots, L.; Engelen, K.; Das, F.; Marchal, K.; Vanderleyden, J.; Spaepen, S. Transcriptome Analysis of the Rhizosphere Bacterium Azospirillumbrasilense Reveals an Extensive Auxin Response. MicrobEcol 2011, 61, 723–728. [Google Scholar] [CrossRef]
- Matsukawa, E.; Nakagawa, Y.; Iimura, Y.; Hayakawa, M. Stimulatory Effect of Indole-3-Acetic Acid on Aerial Mycelium Formation and Antibiotic Production in Streptomyces spp. Actinomycetologica 2007, 21, 32–39. [Google Scholar] [CrossRef]
- Laird, T.S.; Flores, N.; Leveau, J.H.J. Bacterial Catabolism of Indole-3-Acetic Acid. Appl. Microbiol. Biotechnol. 2020, 104, 9535–9550. [Google Scholar] [CrossRef] [PubMed]
- Rico-Jiménez, M.; Roca, A.; Krell, T.; Matilla, M.A. A Bacterial Chemoreceptor That Mediates Chemotaxis to Two Different Plant Hormones. Environ. Microbiol. 2022, 24, 3580–3597. [Google Scholar] [CrossRef] [PubMed]
- Fitzpatrick, C.R.; Salas-González, I.; Conway, J.M.; Finkel, O.M.; Gilbert, S.; Russ, D.; Teixeira, P.J.P.L.; Dangl, J.L. The Plant Microbiome: From Ecology to Reductionism and Beyond. Annu. Rev. Microbiol. 2020, 74, 81–100. [Google Scholar] [CrossRef] [PubMed]
- Jacoby, R.P.; Kopriva, S. Metabolic Niches in the Rhizosphere Microbiome: New Tools and Approaches to Analyse Metabolic Mechanisms of Plant-Microbe Nutrient Exchange. J. Exp. Bot. 2019, 70, 1087–1094. [Google Scholar] [CrossRef] [PubMed]
- Murillo-Roos, M.; Abdullah, H.S.M.; Debbar, M.; Ueberschaar, N.; Agler, M.T. Cross-Feeding Niches among Commensal Leaf Bacteria Are Shaped by the Interaction of Strain-Level Diversity and Resource Availability. ISME J. 2022, 16, 2280–2289. [Google Scholar] [CrossRef]
- Pascale, A.; Proietti, S.; Pantelides, I.S.; Stringlis, I.A. Modulation of the Root Microbiome by Plant Molecules: The Basis for Targeted Disease Suppression and Plant Growth Promotion. Front. Plant Sci. 2020, 10, 1741. [Google Scholar] [CrossRef]
- Song, G.C.; Ryu, S.Y.; Kim, Y.S.; Lee, J.Y.; Choi, J.S.; Ryu, C.M. Elicitation of Induced Resistance against Pectobacteriumcarotovorum and Pseudomonas syringae by Specific Individual Compounds Derived from Native Korean Plant Species. Molecules 2013, 18, 12877–12895. [Google Scholar] [CrossRef]

| Microbes | Microbial Exudates | Mode of Action | References |
|---|---|---|---|
| Azotobacter chroococcum | Exopolysaccharide | Plant growth promotion in Faba bean | [130] |
| Bacillus gibsonii (PM11), B. xiamenensis (PM14) | Exopolysaccharide | Enhanced nutrient availability and plant growth of Linum usitatissimum by minimizing metal-induced stressed conditions | [131] |
| Acinetobacter calcoaceticus (9EU- LRNA-72), Penicillium sp. (EU-FTF-6) | Metabolites containing glycine betaine, proline, sugars, etc. | Increase in chlorophyll synthesis and decrease in lipid peroxidation | [132] |
| Trichoderma harzianum (M 10) | Harzianic acid (siderophore) | Induce expression of resistant genes (CC-NBS-LRR) in tomato | [133] |
| Bacillus amyloliquefaciens (FZB42) | Bacillomycin D (lipopeptide) | Degradation of mycotoxin production and disintegration of plasma membrane of Fusarium graminearum (head blight pathogen of wheat) through the production of reactive oxygen species (ROS) | [134] |
| Aureobasidium pullulans | VOCs (ethanol, 2-methylpropan-1-ol, 3-methylbutan-1-ol, and 2-phenylethanol) | Increases intracellular reactive oxygen species (ROS) accumulation, lipid peroxidation, and content leakage, thereby inhibiting Botrytis cinerea growth | [135] |
| A. pullulans | VOCs | Triggers lipid peroxidation and electrolyte leakage in B. cinerea and Alternaria alternate | [135] |
| B. subtilis (BS2) | Metabolites | Defense enzyme production such as peroxidase (PO), polyphenol oxidase (PPO), chitinase, and phenylalanine | [136] |
| Pseudomonas furukawaii, P. plecoglossicida, P. alcaligenes, P. oleovarans, Leclercia adecarboxylata, Citrobacter youngae, Enterobacter cloacae | Hydroxymate and catecholate | Antagonistic activities against different phytopathogens like Rhizoctonia solani, Phythium sp., Fusarium oxysporum in Phaseolus vulgaris, Helianthus sp., Triticum astivum, Oryza sativa | [137] |
| Microalgae | Polysaccharides | Phytostimulant property in tomato | [138] |
| Arthospira platensis | Polyamines | Regulation of gene expression and protein synthesis for the modulation of signal transduction | [139] |
| Pseudomonas putida (CRN-09), Bacillus subtilis (CRN-16) | Metabolites | Production of PO, PPO, beta 1,3-glucanse, chitinase, and phenylalanine ammonia lyase (PAL) against Macrophomina phaseolina | [140] |
| B. subtilis | VOCs (2,3-butanedione; 3-methylbutyric acid) | Antifungal activity (inhibited hyphal growth) against Mucor circinelloides; Fusarium arcuatisporum; A. iridiaustralis; Colletotrichum fioriniae; and reduced decay of wolfberry fruits | [141] |
| Pseudomonas fluorescens (G20-18) | Bacterial cytokinin | Activates plant resistance against pathogenic P. syringae | [142,143] |
| P. fluorescens(C7R12) | Pyoverdine siderophore | Enhanced root and shoot ratio in Pisum sativum by promoting plant iron nutrition | [144] |
| Bacillus licheniformis (DS3) | Hydroxymate | Biological agents against several fungal pathogens like Aspergillus niger, Alternaria solani, Fusarium solani, and Fusarium oxysporium in Vigna mungo | [145] |
| Ascophyllum nodosum | Complex polysaccharides (fucans and alginates) | The combination treatment of chitosan and A. nodosum liquid sea weed extract (containing complex polysaccharide) reduced the level of mycotoxins deoxynivalenol and sambucinol produced by Fusarium graminearum in wheat grains by inducing defense genes and enzymes. | [146] |
| Microbes | Microbial Exudates | Mode of Action | References |
|---|---|---|---|
| Pseudomonas anguilliseptica (SAW24) | Exopolysaccharide | Enhances biofilm stability under salinity stress and, thus, protecting the plant root system | [147] |
| Azotobacter sp. (AztRMD2) | Exopolysaccharide | Augment soil aggregate stability in rice under drought stress condition | [148] |
| Bacillus endophyticus (J13), B. tequilensis (J12) | Exopolysaccharide, IAA, cytokinin | Alleviation of osmotic stress in Arabidopsis | [149] |
| Bacillus gibsonii (PM11), B. xiamenensis (PM14) | Exopolysaccharide | Enhanced nutrient availability and plant growth of Linum usitatissimum by minimizing metal stress | [131] |
| Leclercia adecarboxylata (MO1) | Metabolites | Salinity stress tolerance in soybean via auxin biosynthesis | [150] |
| Dermacoccus barathri (MT2.1T), D. profundi (MT2.2T), and D. nishinomiyaensis (DSM20448T) | Hydroxamate and catechol-type siderophores | Increased tomato seedling and plant growth under saline condition | [151] |
| Streptomyces acidiscabies (E13) | Desferridoxine E Desferridoxine B Coelichelin | Nickel stress tolerance in cowpea through nickle sequesteration | [152] |
| B. subtilis | Endophytic siderophore | Enhanced growth and survivability of wheat under drought condition | [153] |
| Pseudomonas citronellolis strain (SLP6) | Hydroxymate siderophore | Significantly enhanced chlorophyll content, antioxidant enzyme production, and plant growth in Helianthus annus under salinity stress condition | [154] |
| Halomonas sp. | Exo1exopolysaccharide | In presence of arsenic, Exo1 EPSs favor metal ion sequestration by biosorption due to the negative charge matrix of the EPS and alleviated heavy metal stress in rice | [155] |
| Pseudomonas pseudoalcaligenes | VOCs (dimethyl disulfide, 2,3-butanediol, and 2-pentylfuran) | Drought tolerance in maize plants by reducing electrolyte leakage and malondialdehyde content, and increasing proline and phytohormone content | [156] |
| Halobacillus sp.(ADN1), Halomonas sp.(MAN5),and Halobacillus sp. (MAN6) | Exopolysaccharide | Retention of indole acetic acid and phosphate solubilization capacity under salinity and heavy metal stress (Cd, Ni, Hg, and Ag) to enhance root growth in Sesuvium portulacastrum | [157] |
| Arthrobacter globiformis(MSRC52), Bacillus licheniformis(MSRC76), B. megaterium (MSRC23) | Siderophore and IAA | Tolerance to salinity and high-temperature stress in olive trees | [158] |
| B. velezensis D3 | ACC-deaminase, EPS and siderophore | Improved the growth and physiology of maize under drought stress throughout | [159] |
| B. Cereus | ACC-deaminase and EPS | Mitigation of heat stress in Solanum lycopersicum and improvement of physiological and biochemical traits | [160] |
| Xenobiotic | Microbe | Enzyme | Mechanism of Degradation | References |
|---|---|---|---|---|
| Atrazine | Bjerkandera adusta | Laccases, tyrosimases, manganese peroxidases (MnPs), manganese- independent peroxidases (MiPs), and lignin peroxidases | De-alkylation of atrazine resulting in removal efficiency of upto 92%. | [217] |
| Chlorpyrifos | Cladosporium cladosporioides | Chlorpyrifos hydrolase, pectin methylesterase (PME), and polygalacturonase (PG) | Responsible for pectin degradation by catalyzing the demethoxylation of the homogalacturonan chain of pectin to release methanol and acidic pectin | [218] |
| Atrazine Monocrotophos, DDT | Fusarium spp. | N-acetyltransferae and N-malonyltransferase | Detoxification and degradation of aromatic amines | [219] |
| Aromatic compounds, aliphatic hydrocarbons, PAHs | Trichoderma harzianum, Aspergillus fumigatus, Cunninghamella elegans, Aspergillus niger, Penicillium sp., Cunninghamella elegans, Aspergillus ochraceus, Trametes versicolor, Penicillium sp. RMA1 and RMA2, and Aspergillus sp. RFC-1 | Lactase, lignin peroxidases (LiPs), MnPs, epoxide hydrolases cytochrome P450 monoxygenase, dioxygenases, protease, and lipase | By peripheral degradation pathways, organic pollutants are gradually transformed, and many intermediate products are formed | [220] |
| Lignin, polychlorinated biphenyls (PCBs), petroleum hydrocarbons, PAHs, trinitroluenes, industrial dye effluents, herbicides, and pesticides | Trametes versicolor, Phanerochaete chrysosporium, Rigidoporous lignosus, and Pleurotus ostreatus | Lignin peroxidase, versatile peroxidase, laccase, and manganese peroxidise | Formation of semi-quinone intermediate during the oxidation of lignin-derived hyroquinone by laccase. It cleaves C-C bonds and oxidizes benzyl alcohols to aldehydes or ketones. | [221,222] |
| Organophosphorus pesticide- Profenfos and Quinalphos | Kosakonia oryzae strain VITPSCQ3 | Organophosphorous hydrolase and phosphatase | Hydrolytic cleavage of P–S bond in phosphorodithioate and phosphorothioate and P–O bond in phosphate-containing pesticides | [223] |
| Fipronil (Phenyl-pyrazole insecticide) | Aspergillus glaucus, Bacillus frmus, B. thuringiensis, Bacillus sp., Paracoccus sp., Streptomyces rochei,and Stenotrophomonas acidaminiphila | Ligninolytic enzyme MnPs, the cytochrome P450 enzyme, and esterase | Oxidation, reduction, and hydrolysis | [224] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ansari, M.; Devi, B.M.; Sarkar, A.; Chattopadhyay, A.; Satnami, L.; Balu, P.; Choudhary, M.; Shahid, M.A.; Jailani, A.A.K. Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation. J. Xenobiot. 2023, 13, 572-603. https://doi.org/10.3390/jox13040037
Ansari M, Devi BM, Sarkar A, Chattopadhyay A, Satnami L, Balu P, Choudhary M, Shahid MA, Jailani AAK. Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation. Journal of Xenobiotics. 2023; 13(4):572-603. https://doi.org/10.3390/jox13040037
Chicago/Turabian StyleAnsari, Mariya, B. Megala Devi, Ankita Sarkar, Anirudha Chattopadhyay, Lovkush Satnami, Pooraniammal Balu, Manoj Choudhary, Muhammad Adnan Shahid, and A. Abdul Kader Jailani. 2023. "Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation" Journal of Xenobiotics 13, no. 4: 572-603. https://doi.org/10.3390/jox13040037
APA StyleAnsari, M., Devi, B. M., Sarkar, A., Chattopadhyay, A., Satnami, L., Balu, P., Choudhary, M., Shahid, M. A., & Jailani, A. A. K. (2023). Microbial Exudates as Biostimulants: Role in Plant Growth Promotion and Stress Mitigation. Journal of Xenobiotics, 13(4), 572-603. https://doi.org/10.3390/jox13040037








