Bacterial Cytochrome P450 Involvement in the Biodegradation of Fluorinated Pyrethroids
Abstract
:1. Introduction
2. Materials and Methods
2.1. Chemicals and Reagents
2.2. Soil Microbe Isolation and Cultivation
2.3. Whole Genome Sequencing and Phylogenetic Analysis
2.4. Biodegradation of Pyrethroids and Metabolite Extraction
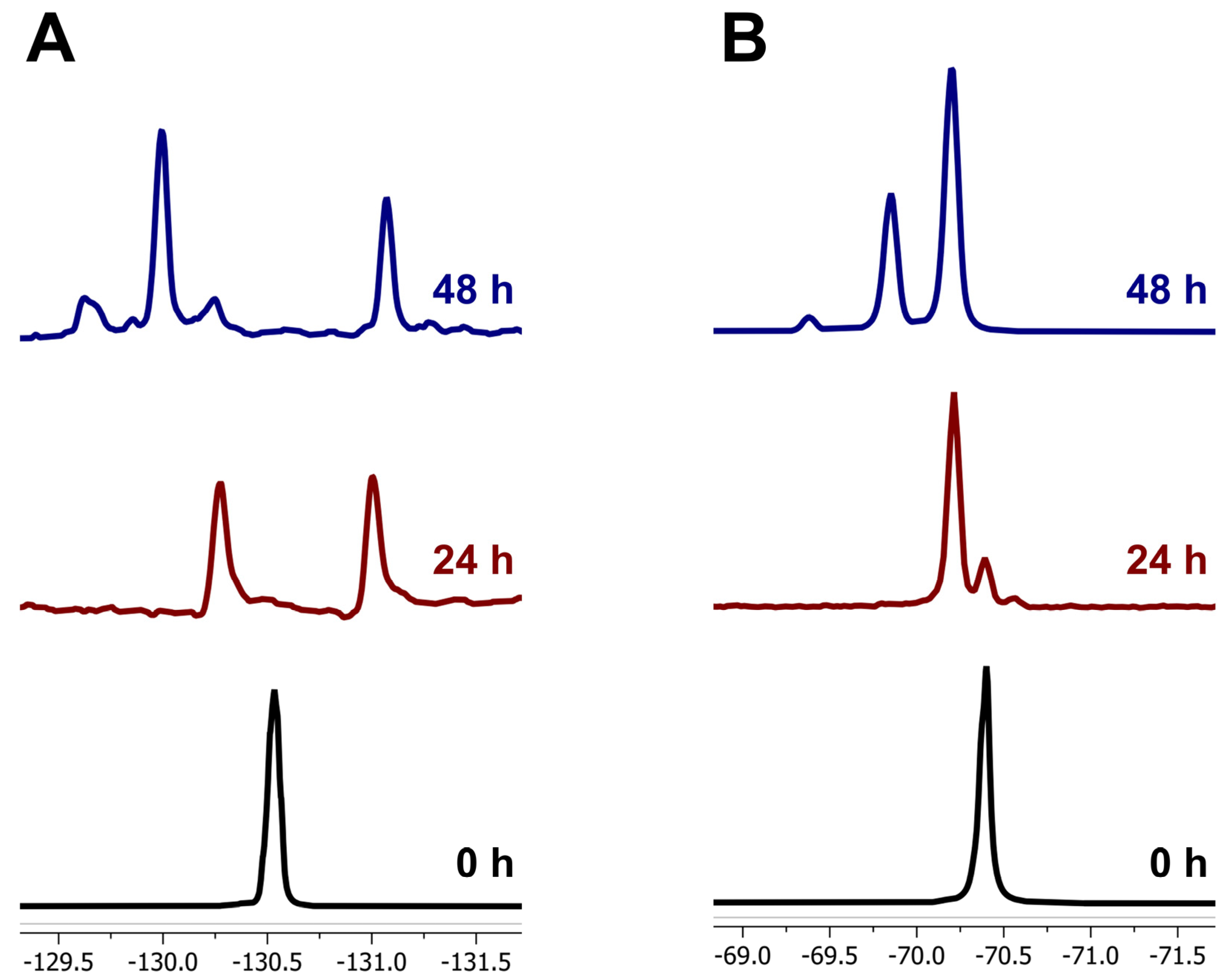
2.5. 19F NMR and GC-MS Analysis
2.6. Inhibition Assays and CYPome Prediction
3. Results
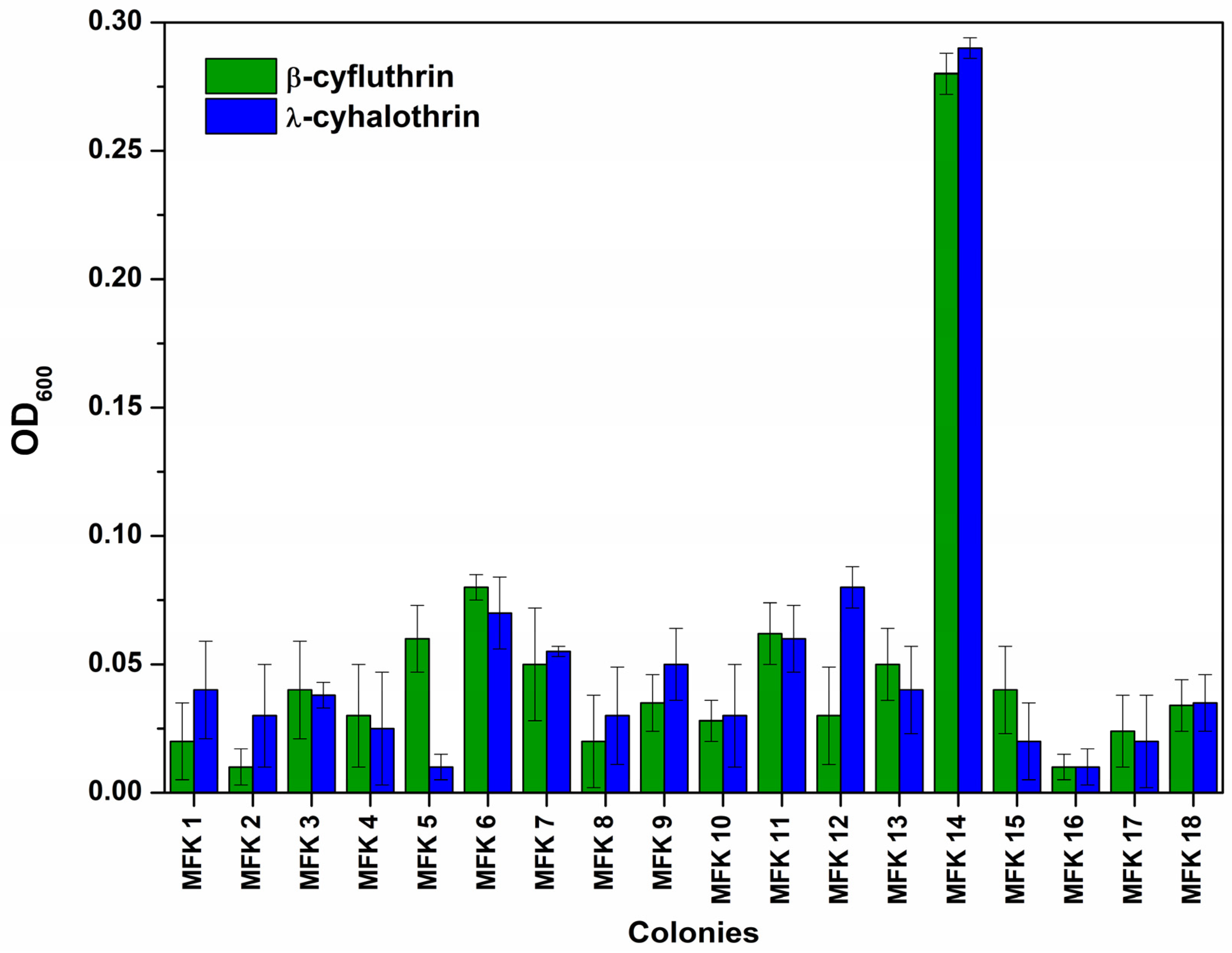
3.1. Pyrethroid Biodegradation by Microbial Consortium Isolated from Garden Soil
3.2. Isolation and Identification of Bacterium Utilising Fluorinated Pyrethroids as a Carbon Source
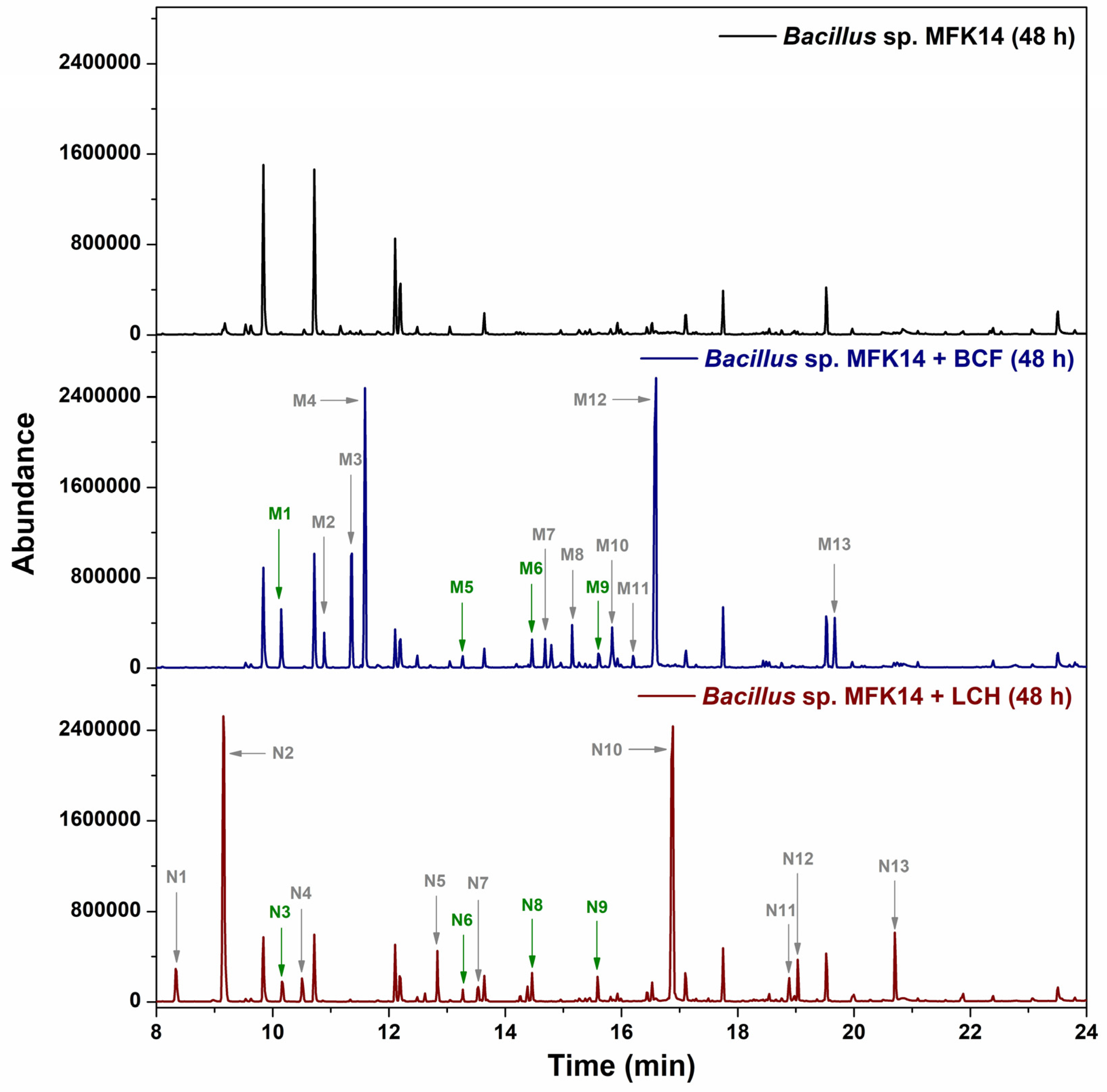
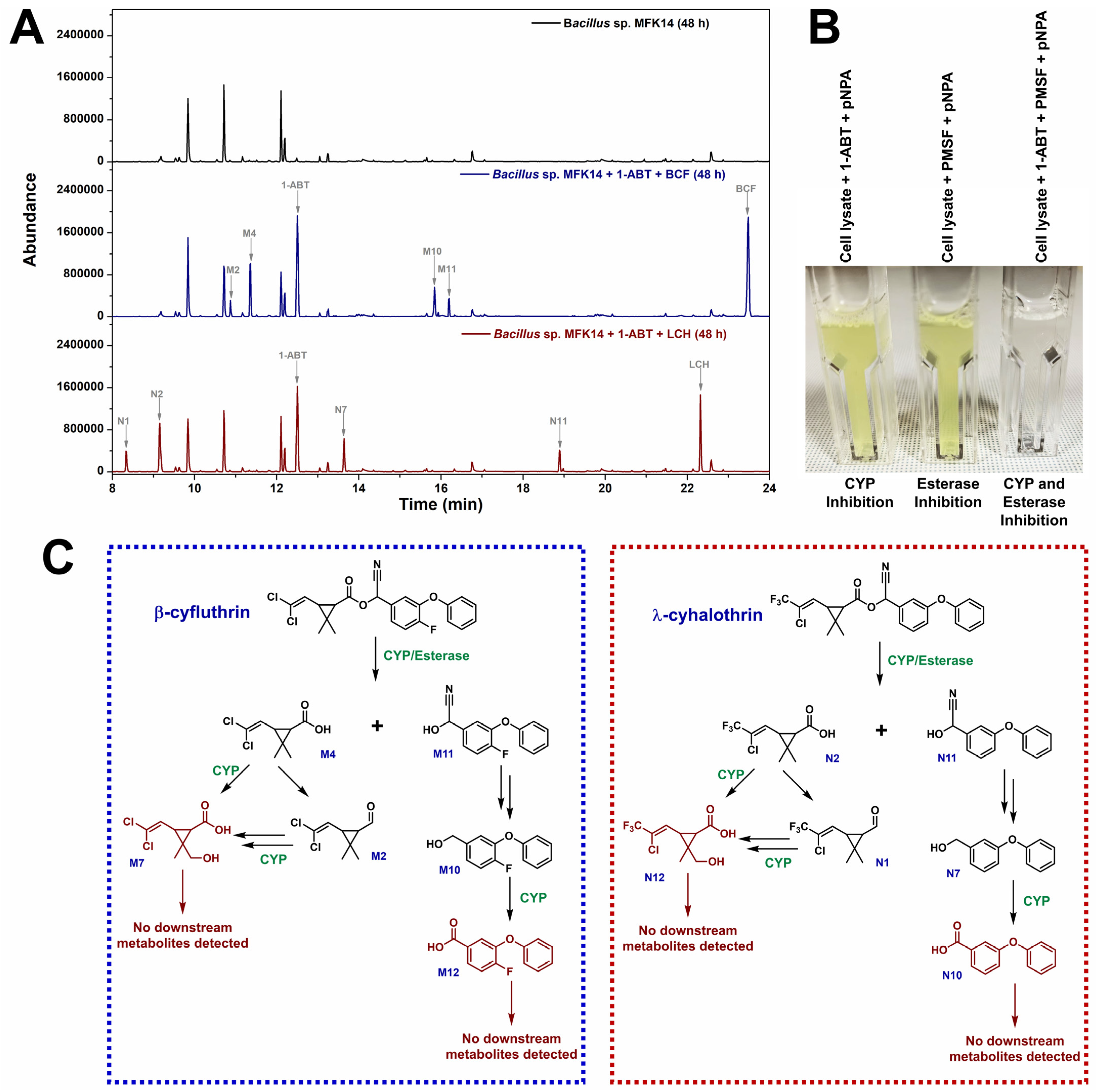
3.3. Metabolite Identification in Pyrethroid Degradation by Bacillus sp. MFK14
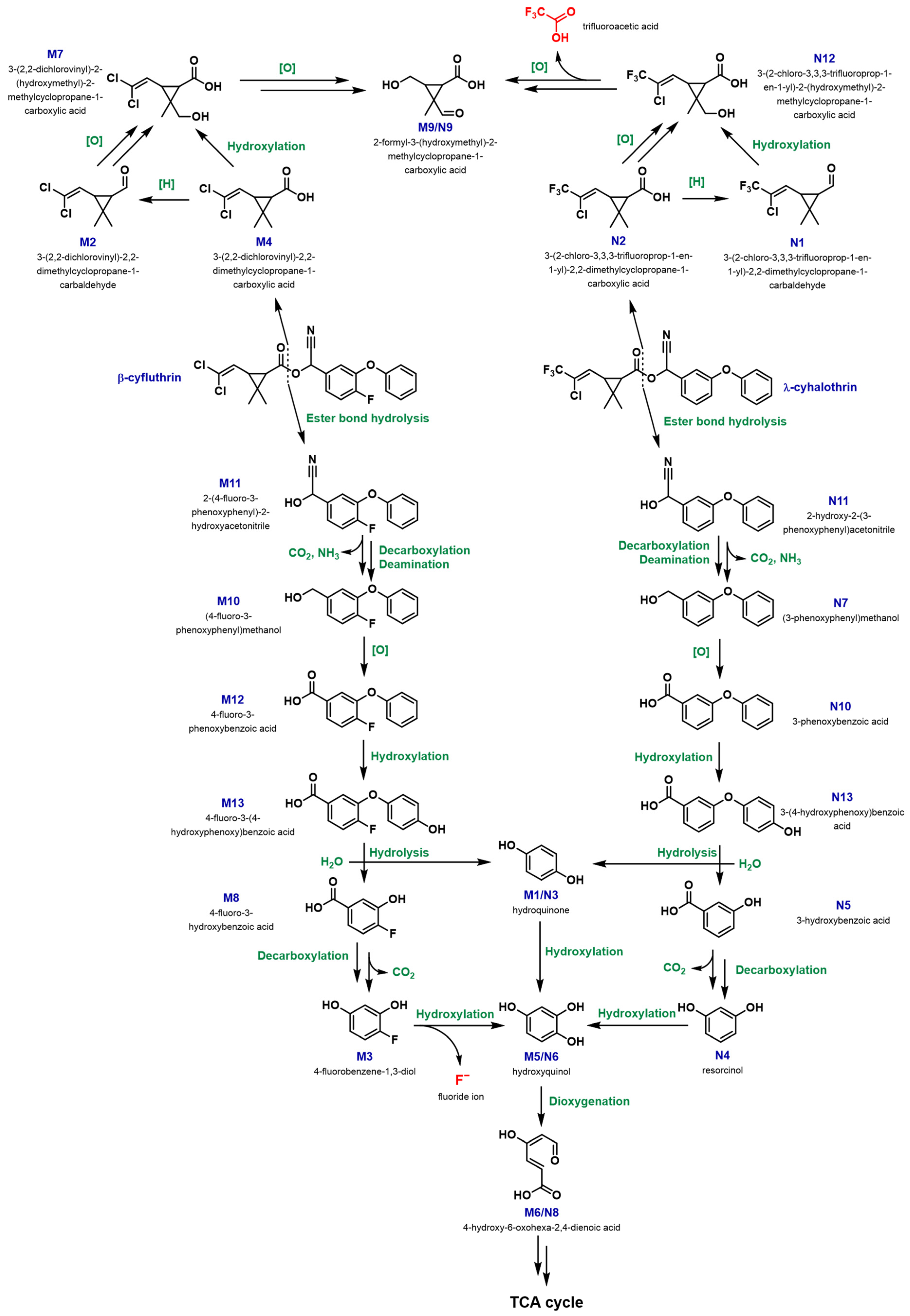
3.3.1. Biodegradation Pathway for β-cyfluthrin
3.3.2. Biodegradation Pathway for λ-cyhalothrin
3.4. CYP Involvement in the Biodegradation of β-cyfluthrin and λ-cyhalothrin
3.5. Predicted CYPome of Bacillus sp. MFK14
4. Discussion
5. Conclusions
6. Future Perspectives
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations
| 1-ABT | 1-aminobenzotriazole |
| 19F NMR | 19F nuclear magnetic resonance spectroscopy |
| BCF | β-cyfluthrin |
| LCH | λ-cyhalothrin |
| CYP | Cytochrome P450 |
| GC-MS | Gas chromatography-mass spectrometry |
| PBS | Phosphate-buffered saline |
| p-NPA | p-Nitrophenyl acetate |
| PMSF | Phenylmethylsulfonyl fluoride |
| TFA | Trifluoroacetic acid |
| TSB | Tryptone soya broth |
| TSA | Tryptone soya agar |
References
- Asem, D.; Kumari, M.; Singh, L.R.; Bhushan, M. Pesticides: Pollution, risks, and abatement measures. In Emerging Aquatic Contaminants, 2nd ed.; Elsevier: Amsterdam, The Netherlands, 2023; pp. 307–326. [Google Scholar]
- Galadima, M.; Singh, S.; Pawar, A.; Khasnabis, S.; Dhanjal, D.S.; Anil, A.G.; Rai, P.; Ramamurthy, P.C.; Singh, J. Toxicity, microbial degradation and analytical detection of pyrethroids: A review. Environ. Adv. 2021, 5, 100105. [Google Scholar] [CrossRef]
- Martínez, M.A.; Rodríguez, J.L.; Lopez-Torres, B.; Martínez, M.; Martínez-Larrañaga, M.R.; Anadón, A.; Ares, I. Oxidative stress and related gene expression effects of cyfluthrin in human neuroblastoma SH-SY5Y cells: Protective effect of melatonin. Environ. Res. 2019, 177, 108579. [Google Scholar] [CrossRef] [PubMed]
- Ham, J.; You, S.; Lim, W.; Song, G. Bifenthrin impairs the functions of Leydig and Sertoli cells in mice via mitochondrion-endoplasmic reticulum dysregulation. Environ. Pollut. 2020, 266, 115174. [Google Scholar] [CrossRef] [PubMed]
- Wang, R.; Wang, F.; Lu, Y.; Zhang, S.; Cai, M.; Guo, D.; Zheng, H. Spatial distribution and risk assessment of pyrethroid insecticides in surface waters of East China Sea estuaries. Environ. Pollut. 2024, 344, 123302. [Google Scholar] [CrossRef]
- Huang, Y.; Chen, S. Pyrethroid-degrading microorganisms and their potential application for the bioremediation of contaminated environments. In Enzymes for Pollutant Degradation; Springer Nature: Singapore, 2022; pp. 119–137. [Google Scholar]
- Li, H.; Ma, Y.; Yao, T.; Ma, L.; Zhang, J.; Li, C. Biodegradation pathway and detoxification of β-cyfluthrin by the bacterial consortium and its bacterial community structure. J. Agric. Food Chem. 2022, 70, 7626–7635. [Google Scholar] [CrossRef]
- Deng, W.; Lin, D.; Yao, K.; Yuan, H.; Wang, Z.; Li, J.; Zou, L.; Han, X.; Zhou, K.; He, L.; et al. Characterization of a novel β-cypermethrin-degrading Aspergillus niger YAT strain and the biochemical degradation pathway of β-cypermethrin. Appl. Microbiol. Biotechnol. 2015, 99, 8187–8198. [Google Scholar] [CrossRef]
- Xiao, Y.; Chen, S.; Gao, Y.; Hu, W.; Hu, M.; Zhong, G. Isolation of a novel beta-cypermethrin degrading strain Bacillus subtilis BSF01 and its biodegradation pathway. Appl. Microbiol. Biotechnol. 2015, 99, 2849–2859. [Google Scholar] [CrossRef]
- Khan, M.F.; Murphy, C.D. Cunninghamella spp. produce mammalian-equivalent metabolites from fluorinated pyrethroid pesticides. AMB Express 2021, 11, 101. [Google Scholar] [CrossRef]
- Khan, M.F.; Murphy, C.D. Cytochrome P450 5208A3 is a promiscuous xenobiotic biotransforming enzyme in Cunninghamella elegans. Enzyme Microb. Technol. 2022, 161, 110102. [Google Scholar] [CrossRef]
- Khan, M.F.; Hof, C.; Niemcova, P.; Murphy, C.D. Biotransformation of fluorinated drugs and xenobiotics by the model fungus Cunninghamella elegans. Methods Enzymol. 2024, 696, 251–285. [Google Scholar]
- Tang, J.; Liu, B.; Chen, T.T.; Yao, K.; Zeng, L.; Zeng, C.Y.; Zhang, Q. Screening of a beta-cypermethrin-degrading bacterial strain Brevibacillus parabrevis BCP-09 and its biochemical degradation pathway. Biodegradation 2018, 29, 525–541. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.F.; Chowdhary, S.; Koksch, B.; Murphy, C.D. Biodegradation of amphipathic fluorinated peptides reveals a new bacterial defluorinating activity and a new source of natural organofluorine compounds. Environ. Sci. Technol. 2023, 57, 9762–9772. [Google Scholar] [CrossRef] [PubMed]
- Khan, M.F.; Murphy, C.D. Bacterial degradation of the anti-depressant drug fluoxetine produces trifluoroacetic acid and fluoride ion. Appl. Microbiol. Biotechnol. 2021, 105, 9359–9369. [Google Scholar] [CrossRef] [PubMed]
- Hof, C.; Khan, M.F.; Murphy, C.D. Endogenous production of 2-phenylethanol by Cunninghamella echinulata inhibits biofilm growth of the fungus. Fungal Biol. 2023, 127, 1384–1388. [Google Scholar] [CrossRef]
- Singh, S.; Mukherjee, A.; Jaiswal, D.K.; de Araujo Pereira, A.P.; Prasad, R.; Sharma, M.; Kuhad, R.C.; Shukla, A.C.; Verma, J.P. Advances and future prospects of pyrethroids: Toxicity and microbial degradation. Sci. Total Environ. 2022, 829, 154561. [Google Scholar] [CrossRef]
- Cycoń, M.; Piotrowska-Seget, Z. Pyrethroid-degrading microorganisms and their potential for the bioremediation of contaminated soils: A review. Front. Microbiol. 2016, 7, 1463. [Google Scholar] [CrossRef]
- Birolli, W.G.; da Silva, B.F.; Rodrigues Filho, E. Biodegradation of the pyrethroid cypermethrin by bacterial consortia collected from orange crops. Environ. Res. 2022, 215, 114388. [Google Scholar] [CrossRef]
- Liu, J.; Ding, Y.; Ma, L.; Gao, G.; Wang, Y. Combination of biochar and immobilized bacteria in cypermethrin-contaminated soil remediation. Int. Biodeter. Biodegr. 2017, 120, 15–20. [Google Scholar] [CrossRef]
- Zhang, H.; Zhang, Y.; Hou, Z.; Wang, X.; Wang, J.; Lu, Z.; Zhao, X.; Sun, F.; Pan, H. Biodegradation potential of deltamethrin by the Bacillus cereus strain Y1 in both culture and contaminated soil. Int. Biodeter. Biodegr. 2016, 106, 53–59. [Google Scholar] [CrossRef]
- Chen, S.; Dong, Y.H.; Chang, C.; Deng, Y.; Zhang, X.F.; Zhong, G.; Song, H.; Hu, M.; Zhang, L.H. Characterization of a novel cyfluthrin-degrading bacterial strain Brevibacterium aureum and its biochemical degradation pathway. Bioresour. Technol. 2013, 132, 16–23. [Google Scholar] [CrossRef]
- Chen, S.; Deng, Y.; Chang, C.; Lee, J.; Cheng, Y.; Cui, Z.; Zhou, J.; He, F.; Hu, M.; Zhang, L.H. Pathway and kinetics of cyhalothrin biodegradation by Bacillus thuringiensis strain ZS-19. Sci. Rep. 2015, 5, 8784. [Google Scholar] [CrossRef] [PubMed]
- Wang, T.; Hu, C.; Zhang, R.; Sun, A.; Li, D.; Shi, X. Mechanism study of cyfluthrin biodegradation by Photobacterium ganghwense with comparative metabolomics. Appl. Microbiol. Biotechnol. 2019, 103, 473–488. [Google Scholar] [CrossRef] [PubMed]
- Hu, G.P.; Zhao, Y.; Song, F.Q.; Liu, B.; Vasseur, L.; Douglas, C.; You, M.S. Isolation, identification and cyfluthrin-degrading potential of a novel Lysinibacillus sphaericus strain, FLQ-11-1. Res. Microbiol. 2014, 165, 110–118. [Google Scholar] [CrossRef]
- Tian, J.W.; Long, X.F.; Zhang, S.; Qin, Q.M.; Gan, L.Z.; Tian, Y.Q. Screening cyhalothrin degradation strains from locust epiphytic bacteria and studying Paracoccus acridae SCU-M53 cyhalothrin degradation process. Environ. Sci. Pollut. Res. 2018, 25, 11505–11515. [Google Scholar] [CrossRef]
- Palmer-Brown, W.; de Melo Souza, P.L.; Murphy, C.D. Cyhalothrin biodegradation in Cunninghamella elegans. Environ. Sci. Pollut. Res. Int. 2019, 26, 1414–1421. [Google Scholar] [CrossRef]
- Birolli, W.G.; Vacondio, B.; Alvarenga, N.; Seleghim, M.H.R.; Porto, A.L.M. Enantioselective biodegradation of the pyrethroid (+/)-lambda-cyhalothrin by marine-derived fungi. Chemosphere 2018, 197, 651–660. [Google Scholar] [CrossRef]
- Bhatt, P.; Bhatt, K.; Huang, Y.; Lin, Z.; Chen, S. Esterase is a powerful tool for the biodegradation of pyrethroid insecticides. Chemosphere 2020, 244, 125507. [Google Scholar] [CrossRef]
- Hu, W.; Lu, Q.; Zhong, G.; Hu, M.; Yi, X. Biodegradation of pyrethroids by a hydrolyzing carboxylesterase EstA from Bacillus cereus BCC01. Appl. Sci. 2019, 9, 477. [Google Scholar] [CrossRef]
- Gangola, S.; Sharma, A.; Bhatt, P.; Khati, P.; Chaudhary, P. Presence of esterase and laccase in Bacillus subtilis facilitates biodegradation and detoxification of cypermethrin. Sci. Rep. 2018, 8, 12755. [Google Scholar] [CrossRef]
- Hedges, L.; Brown, S.; MacLeod, A.K.; Moreau, M.; Yoon, M.; Creek, M.R.; Osimitz, T.G.; Lake, B.G. Metabolism of bifenthrin, β-cyfluthrin, λ-cyhalothrin, cyphenothrin and esfenvalerate by rat and human cytochrome P450 and carboxylesterase enzymes. Xenobiotica 2020, 50, 1434–1442. [Google Scholar] [CrossRef]
- Harkey, A.; Kim, H.J.; Kandagatla, S.; Raner, G.M. Defluorination of 4-fluorophenol by cytochrome P450BM3-F87G: Activation by long chain fatty aldehydes. Biotechnol. Lett. 2012, 34, 1725–1731. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Y.; Zhang, W.; Li, J.; Pang, S.; Mishra, S.; Bhatt, P.; Zeng, D.; Chen, S. Emerging technologies for degradation of dichlorvos: A review. Int. J. Environ. Res. Public Health 2021, 18, 5789. [Google Scholar] [CrossRef] [PubMed]
- Schmidt, S.; Wittich, R.M.; Erdmann, D.; Wilkes, H.; Francke, W.; Fortnagel, P. Biodegradation of diphenyl ether and its monohalogenated derivatives by Sphingomonas sp. strain SS3. Appl. Environ. Microbiol. 1992, 58, 2744–2750. [Google Scholar] [CrossRef]
- Gu, C.; Jin, Z.; Fan, X.; Ti, Q.; Yang, X.; Sun, C.; Jiang, X. Comparative evaluation and prioritization of key influences on biodegradation of 2, 2′, 4, 4′-tetrabrominated diphenyl ether by bacterial isolate B. xenovorans LB400. J. Environ. Manag. 2023, 331, 117320. [Google Scholar] [CrossRef]
- Hundt, K.; Jonas, U.; Hammer, E.; Schauer, F. Transformation of diphenyl ethers by Trametes versicolor and characterization of ring cleavage products. Biodegradation 1999, 10, 279–286. [Google Scholar] [CrossRef]

| CYP Name | Protein ID * | Source Organism | Protein Identity of CYP Orthologs (%) in Bacillus sp. MFK14 |
|---|---|---|---|
| CYP102A1 | P14779 | Bacillus megaterium | 65.0 |
| CYP102A2 | O08394 | Bacillus subtilis | 98.5 |
| CYP102A3 | O08336 | Bacillus subtilis | 99.3 |
| CYP102A4 | A0A6L7HJT4 | Bacillus anthracis | 75.2 |
| CYP102A5 | Q81BF4 | Bacillus cereus | 75.5 |
| CYP102A7 | NA | Bacillus licheniformis | 61.8 |
| CYP102A8 | NA | Bacillus thuringiensis | 75.8 |
| CYP102A9 | NA | Bacillus weihenstephanensis | 75.0 |
| CYP102A15 | NA | Bacillus pumilus | 60.7 |
| CYP106A1 | P14762 | Bacillus megaterium | 40.8 |
| CYP106A2 | Q06069 | Bacillus megaterium | 42.0 |
| CYP106B1 | A0A6L7HJD6 | Bacillus anthracis | 47.1 |
| CYP107DY1 | D5E3H2 | Bacillus subtilis | 42.8 |
| CYP107H1 | P53554 | Bacillus subtilis | 98.9 |
| CYP107J1 | O08469 | Bacillus subtilis | 94.6 |
| CYP107J2 | A0A6L7H515 | Bacillus anthracis | 57.6 |
| CYP107J3 | Q81CY0 | Bacillus cereus | 57.6 |
| CYP107K1 | O31785 | Bacillus subtilis | 98.9 |
| CYP109A1 | P27632 | Bacillus subtilis | 42.1 |
| CYP109B1 | O34374 | Bacillus subtilis | 93.0 |
| CYP134A1 | O34926 | Bacillus subtilis | 96.8 |
| CYP152A1 | O31440 | Bacillus subtilis | 95.2 |
| CYP197A1 | Q9KFA6 | Bacillus halodurans | 27.7 |
| Microorganism | Type | Concentration (mg/L) | Degradation (%) | Time (days) | Temperature (°C) | Reference |
|---|---|---|---|---|---|---|
| β-Cyfluthrin | ||||||
| Bacillus sp. MFK14 | Bacteria | 100 | 100 | 2 | 30 | Current study |
| Photobacterium ganghwense 6046 | Bacteria | 100 | 82.8 | 3 | 30 | [24] |
| Brevibacterium aureum DG-12 | Bacteria | 50 | 88.6 | 5 | 27 | [22] |
| Lysinibacillus sphaericus FLQ-11-1 | Bacteria | 50 | 80.4 | 5 | 35 | [25] |
| Cunninghamella spp. | Fungus | 100 | NA 1 | 5 | 28 | [10] |
| λ-cyhalothrin | ||||||
| Bacillus sp. MFK14 | Bacteria | 100 | 100 | 2 | 30 | Current study |
| Bacillus thuringiensis ZS-19 | Bacteria | 100 | 100 | 3 | 30 | [23] |
| Paracoccus acridae SCU-M53 | Bacteria | 75 | 79.84 | 2 | 28 | [26] |
| Cunninghamella elegans DSM1908 | Fungus | 100 | NA 1 | 5 | 28 | [27] |
| Aspergillus sp. CBMAI 1829 | Fungus | 100 | 44.8 | 14 | 32 | [28] |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khan, M.F.; Liao, J.; Liu, Z.; Chugh, G. Bacterial Cytochrome P450 Involvement in the Biodegradation of Fluorinated Pyrethroids. J. Xenobiot. 2025, 15, 58. https://doi.org/10.3390/jox15020058
Khan MF, Liao J, Liu Z, Chugh G. Bacterial Cytochrome P450 Involvement in the Biodegradation of Fluorinated Pyrethroids. Journal of Xenobiotics. 2025; 15(2):58. https://doi.org/10.3390/jox15020058
Chicago/Turabian StyleKhan, Mohd Faheem, Jun Liao, Zhenyang Liu, and Gaurav Chugh. 2025. "Bacterial Cytochrome P450 Involvement in the Biodegradation of Fluorinated Pyrethroids" Journal of Xenobiotics 15, no. 2: 58. https://doi.org/10.3390/jox15020058
APA StyleKhan, M. F., Liao, J., Liu, Z., & Chugh, G. (2025). Bacterial Cytochrome P450 Involvement in the Biodegradation of Fluorinated Pyrethroids. Journal of Xenobiotics, 15(2), 58. https://doi.org/10.3390/jox15020058








