Production of Ribosomal Protein S12/Renilla Luciferase Fusion and Development of a Bioluminescent Method for Detection of Aminoglycosides in Pork and Studying Its Recognition Mechanism
Abstract
:1. Introduction
2. Materials and Methods
2.1. Reagents
2.2. Expression of RpsL12–Rluc Fusion Protein
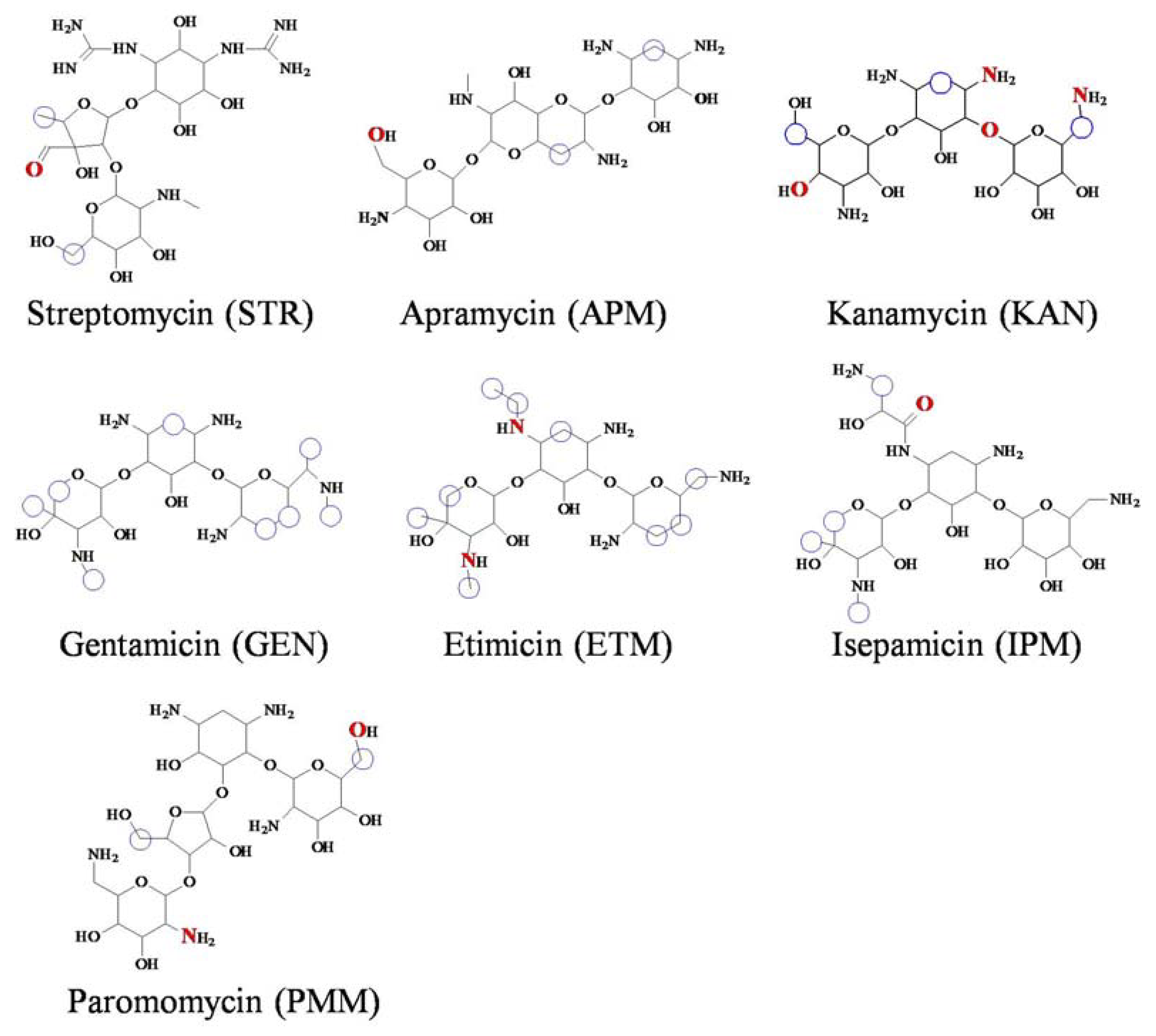
2.3. Recognition Mechanisms for AGs
2.4. Surface Plasmon Resonance (SPR)
2.5. Preparation of Coating Conjugate
2.6. Development of the Bioluminescent Method
2.7. Method Application
3. Results and Discussions
3.1. Characterization of the RpsL12–Rluc Fusion Protein
3.2. Recognition Mechanisms for AGs
3.3. Characterization of the Coating Conjugate
3.4. Optimization of Method Parameters
3.5. Method Performances
3.6. Sample Determination
3.7. Comparison with Related Immunoassays
4. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
References
- Selimoglu, E. Aminoglycoside-induced ototoxicity. Curr. Pharm. Des. 2007, 3, 119–126. [Google Scholar] [CrossRef]
- GB 31650-2019; National Food Safety Standard—Maximum Residue Limits for Veterinary Drugs in Foods. Ministry of Agriculture of China: Beijing, China, 2019.
- Frouk, F.; Azzazy, H.M.E.; Niessen, W.M.A. Challenges in the determination of aminoglycoside antibiotics, a review. Anal. Chim. Acta 2015, 890, 21–43. [Google Scholar] [CrossRef]
- Hari, R.; Taherunnisa, S.; Raut, S.Y.; Mutalik, S.; Koteshwara, K.B. Challenges in the development of analytical test procedure for aminoglycosides: A critical review. J. App. Pharm. Sci. 2019, 9, 145–152. [Google Scholar]
- Abuknesha, R.A.; Luk, C. Enzyme immunoassays for the analysis of streptomycin in milk, serum and water: Development and assessment of a polyclonal antiserum and assay procedures using novel streptomycin derivatives. Analyst 2005, 130, 964–970. [Google Scholar] [CrossRef]
- Chen, Y.; Wang, Z.; Wang, Z.; Tang, S.; Zhu, Y.; Xiao, X. Rapid enzyme-linked immunosorbent assay and colloidal gold immunoassay for kanamycin and tobramycin in swine tissues. J. Agric. Food Chem. 2008, 56, 2944–2952. [Google Scholar] [CrossRef]
- Ho, T.Y.J.; Chan, C.; Chan, K.; Wang, Y.C.; Lin, J.; Chang, C.; Chen, C. Development of a novel bead-based 96-well filtration plate competitive immunoassay for the detection of gentamycin. Biosens. Bioelectron. 2013, 49, 126–132. [Google Scholar] [CrossRef]
- Xu, F.; Jiang, W.; Zhou, J.; Wen, K.; Wang, Z.; Jiang, H.; Ding, S. Production of monoclonal antibody and development of a new immunoassay for apramycin in food. J. Agric. Food Chem. 2014, 62, 3108–3113. [Google Scholar] [CrossRef] [PubMed]
- Jiang, L.; Wei, D.; Zeng, K.; Shao, J.; Zhu, F.; Du, D. An enhanced direct competitive immunoassay for the detection of kanamycin and tobramycin in milk using multienzyme-particle amplification. Food Anal. Method. 2018, 11, 2066–2075. [Google Scholar] [CrossRef]
- Wei, D.; Meng, H.; Zeng, K.; Huang, Z. Visual dual dot immunoassay for the simultaneous detection of kanamycin and streptomycin in milk. Anal. Methods 2019, 11, 70–77. [Google Scholar] [CrossRef]
- Song, E.; Yu, M.; Wang, Y.; Hu, W.; Cheng, D.; Swihart, M.T.; Song, Y. Multi-color quantumdot-based fluorescence immunoassay array for simultaneous visual detection of multiple antibiotic residues in milk. Biosens. Bioelectron. 2015, 72, 320–325. [Google Scholar] [CrossRef] [PubMed]
- Beloglazova, N.V.; Shmelin, P.S.; Eremin, S.A. Sensitive immunochemical approaches for quantitative (FPIA) and qualitative (lateral flow tests) determination of gentamicin in milk. Talanta 2016, 149, 217–224. [Google Scholar] [CrossRef] [PubMed]
- Li, C.; Zhang, Y.; Eremin, S.A.; Yakup, O.; Yao, G.; Zhang, X. Detection of kanamycin and gentamicin residues in animal-derived food using IgY antibody based ic-ELISA and FPIA. Food Chem. 2017, 227, 48–54. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Luo, P.J.; Zhang, J.B.; Wang, H.L.; Chen, X.; Wu, N.; Zhao, Y.F.; Wang, X.M.; Zhang, H.; Zhang, J.Y.; Zhu, L.; et al. Rapid and sensitive chemiluminescent enzyme immunoassay for the determination of neomycin residues in milk. Biomed. Environ. Sci. 2016, 29, 374–378. [Google Scholar] [PubMed]
- Hendrickson, O.D.; Byzova, N.A.; Zvereva, E.A.; Zherdev, A.V.; Dzantiev, B.B. Sensitive lateral flow immunoassay of an antibiotic neomycin. J. Food Sci. Technol. 2021, 58, 292–301. [Google Scholar] [CrossRef] [PubMed]
- Sun, Y.; Yang, J.; Yang, S.; Sang, Q.; Teng, M.; Li, Q.; Deng, R.; Feng, L.; Hu, X.; Zhang, G. Development of an immunochromatographic lateral flow strip for the simultaneous detection of aminoglycoside residues in milk. RSC Adv. 2018, 8, 9580. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wei, D.; Zhang, X.; Chen, B.; Zeng, K. Using bimetallic Au@Pt nanozymes as a visual tag and as an enzyme mimic in enhanced sensitive lateral-flow immunoassays: Application for the detection of streptomycin. Anal. Chim. Acta. 2020, 1126, 106–113. [Google Scholar] [CrossRef]
- Ahmed, S.; Ning, J.; Cheng, G.; Ahmad, I.; Li, J.; Mingyue, L.; Qu, W.; Iqbal, M.; Shabbir, M.A.B.; Yuan, Z. Receptor-based screening assays for the detection of antibiotics residues-A review. Talanta 2017, 166, 176–186. [Google Scholar] [CrossRef]
- Santillo, M.F. Trends using biological target-based assays for drug detection in complex sample matrices. Anal. Bioanal. Chem. 2020, 412, 3975–3982. [Google Scholar] [CrossRef]
- He, T.; Cui, P.L.; Liu, J.; Feng, C.; Wang, J.P. Production of a natural dihydropteroate synthase and development of a signal amplified pseudo immunoassay for determination of sulfonamides in pork. J. Agric. Food Chem. 2022, 70, 3023–3032. [Google Scholar] [CrossRef]
- Carter, A.P.; Clemons, W.M.; Brodersen, D.E.; Morgan-Warren, R.J.; Wimberly, B.T.; Ramakrishnan, V. Functional insights from the structure of the 30S ribosomal subunit and its interactions with antibiotics. Nature 2000, 407, 340–348. [Google Scholar] [CrossRef] [PubMed]
- Magnet, S.; Blanchard, J.S. Molecular insights into aminoglycoside action and resistance. Chem. Rev. 2005, 105, 477–497. [Google Scholar] [CrossRef]
- Fosso, M.Y.; Li, Y.; Garneau-Tsodikova, S. New trends in the use of aminoglycosides. MedChemCommun. 2014, 5, 1075–1091. [Google Scholar] [CrossRef] [Green Version]
- Sreevatsan, S.; Pan, X.; Stockbauer, K.E.; Williams, D.L.; Kreiswirth, B.N.; Musser, J.M. Characterization of rpsL and rrs mutations in streptomycin-resistant Mycobacterium tuberculosis isolates from diverse geographic localities. Antimicrob. Agents. Chemother. 1996, 40, 1024–1026. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Suriyanarayanan, B.; Lakshmi, P.P.; Santhosh, R.S.; Dhevendaran, K.; Priya, B.; Krishna, S. Streptomycin affinity depends on 13 amino acids forming a loop in homology modeled ribosomal S12 protein (rpsL gene) of Lysinibacillus sphaericus DSLS5 associated with marine sponge (Tedania anhelans). J. Biomol. Struct. Dyn. 2016, 34, 1190–1200. [Google Scholar] [CrossRef]
- Xia, W.Q.; Zhang, L.; Wang, J.P. Development of a fluorescence polarization assay for multi-determination of 10 aminoglycosides in pork muscle sample based on ribosomal protein S12 and studying its recognition mechanism. Foods 2022, 11, 3196. [Google Scholar] [CrossRef]
- Zhang, Z.; Lai, J.; Wu, K.; Huang, X.; Guo, S.; Zhang, L.; Liu, J. Peroxidase-catalyzed chemiluminescence system and its application inimmunoassay. Talanta 2018, 180, 260–270. [Google Scholar] [CrossRef] [PubMed]
- Bellah, S.F.; MensahSedzro, D.; Akbar, H.; Billah, S.M.S. Structure, enzymatic mechanism of action, applications, advantages, disadvantages and modifications of luciferase enzyme. J. Gene. Engg. Bio. Res. 2019, 1, 1–6. [Google Scholar]
- Lomakina, G.Y.; Istrate, A.; Rudenko, N.V.; Ugarova, N.N. Synthesis and application of firefly luciferase antibody conjugates in a bioluminescent immunoassay of Salmonella cells. Mosc. Univ. Chem. Bull. 2014, 69, 49–55. [Google Scholar] [CrossRef]
- Yu, S.; Li, Z.; Li, J.; Zhao, S.; Wu, S.; Liu, H.; Bi, X.; Li, D.; Dong, J.; Duan, S.; et al. Generation of dual functional nanobody-nanoluciferase fusion and its potential in bioluminescence enzyme immunoassay for trace glypican-3 in serum. Sensor. Actuat B. 2021, 336, 129717. [Google Scholar] [CrossRef]
- Wang, G.; Mashimo, Y.; Kobatake, E.; Mie, M. Development of an enhanced immunoassay based on protein nanoparticles displaying an IgG-binding domain and luciferase. Anal. Bioanal. Chem. 2022, 414, 2079–2088. [Google Scholar] [CrossRef]
- Oyama, H.; Morita, I.; Kiguchi, Y.; Miyake, S.; Moriuchi, A.; Akisada, T.; Niwa, T.; Kobayashi, N. Gaussia luciferase as a genetic fusion partner with antibody fragments for sensitive immunoassay monitoring of clinical biomarkers. Anal. Chem. 2015, 87, 12387–12395. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.; Zhang, X.; Wang, Z.; Jiang, H.; Lv, Z.; Shen, J.; Xia, G.; Wen, K. Universal simultaneous multiplex ELISA of small molecules in milk based on dual luciferases. Anal. Chim. Acta. 2018, 1001, 125–133. [Google Scholar] [CrossRef]
- Ren, W.; Li, Z.; Xu, Y.; Wan, D.; Barnych, B.; Li, Y.; Tu, Z.; He, Q.; Fu, J.; Hammock, B.D. One-step ultrasensitive bioluminescent enzyme immunoassay based on nanobody/nanoluciferase fusion for detection of aflatoxin B1 in cereal. J. Agric. Food Chem. 2019, 67, 5221–5229. [Google Scholar] [CrossRef] [PubMed]
- Li, Z.; Wang, Y.; Vasylieva, N.; Wan, D.; Yin, Z.; Dong, J.; Hammock, B.D. An ultrasensitive bioluminescent enzyme immunoassay based on nanobody/ nanoluciferase heptamer fusion for the detection of tetrabromobisphenol A in sediment. Anal. Chem. 2020, 92, 10083–10090. [Google Scholar] [CrossRef]
- Wang, G.; Zhang, H.C.; Liu, J.; Wang, J.P. A receptor-based chemiluminescence enzyme linked immunosorbent assay for determination of tetracyclines in milk. Anal. Biochem. 2019, 564–565, 40–46. [Google Scholar] [CrossRef]
- He, T.; Liu, J.; Wang, J.P. Development of a dihydropteroate synthase based fluorescence polarization assay for detection of sulfonamides and studying its recognition mechanism. J. Agric. Food Chem. 2021, 69, 13953–13963. [Google Scholar] [CrossRef] [PubMed]







| Drug | Binding Energy (kcal/mol) | Contact Amino Acid | Ka (1/Ms) | Kd (1/s) | Kd (1/s) | KA | IC50 (ng/mL) | LOD (ng/mL) | |
|---|---|---|---|---|---|---|---|---|---|
| Hydrogen Bond | Hydrophobic Interaction | ||||||||
| STR | 6.02 | GLY69 | GLY68 VAL87 ASP89 HIS96 TYR117 | 2.05 × 10³ | 5.19 × 10−5 | 2.53 × 10−8 | 25.235 | 43.6 | 0.73 |
| APM | 5.99 | TYR66 | TYR66 GLY68 VAL87 TYR117 | 1.90 × 102 | 1.27 × 10−4 | 6.67 × 10−7 | 20.515 | 63.1 | 0.86 |
| GEN | 6.29 | -- | PRO42 ASN46 ALA48 ARG50 TYR66 GLY68 VAL87 HIS96 LYS116 TYR117 | 4.46 × 102 | 2.57 × 10−5 | 5.77 × 10−8 | 24.048 | 57.9 | 0.68 |
| KAN | 5.6 | TYR66 GLU70 ARG86 LYS88 TYR117 | GLY68 GLU70 LYS88 ASP89 TYR117 | 4.98 × 102 | 2.83 × 10−4 | 5.69 × 10−7 | 20.746 | 60.1 | 0.84 |
| ETM | 5.44 | HIS96 TYR117 | ARG50 TYR66 GLY68 GLY69 GLU70 ARG83 ARG86 LYS88 HIS96 TYR117 | 8.82 × 101 | 1.52 × 10−2 | 1.72 × 10−4 | 12.502 | 75.5 | 1.1 |
| IPM | 6.32 | LYS116 | GLY68 VAL87 LYS88 ASP89 TYR117 | 4.37 × 10³ | 1.70 × 10−3 | 3.90 × 10−7 | 21.290 | 52.6 | 0.51 |
| PMM | 5.61 | ARG83 ASP89 | LYS88 TYR117 | 4.61 × 102 | 9.97 × 10−5 | 2.16 × 10−7 | 22.140 | 67.5 | 0.82 |
| Analyte | Fortified (ng/g) | Intra-Day | Inter-Day | Analyte | Fortified (ng/g) | Intra-Day | Inter-Day | ||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Recovery (%) | CV (%) | Recovery (%) | CV (%) | Recovery (%) | CV (%) | Recovery (%) | CV (%) | ||||
| STR | 10 | 78.4 | 5.9 | 82.7 | 9.6 | IPM | 10 | 80.5 | 6.8 | 77.5 | 8.9 |
| 100 | 87.5 | 8.8 | 89.9 | 11.5 | 50 | 87.4 | 8.7 | 81.7 | 9.9 | ||
| 600 | 88.3 | 7.7 | 82.0 | 9.0 | 100 | 83.2 | 8.1 | 94.3 | 9.3 | ||
| APM | 10 | 89.5 | 7.8 | 90.4 | 8.9 | PMM | 10 | 89.1 | 7.7 | 85.7 | 8.8 |
| 50 | 84.6 | 8.5 | 94.2 | 8.4 | 50 | 96.2 | 7.4 | 89.1 | 11.7 | ||
| 100 | 93.8 | 6.3 | 91.8 | 10.7 | 100 | 91.5 | 8.6 | 82.4 | 12.9 | ||
| GEN | 10 | 79.4 | 9.4 | 78.5 | 9.5 | ETM | 10 | 73.8 | 8.3 | 91.9 | 10.2 |
| 50 | 83.8 | 8.3 | 86.7 | 8.9 | 50 | 95.7 | 7.9 | 94.8 | 8.6 | ||
| 100 | 88.4 | 8.8 | 83.7 | 8.7 | 100 | 91.6 | 7.2 | 90.7 | 8.0 | ||
| KAN | 10 | 80.6 | 7.4 | 93.7 | 12.8 | ||||||
| 50 | 84.8 | 9.1 | 92.7 | 10.7 | |||||||
| 100 | 92.7 | 9.4 | 88.6 | 8.4 | |||||||
| Recognition Element | Immunoassay | Detection Spectrum | Test Time (after Add Sample) | Limit of Detection(ng/g) | Ref. |
|---|---|---|---|---|---|
| Streptomycin pAb | ELISA | 1 drug | 30 min | 1 | [5] |
| Kanamycin mAb | ELISA | 2 AGs | 30 min | 0.9–1.8 | [6] |
| Gentamycin pAb | ELISA | 1 drug | 2 h | 14.16 | [7] |
| Kanamycin mAb | ELISA | 2 AGs | 100 min | 0.022 | [9] |
| Streptomycin mAb | ELISA | 2 AGs | 40 min | 0.09–1.37 | [10] |
| Streptomycin mAb | fluorescence ELISA | 1 drug | 60 min | 0.005 | [11] |
| Commercial Ab | chemiluminescent ELISA | 1 drug | 60 min | 9.4 | [14] |
| E. coli RpsL12 | bioluminescent method | 7 AGs | 30 min | 0.51–1.1 | This study |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xia, W.; Zhang, L.; Wang, J. Production of Ribosomal Protein S12/Renilla Luciferase Fusion and Development of a Bioluminescent Method for Detection of Aminoglycosides in Pork and Studying Its Recognition Mechanism. Foods 2023, 12, 284. https://doi.org/10.3390/foods12020284
Xia W, Zhang L, Wang J. Production of Ribosomal Protein S12/Renilla Luciferase Fusion and Development of a Bioluminescent Method for Detection of Aminoglycosides in Pork and Studying Its Recognition Mechanism. Foods. 2023; 12(2):284. https://doi.org/10.3390/foods12020284
Chicago/Turabian StyleXia, Wanqiu, Lei Zhang, and Jianping Wang. 2023. "Production of Ribosomal Protein S12/Renilla Luciferase Fusion and Development of a Bioluminescent Method for Detection of Aminoglycosides in Pork and Studying Its Recognition Mechanism" Foods 12, no. 2: 284. https://doi.org/10.3390/foods12020284
APA StyleXia, W., Zhang, L., & Wang, J. (2023). Production of Ribosomal Protein S12/Renilla Luciferase Fusion and Development of a Bioluminescent Method for Detection of Aminoglycosides in Pork and Studying Its Recognition Mechanism. Foods, 12(2), 284. https://doi.org/10.3390/foods12020284






