Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area and Soil Sampling
2.2. Analysis of Soil Chemical and Biological Properties
2.3. DNA Extraction, High-throughput Sequencing and Data Analysis
2.4. Statistical Analysis
3. Results
3.1. Soil Biochemical Properties
3.2. Compositions and Structures of Bacterial Communities
3.3. Relationships between Bacterial Communities and Soil Biochemical Properties
4. Discussion
4.1. Effect of Vegetation Restoration on Soil Biochemical Properties
4.2. Effect of Vegetation Restoration on Bacterial Communities
4.3. Response of Soil Bacterial Communities to Soil Biochemical Properties
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Wang, L.; Zhang, W.; Guo, G.; Qian, K.; Huang, X. Selection experiments for the optimum combination of AMF-plant-substrate for the restoration of coal mines. Min. Sci. Technol. 2009, 19, 479–482. [Google Scholar] [CrossRef]
- Li, Z.; Liu, W.Z.; Zhang, X.C.; Zheng, F.L. Assessing the site-specific impacts of climate change on hydrology, soil erosion and crop yields in the Loess Plateau of China. Clim. Chang. 2011, 105, 223–242. [Google Scholar] [CrossRef]
- Ingram, L.J.; Schuman, G.E.; Stahl, P.D.; Spackman, L.K. Microbial respiration and organic carbon indicate nutrient cycling recovery in reclaimed soils. Soil Sci. Soc. Am. J. 2005, 69, 1737–1745. [Google Scholar] [CrossRef]
- Li, Y.; Wen, H.; Chen, L.; Yin, T. Succession of bacterial community structure and diversity in soil along a chronosequence of reclamation and re-vegetation on coal mine spoils in China. PLoS ONE 2014, 9, e115024. [Google Scholar] [CrossRef] [PubMed]
- Jason, G.; Murray, W.; John, D. Computational improvements reveal great bacterial diversity and high metal toxicity in soil. Science 2005, 309, 1387. [Google Scholar] [CrossRef]
- Steenwerth, K.L.; Jackson, L.E.; Calderón, F.J.; Stromberg, M.R.; Scow, K.M. Soil microbial community composition and land use history in cultivated and grassland ecosystems of coastal California. Soil Biol. Biochem. 2003, 34, 1599–1611. [Google Scholar] [CrossRef]
- Leff, J.W.; Jones, S.E.; Prober, S.M.; Albert, B.; Borer, E.T.; Firn, J.L.; W Stanley, H.; Hobbie, S.E.; Hofmockel, K.S.; Knops, J.M.H. Consistent responses of soil microbial communities to elevated nutrient inputs in grasslands across the globe. Proc. Natl. Acad. Sci. USA 2015, 112, 10967. [Google Scholar] [CrossRef] [PubMed]
- Delgado-Baquerizo, M.; Eldridge, D.J.; Ochoa, V.; Gozalo, B.; Singh, B.K.; Maestre, F.T. Soil microbial communities drive the resistance of ecosystem multifunctionality to global change in drylands across the globe. Ecol. Lett. 2017, 20, 1295. [Google Scholar] [CrossRef] [PubMed]
- Cui, Y.; Fang, L.; Guo, X.; Xia, W.; Wang, Y.; Zhang, Y.; Zhang, X. Responses of soil bacterial communities, enzyme activities and nutrients to agricultural-to-natural ecosystem conversion in the Loess Plateau, China. J. Soils Sediment. 2018, 1–14. [Google Scholar] [CrossRef]
- Chen, D.; Xing, W.; Lan, Z.; Saleem, M.; Wu, Y.; Hu, S.; Bai, Y. Direct and indirect effects of nitrogen enrichment on soil organisms and carbon and nitrogen mineralization in a semi-arid grassland. Funct. Ecol. 2019, 33175–33187. [Google Scholar] [CrossRef]
- Burke, D.J.; Weintraub, M.N.; Hewins, C.R.; Kalisz, S. Relationship between soil enzyme activities, nutrient cycling and soil fungal communities in a northern hardwood forest. Soil Biol. Biochem. 2011, 43, 795–803. [Google Scholar] [CrossRef]
- Gómez-Sagasti, M.T.; Alkorta, I.; Becerril, J.M.; Epelde, L.; Anza, M.; Garbisu, C. Microbial Monitoring of the Recovery of Soil Quality During Heavy Metal Phytoremediation. Water Air Soil Pollut. 2012, 223, 3249–3262. [Google Scholar] [CrossRef]
- Masto, R.E.; Sheik, S.; Nehru, G.; Selvi, V.A.; George, J.; Ram, L.C. Assessment of environmental soil quality around Sonepur Bazari mine of Raniganj coalfield, India. Solid Earth 2015, 6, 811–821. [Google Scholar] [CrossRef] [Green Version]
- Liu, X.; Bai, Z.; Zhou, W.; Cao, Y.; Zhang, G. Changes in soil properties in the soil profile after mining and reclamation in an opencast coal mine on the Loess Plateau, China. Ecol. Eng. 2017, 98, 228–239. [Google Scholar] [CrossRef]
- Sheoran, V.; Sheoran, A.S.; Poonia, P. Soil reclamation of abandoned mine land by revegetation: A review. Int. J. Soil Sediment. Water 2010, 3, 1–4. [Google Scholar]
- Zhou, W.; Yang, K.; Bai, Z.; Cheng, H.; Liu, F. The development of topsoil properties under different reclaimed land uses in the Pingshuo opencast coalmine of Loess Plateau of China. Ecol. Eng. 2017, 100, 237–245. [Google Scholar] [CrossRef]
- Huang, L.; Zhang, P.; Hu, Y.; Zhao, Y. Vegetation and soil restoration in refuse dumps from open pit coal mines. Ecol. Eng. 2016, 94, 638–646. [Google Scholar] [CrossRef]
- Kaschuk, G.; Alberton, O.; Hungria, M. Three decades of soil microbial biomass studies in Brazilian ecosystems: Lessons learned about soil quality and indications for improving sustainability. Soil Biol. Biochem. 2010, 42, 1–13. [Google Scholar] [CrossRef]
- Liu, J.; Sui, Y.; Yu, Z.; Shi, Y.; Chu, H.; Jin, J.; Liu, X.; Wang, G. High throughput sequencing analysis of biogeographical distribution of bacterial communities in the black soils of northeast China. Soil Biol. Biochem. 2014, 70, 113–122. [Google Scholar] [CrossRef]
- Zhen, Q.; Ma, W.; Li, M.; He, H.; Zhang, X.; Wang, Y. Effects of vegetation and physicochemical properties on solute transport in reclaimed soil at an opencast coal mine site on the Loess Plateau, China. Catena 2015, 133, 403–411. [Google Scholar] [CrossRef]
- Zhang, P.; Zhao, Y.; Huang, L.; Yigang, H.U.; Han, X. Effect of revegetation on soil extracellular enzyme activity in the dumping site of an open-pit coal mine in Heidaigou. Acta Ecol. Sin. 2016, 36, 2715–2723. (In Chinese) [Google Scholar]
- Nelson, D.W.; Sommers, L.E.; Sparks, D.L.; Page, A.L.; Helmke, P.A.; Loeppert, R.H.; Soltanpour, P.N.; Tabatabai, M.A.; Johnston, C.T.; Sumner, M.E. Total carbon, organic carbon and organic matter. Methods Soil Anal. 1996, 9, 961–1010. [Google Scholar] [CrossRef]
- Walmsley, D.; Cornforth, I.S. Methods of measuring available nutrients in West Indian soils. Plant Soil 1973, 39, 93–101. [Google Scholar] [CrossRef]
- Olsen, S.R.; Cole, C.V.; Watanabe, F.S.; Dean, L.A. Estimation of Available Phosphorus in Soils by Extraction with Sodium Biocarbonate; USDA Circular No. 939; USDA: Washington, DC, USA, 1954.
- Guan, S.Y. Study Way of Soil Enzymes; Agriculture Press: Beijing, China, 1986. [Google Scholar]
- Alef, K.; Nannipieri, P. Methods in Applied Soil Microbiology and Biochemistry; Academic Press: Cambridge, MA, USA, 1995. [Google Scholar]
- Brookes, P.C.; Landman, A.; Pruden, G.; Jenkinson, D.S. Chloroform fumigation and the release of soil nitrogen: A rapid direct extraction method to measure microbial biomass nitrogen in soil. Soil Biol. Biochem. 1985, 17, 837–842. [Google Scholar] [CrossRef]
- Cui, Y.; Fang, L.; Guo, X.; Wang, X.; Wang, Y.; Li, P.; Zhang, Y.; Zhang, X. Responses of soil microbial communities to nutrient limitation in the desert-grassland ecological transition zone. Sci. Total Environ. 2018, 642, 45–55. [Google Scholar] [CrossRef]
- Bolger, A.M.; Marc, L.; Bjoern, U. Trimmomatic: A flexible trimmer for Illumina sequence data. Bioinformatics 2014, 30, 2114–2120. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Thomas, R.; Hall, J.R.; Martin, H.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009. [CrossRef]
- Wang, Q.; Garrity, G.M.; Tiedje, J.M.; Cole, J.R. Naïve bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy. Appl. Environ. Microbiol. 2007, 73, 5261–5267. [Google Scholar] [CrossRef]
- Chen, H.; Boutros, P.C. VennDiagram: A package for the generation of highly-customizable Venn and Euler diagrams in R. BMC Bioinform. 2011, 12, 35. [Google Scholar] [CrossRef]
- Wang, H.; Wang, J.; Cao, Y.; Yuanqing, L.U.; Qin, Q.; Wang, Y. Effect of soil and topography on vegetation restoration in an opencast coal mine dump in a loess area. Sci. Rep. 2016, 6. [Google Scholar] [CrossRef]
- Yang, H.; Zhang, F.; Chen, Y.; Xu, T.; Cheng, Z.; Liang, J. Assessment of Reclamation Treatments of Abandoned Farmland in an Arid Region of China. Sustainability 2016, 8, 1183. [Google Scholar] [CrossRef]
- Wei, X.; Huang, M.; Shao, M.; Li, L.; Zhang, X.; Horton, R. Shrubs increase soil resources heterogeneity along semiarid grass slopes in the Loess Plateau. J. Arid Environ. 2013, 88, 175–183. [Google Scholar] [CrossRef]
- Reynolds, J.F.; Virginia, R.A.; Kemp, P.R.; Soyza, A.G.D.; Tremmel, D.C. Impact of Drought on Desert Shrubs: Effects of Seasonality and Degree of Resource Island Development. Ecol. Monogr. 1999, 69, 69–106. [Google Scholar] [CrossRef]
- Luca, R.; Francesco, L.; Paolo, D.O. Fertility Island Formation and Evolution in Dryland Ecosystems. Ecol. Soc. 2008, 13. [Google Scholar] [CrossRef]
- Saleem, M.; Pervaiz, Z.H.; Traw, M.B. Theories, mechanisms and patterns of microbiome species coexistence in an era of climate change. In Microbiome Community Ecology; Saleem, M., Ed.; Springer: New York, NY, USA, 2015; pp. 15–53. [Google Scholar]
- Li, J.H.; Fang, X.W.; Jia, J.J.; Wang, G. Effect of legume species introduction to early abandoned field on vegetation development. Plant Ecol. 2007, 191, 1–9. [Google Scholar] [CrossRef]
- Buckley, D.H.; Schmidt, T.M. The structure of microbial communities in soil and the lasting impact of cultivation. Microb. Ecol. 2001, 42, 11–21. [Google Scholar] [CrossRef]
- Jangid, K. Land-use history has a stronger impact on soil microbial community composition than aboveground vegetation and soil properties. Soil Biol. Biochem. 2011, 43, 2184–2193. [Google Scholar] [CrossRef]
- Jangid, K.; Williams, M.A.; Franzluebbers, A.J.; Blair, J.M.; Coleman, D.C.; Whitman, W.B. Development of soil microbial communities during tallgrass prairie restoration. Soil Biol. Biochem. 2010, 42, 302–312. [Google Scholar] [CrossRef]
- Li, Y.; Chen, L.; Wen, H. Changes in the composition and diversity of bacterial communities 13 years after soil reclamation of abandoned mine land in eastern China. Ecolo. Res. 2015, 30, 357–366. [Google Scholar] [CrossRef]
- Rastogi, G.; Osman, S.; Vaishampayan, P.A.; Andersen, G.L.; Stetler, L.D.; Sani, R.K. Microbial Diversity in Uranium Mining-Impacted Soils as Revealed by High-Density 16S Microarray and Clone Library. Microb. Ecol. 2010, 59, 94–108. [Google Scholar] [CrossRef]
- Qiu, M.; Zhang, R.; Xue, C.; Zhang, S.; Li, S.; Zhang, N.; Shen, Q. Application of bio-organic fertilizer can control Fusarium wilt of cucumber plants by regulating microbial community of rhizosphere soil. Biol. Fertil. Soils 2012, 48, 807–816. [Google Scholar] [CrossRef]
- Melanie, K.; Brigitte, H.; Thomas, R.; Christelle, M.; Michael, S.; Sophie, Z.B.; Robert, J.; Andreas, S.; Angela, S. Effects of season and experimental warming on the bacterial community in a temperate mountain forest soil assessed by 16S rRNA gene pyrosequencing. FEMS Microbiol. Ecol. 2012, 82, 551–562. [Google Scholar] [CrossRef] [Green Version]
- Wallenstein, M.; Mcmahon, S.; Schimel, J. Bacterial and fungal community structure in Arctic tundra tussock and shrub soils. FEMS Microbiol. Ecol. 2010, 59, 428–435. [Google Scholar] [CrossRef] [PubMed]
- Dedysh, S.N.; Liesack, W.; Ricke, P. NifH and NifD phylogenies:an evolutionary basis for understanding nitrogen fixation capabilities of methanotrophic bacteria. Microbiology 2004, 150, 1301. [Google Scholar] [CrossRef]
- Janssen, P.H. Identifying the dominant soil bacterial taxa in libraries of 16S rRNA and 16S rRNA genes. Appl. Environ. Microbiol. 2006, 72, 1719–1728. [Google Scholar] [CrossRef]
- Naether, A.; Foesel, B.U.; Naegele, V.; Wüst, P.K.; Weinert, J.; Bonkowski, M.; Alt, F.; Oelmann, Y.; Polle, A.; Lohaus, G.; et al. Environmental Factors Affect Acidobacterial Communities below the Subgroup Level in Grassland and Forest Soils. Appl. Environ. Microbiol. 2012, 78, 7398–7406. [Google Scholar] [CrossRef] [Green Version]
- Rojas, C.; Gutierrez, R.M.; Bruns, M.A. Bacterial and eukaryal diversity in soils forming from acid mine drainage precipitates under reclaimed vegetation and biological crusts. Appl. Soil Ecol. 2016, 105, 57–66. [Google Scholar] [CrossRef] [Green Version]
- Liu, C.; Zuo, W.; Zhao, Z. Bacterial diversity of different successional stage forest soils in Dinghushan. Acta Microbiol. Sin. 2012, 52, 1489. (In Chinese) [Google Scholar]
- Sui, X.; Zhang, R.T.; Zhong, H.X.; Nan, X.U.; Wang, J.F.; Liu, Y.Z.; Yuan, H.F.; Hong-Wei, N.I. Study on Bacterial Diversity of Deyeuxia angustifolia Wetland by Application of High-throughput Sequencing Technology in Sanjiang Plain. Soils 2015, 47, 919–925. (In Chinese) [Google Scholar]
- Zhang, C.; Liu, G.; Sha, X.; Wang, G. Soil bacterial community dynamics reflect changes in plant community and soil properties during the secondary succession of abandoned farmland in the Loess Plateau. Soil Biol. Biochem. 2016, 97, 40–49. [Google Scholar] [CrossRef]
- Peacock, A.D.; Macnaughton, S.J.; Cantu, J.M.; Dale, V.H.; White, D.C. Soil microbial biomass and community composition along an anthropogenic disturbance gradient within a long-leaf pine habitat. Ecol. Indic. 2002, 1, 113–121. [Google Scholar] [CrossRef]
- Noah, F.; Bradford, M.A.; Jackson, R.B. Toward an ecological classification of soil bacteria. Ecology 2007, 88, 1354–1364. [Google Scholar] [CrossRef]
- Nemergut, D.R.; Cleveland, C.C.; Wieder, W.R.; Washenberger, C.L.; Townsend, A.R. Plot-scale manipulations of organic matter inputs to soils correlate with shifts in microbial community composition in a lowland tropical rain forest. Soil Biol. Biochem. 2011, 42, 2153–2160. [Google Scholar] [CrossRef]



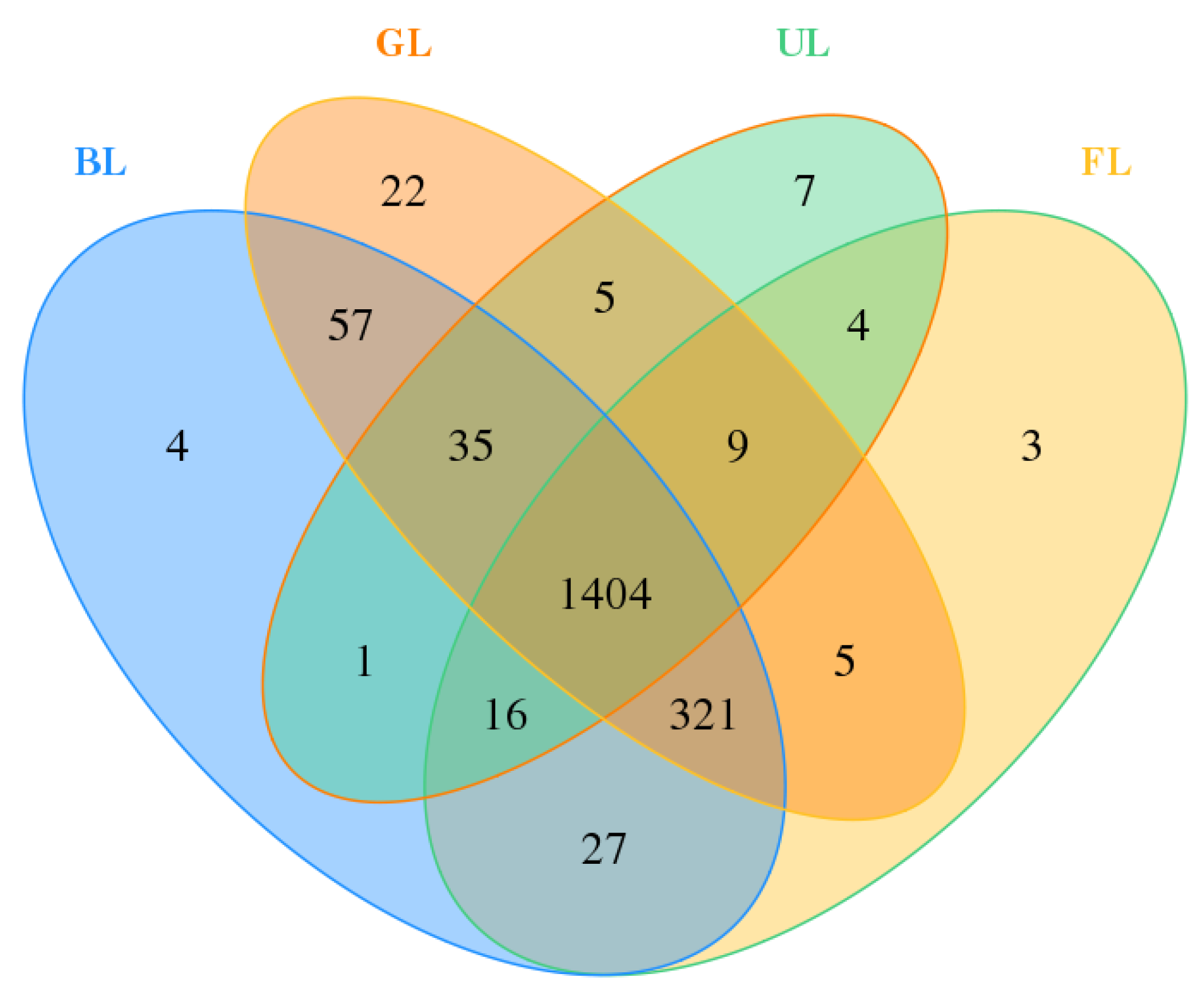

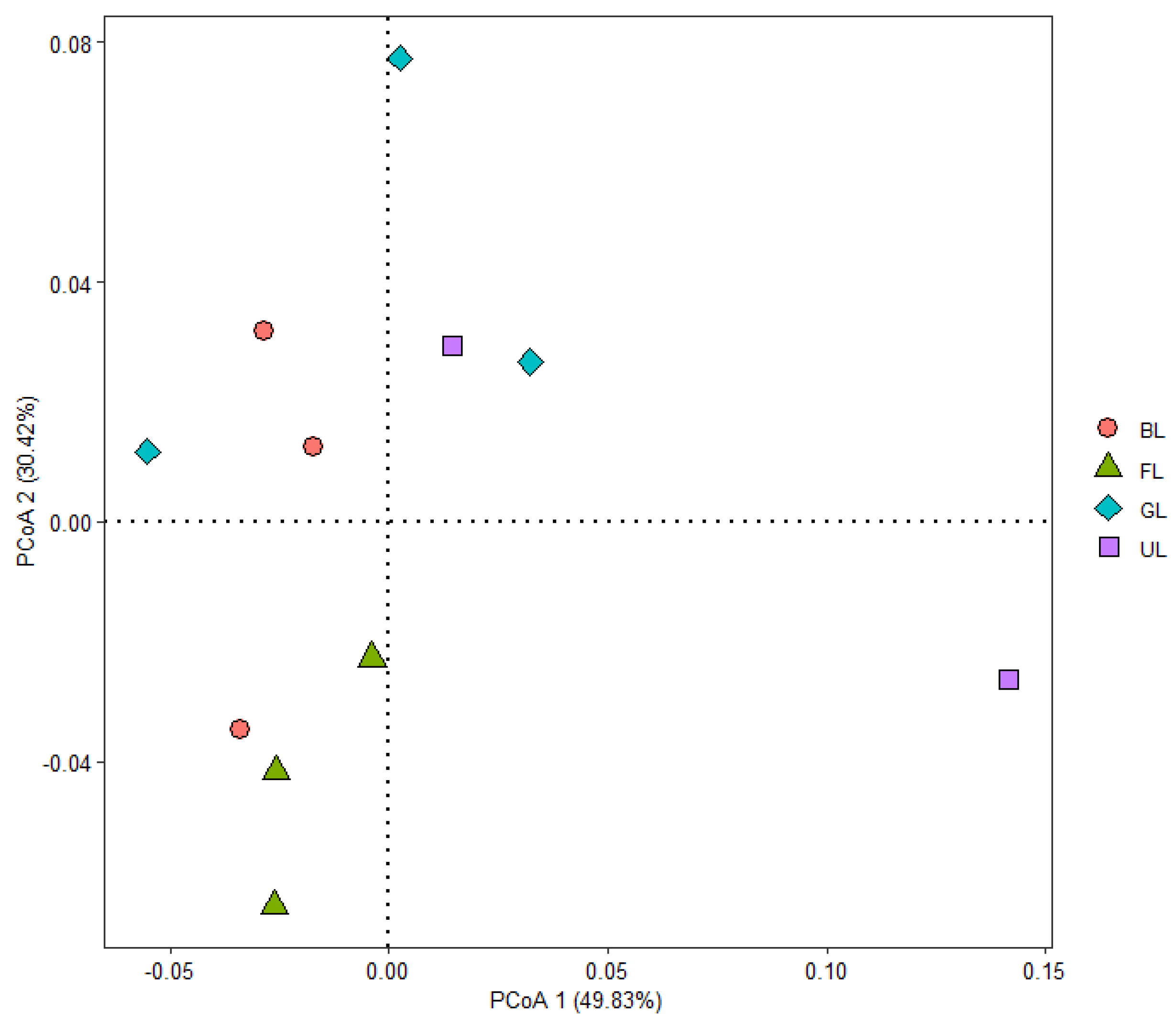


| Site | OTUs | ACE | Chao1 | Shannon | Simpson | Coverage |
|---|---|---|---|---|---|---|
| BL | 1664 ± 56a | 1749 ± 46a | 1773 ± 35a | 6.26 ± 0.07a | 0.006 ± 0b | 0.994 ± 0a |
| FL | 1579 ± 50a | 1668 ± 30a | 1675 ± 32a | 6.03 ± 0.04a | 0.009 ± 0ab | 0.994 ± 0a |
| GL | 1492 ± 10a | 1593 ± 34a | 1624 ± 44a | 6.21 ± 0.05a | 0.006 ± 0b | 0.993 ± 0a |
| UL | 1118 ± 264b | 1331 ± 192b | 1335 ± 193b | 5.59 ± 0.50b | 0.012 ± 0a | 0.986 ± 0a |
| F | 11.288 | 11.628 | 12.354 | 5.479 | 4.477 | 2.792 |
| P | 0.005 | 0.004 | 0.003 | 0.030 | 0.047 | 0.119 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Li, P.; Zhang, X.; Hao, M.; Cui, Y.; Zhu, S.; Zhang, Y. Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China. Sustainability 2019, 11, 2295. https://doi.org/10.3390/su11082295
Li P, Zhang X, Hao M, Cui Y, Zhu S, Zhang Y. Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China. Sustainability. 2019; 11(8):2295. https://doi.org/10.3390/su11082295
Chicago/Turabian StyleLi, Pengfei, Xingchang Zhang, Mingde Hao, Yongxing Cui, Shilei Zhu, and Yanjiang Zhang. 2019. "Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China" Sustainability 11, no. 8: 2295. https://doi.org/10.3390/su11082295
APA StyleLi, P., Zhang, X., Hao, M., Cui, Y., Zhu, S., & Zhang, Y. (2019). Effects of Vegetation Restoration on Soil Bacterial Communities, Enzyme Activities, and Nutrients of Reconstructed Soil in a Mining Area on the Loess Plateau, China. Sustainability, 11(8), 2295. https://doi.org/10.3390/su11082295





