Abstract
Huperzia serrata is a Huperzine A (HupA)-producing herb. HupA is a potent, reversible, highly specific, centrally active and selective acetylcholinesterase inhibitor, and is commonly used to improve Alzheimer’s disease.At present, H. serrata resources have been overexploited and their artificial cultivation has lagged behind the need, so the market demand cannot be met. In this study, the diversity of the endophytic fungi in H. serrata was studied by high-throughput sequencing and traditional culture methods. Furthermore, the ability of the isolated endophytic fungi to produce acetylcholinesterase-inhibiting activity was evaluated. The results related to the diversity of fungi between high-throughput sequencing and traditional culture methods were compared. With high-throughput sequencing, five phyla, 22 classes, 55 orders, 120 families, and 178 genera of fungi were detected in Hubei Province, and five phyla, 22 classes, 54 orders, 124 families, and 196 genera were detected in Fujian Province. After cultivation with traditional culture methods, two phyla, three classes, five orders, six families, and six genera were detected in Hubei, and one phylum, three classes, five orders, five families, and six genera in Fujian. The endophytic fungi of H. serrata are highly diverse, and the results of high-throughput sequencing comprehensively reflect their community compositions. When the acetylcholinesterase-inhibiting activity of the isolated and cultured endophytic fungi was determined, five and three endophytic fungi from Hubei and Fujian Provinces, respectively, had good inhibitory activity (inhibition rate > 50%). Most of them were separated from leaf tissue. The strain HBR-1 and strain FJL-5 had the strongest inhibitory effects on AChE, with inhibition rates of 72.34% and 60.54%, respectively. Although only a proportion of the endophytic fungi was isolated with traditional culture methods, metabolites with broad potential applications can be isolated from these fungal cultures. The combination of high-throughput sequencing and traditional culture methods can be used to isolate and purify endophytic fungi in a targeted manner, gain many endophytic fungi, and enrich the endophytic fungi resource library.
1. Introduction
Huperzia serrata (Thunb. ex Murray) Trev. is a commonly used Chinese herbal medicine. It is a fern belonging to the subfamily Huperzioideae and the genus Huperzia Bemh. The taxonomic status of huperaceae is still partly confused [1]. Huperzia is distributed worldwide (Asia, Oceania, Central America, etc.) and mainly grows in moist forests and rock cracks at altitudes of 300–2700 m in China [2]. The whole plant has long been used in Chinese medicine to treat various ailments [3], including contusions, strains, schizophrenia, myasthenia gravis, abdominal distension, hematuria, and organophosphate poisoning [4]. Huperzia plants are rich in alkaloids [5]. Huperzine A (HupA) was the first alkaloid isolated from H. serrata, in the 1980s. It is a highly efficient acetylcholinesterase (AChE) inhibitor, with low toxicity and few side effects. It has an extremely significant effect on improving memory and treating early-stage Alzheimer’s disease, with less toxicity and side effects than other treatments, and it has broad market potential [6]. H. serrata has attracted intense attention worldwide since its marked anticholinesterase activity was discovered by Chinese scientists. Huperzine A from Huperzia is approved for use as a food additive in the United States [7]. H. serrata is a perennial herb, with slow growth and low biomass, and wild resources are limited. Moreover, this rare wild resource is being driven to extinction by its predatory exploitation, environmental degradation, and other factors.
Plant endophytes are those bacteria, actinomycetes, and fungi that live in various tissues and organs of healthy plants at a certain stage or all stages of their life histories. The infected host generally shows no external symptoms [8,9,10]. Endophytes can be isolated from plant tissues after strict surface disinfection with histological methods, or their DNA can be directly amplified from the tissues to confirm their endogenesis [11]. Endophytes can promote plant growth by altering the plant physiology, including by regulating osmotic pressure, changing stomata size and morphology, modifying the root metabolism, and increasing the absorption of certain minerals [12]. Endophytes can also produce metabolites, such as glucanase, chitinase [13], hydrolase, and antibiotics, which can be used as biological control agents [14]. Plant endophytes have also been shown to have repair functions and increase plant resistance to stress [15]. Some endophytic fungi produce compounds similar to those produced by their hosts [16,17]. Others are potential sources of natural products, such as antibiotics and anticancer agents [18]. Endophytes are recognized as reservoirs of novel compounds of great value in agriculture, industry, and medicine [19].
Studies of the diversity of endophytic fungi have mainly sought to obtain microbial cultures through separation and culturing, and the subsequent identification and classification of the isolated strains. However, given the limitations of experimental conditions, it is impossible to provide the same environments in which microorganisms naturally exist, so large numbers of microorganisms cannot be isolated with traditional methods of separation and culture. However, the rapid development of high-throughput sequencing technology has provided a more effective way to study the diversity of plant endophytic fungi, and can comprehensively and objectively determine the microbial colony structure and the relative compositions of microbial communities in a target environment.
The endophytic fungi of H. serrata have mainly been studied with traditional culture methods, and few researchers have used high-throughput sequencing to determine the diversity of this endophytic fungi [20,21]. To more comprehensively understand the diversity of endophytic fungi in H. serrata, we used high-throughput sequencing and traditional culture methods to study plants from Fujian and Hubei Provinces. We first isolated and identified the endophytic fungi in the roots, stems, and leaves of plants from the two provinces. We then studied the microbial diversity in the roots, stems, and leaves of the plants and the corresponding rhizosphere soil, and compared the differences in the diversity of endophytic fungi determined with high-throughput sequencing and traditional culture methods. We also evaluated the ability of the isolated endophytic fungi to produce AchE inhibitors.
2. Materials and Methods
2.1. Materials
Healthy H. serrata plants were collected in May 2019, from Hubei (29.93° N, 109.27° E) and Fujian (27.11° N, 119.28° E) Provinces, China. All samples were immediately transferred to the laboratory in an icebox. Six plants were collected in total, three samples each in Hubei and Fujian Province. The plants’ roots, stems and leaves were screened for endophytic fungi within 24 h.
2.2. Sample Processing
The samples were sterilized according to Beckers et al. [22], with some modification. Briefly, the root samples were shaken and the fallen soil particles removed. The remaining soil was separated from the root tissue as the “rhizosphere soil” compartment. To eliminate any epiphytic microorganisms, the plants were (a) rinsed with sterile water for 10 min, (b) rinsed with 70% ethanol for 2 min, (c) rinsed three times with sterile water, (d) soaked in sodium hypochlorite solution (2.5% active Cl− with 0.1% Tween 80) for 5 min, (e) rinsed in 70% ethanol for 1 min, and finally (f) rinsed five times with sterile water. In order to confirm the surface disinfection process was successful, sterility was checked with potato dextrose agar (PDA) [23]. The samples were cut into 0.5 cm2 sections, and some were transferred to a Petri dish containing potato dextrose agar (PDA) or Czapek–Dox medium. The samples were incubated at 28 ℃ for 7 days and the endophytic fungi separated according to their different morphological characteristics. Tip mycelia were picked and transferred to a solid medium of PDA or Czapek–Dox medium and incubated at 28 ℃. After colony growth, the previous steps were repeated until pure culture was obtained. The fungal isolates were stored in medium slants at 4 °C or as spore suspensions in 15% (v/v) glycerol at −80 ℃.
2.3. DNA Extraction
An aliquot (1.5 mL) of each homogenized plant material was centrifuged (13,000× g, 10 min). The supernatants were discarded, and DNA was extracted from the plant tissues with the E.Z.N.A. HP Plant DNA Mini Kit (Omega, Guangzhou, China), according to the manufacturer’s instructions; DNA was extracted from the rhizosphere soil with the FastDNA™ Spin Kit for Soil (MP Medicals, Shanghai, China), according to the protocol provided by the manufacturer, and DNA was extracted from the isolated endophytic fungi with the E.Z.N.A.™ HP Fungal DNA Kit (Omega), according to the manufacturer’s protocol.
2.4. PCR Amplification and High-Throughput Sequencing
The DNA from the rhizosphere soil, roots, stems, and leaves was amplified with optimal primers ITS1F (5′-CTTGGTCATTTAGAGGAAGTAA-3′) and ITS2R (5′-GCTGCGTTCTTCATCGATGC-3′) [24] and fungal libraries were created. The PCR products were purified with the E.Z.N.A.® Gel Extraction Kit (Omega). The DNA content was determined with a Qubit 3 fluorometer (Thermo Fisher Scientific Inc., Waltham, MA, USA). The gel extraction products were then mixed in equal amounts. Finally, the PCR products were sequenced (PE250) with the Illumina HiSeq 2500 high-throughput sequencing platform (Novogene Co., Ltd., Beijing, China). The barcode and primer sequences of original data were removed and then spliced. The spliced sequences were strictly filtered and the chimeric sequences removed to obtain the final valid data.
The DNA extracted from the isolated endophytic fungi was amplified with PCR with primers ITS1 (5′-TCCGTAGGTGAACCTGCGG-3′) and ITS4 (5′-TCCTCCGCTTATTGATATGC-3′), and then sequenced (Sangon Biotech Co., Ltd., Shanghai, China).
2.5. Sequence Availability
The microbiome sequences have been deposited in the National Center for Biotechnology Information (NCBI) database under BioProject accession number PRJNA655213 and under the Sequence Read Archive (SRA) accession numbers SRR12401455–SRR12401478.
2.6. Diversity Analysis of Endophytic Fungi
All effective tags of all the samples were clustered with the UCLUST [25], and sequences with ≥97% similarity were assigned to the same operational taxonomic unit (OTU). Rarefaction curves were computed with QIIME (version 1.8.0) [26]. The ITS sequences of the endophytic fungi were compared to the data available in NCBI using BLAST searches to estimate the phylogenetic relationships of the endophytic fungi. A phylogenetic analysis was performed with MEGA version 6.06 [27]. Phylogenetic trees were constructed with the neighbor joining (NJ) method. A principal coordinates analysis (PCoA) and a cluster analysis based on the unweighted pair-group method with arithmetic means (UPGMA) were conducted in R (version 2.15.3). The diversity of the endophytic fungi in H. serrata was determined with the Shannon index and Simpson index.
Shannon index:
Simpson index:
In the formulae, N represents the sum of the number of individuals of all strains; Pi = Ni/N, where Ni is the number of individuals from the ith genus in the different tissues of the plants, and N is the sum of the number of individual strains of all genera.
2.7. Fermentation and Preparation of Endophytic Fungal Extracts
Potato dextrose broth (PDB) liquid medium was inoculated with the individual isolated endophytic fungi. They were cultured at 160 rpm at 28 ℃ for 14 days in a rotary shaker. The mycelial pellets were collected by centrifugation at 13,000× g for 10 min, freeze-dried, and then ground in a mortar with liquid nitrogen. Each sample of raw material (1.0 g) was extracted with 75% ethanol (50 mL) for 30 min; this was repeated three times and the extracts filtered. The filtrates were combined and evaporated under reduced pressure. The dry residue was dissolved in 50 mL of 2.5% hydrochloric acid, and purified by shaking twice with chloroform and then with ethylic ether. The aqueous phase was rendered basic with a 25% ammonia solution (pH 9), salted out with sodium chloride, and then extracted thoroughly with chloroform. The chloroform-combined extract was evaporated to dryness. The dried residue was dissolved in 10 mL of (HPLC-grade) methanol, and the methanol extract was filtered through a 0.45 μm filter [28].
2.8. Determination of AchE Inhibitory Activity in Endophytic Fungal Fermentation Products
The in vitro AchE-inhibiting activities of the endophytic fungal extracts were determined with the method of Atta-ur-Rahman et al. [29], and compared with that of authentic HupA. The preincubation samples contained 250 μL of phosphate buffer (pH 7.7), which consisted of 15 μL of fungal extract or authentic HupA, 80 μL of dithiobis nitrobenzoic acid (DTNB; 3.96 mg DTNB and 1.5 mg sodium bicarbonate dissolved in 10 mL phosphate buffer (pH 7.7)), and 10 μL of AChE (0.22 U·mL−1). The mixture was incubated at 25 ℃ for 5 min. After preincubation, 15 μL of the substrate acetylthiocholine iodide (10.85 mg in 5 mL of phosphate buffer) was added and incubated again for 5 min. Color development was measured in a 96-well microtiter plate at a wavelength of 412 nm in an iMark™ Microplate Absorbance Reader (Bio-Rad, Hercules, CA, USA). Percentage inhibition was calculated with the formula: ([control absorbance − sample absorbance]/control absorbance) × 100%.
3. Results
3.1. Analysis of Endophytic Fungal Community Structures in H. serrata with High-Throughput Sequencing
3.1.1. Alpha Rarefaction Curves and Alpha Diversity
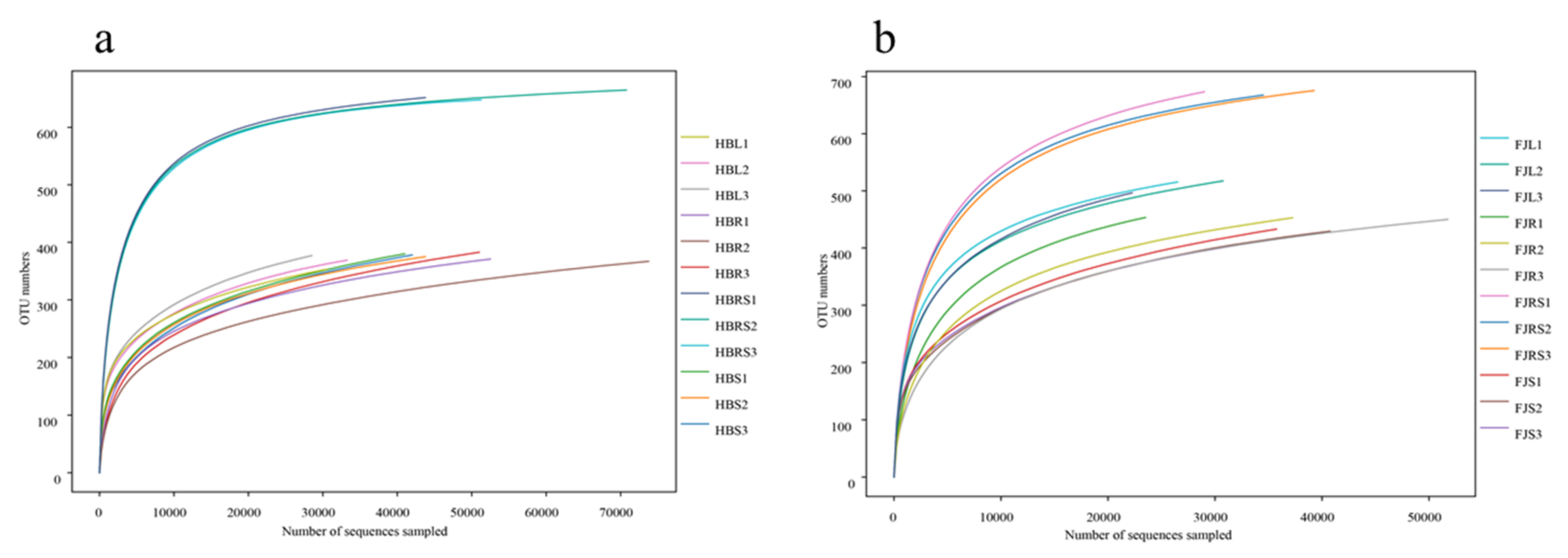
The abundances of the plant endophytic fungal communities in the samples from both Fujian and Hubei Provinces were lower than that of the corresponding rhizosphere soil flora (Figure 1). The abundance of the endophytic fungi in the roots, stems, and leaves was similar. The rhizosphere soil samples from Hubei contained 648, 652 and 665 operational taxonomic units (OTUs), and those from Fujian contained 668, 674, and 676 operational taxonomic units (OTUs). There was a combined total of 375, 378 and 389 OTUs in the root, 367, 371 and 383 OTUs in the stem, and 353, 369 and 377 OTUs in the leaf samples from Hubei, while there was a combined total of 450, 453 and 454 OTUs in the root, 416, 430 and 433 OTUs in the stem, and 497, 516 and 518 in the leaf samples from Fujian.
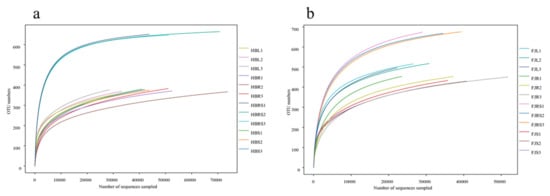
Figure 1.
Rarefaction curves of fungi in rhizosphere soil and of endophytic fungi in different tissue. (a) Rarefaction curves of Huperzia serrata samples from Hubei Province. (b) Rarefaction curves of Huperzia serrata samples from Fujian Province. Note: HBL: H. serrata leaf, HBR: H. serrata root, HBS: H. serrata stem, HBRS: rhizosphere soil; FJL: H. serrata leaf, FJR: H. serrata root, FJS: H. serrata stem, FJRS: rhizosphere soil. The numbers 1, 2, and 3 in the names are used to indicate that the same tissue sample is repeated three times.
3.1.2. Beta Diversity
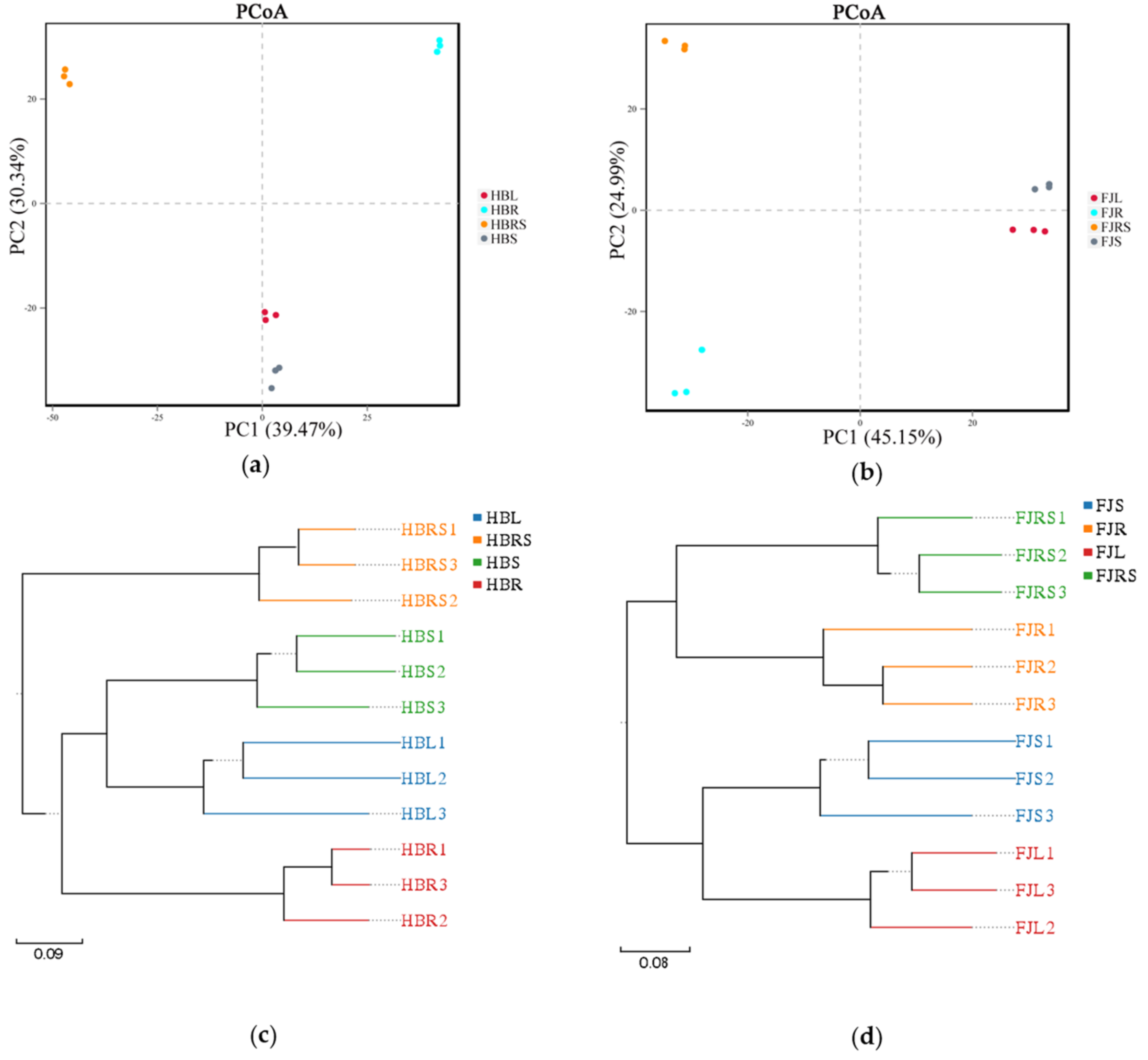
The fungal diversity of each plant tissue and rhizosphere soil was analyzed with a principal coordinates analysis (PCoA) and a cluster analysis. The results of PCoA based on the OTU level are shown in Figure 2. The first principal coordinate (PC1) in the sample from Hubei showed a representative contribution rate of 39.47% to the microbial fungi detected, and the representative contribution of the second principal coordinate (PC2) was 30.34% (Figure 2a). In the sample from Fujian, the representative contribution rate of PC1 to the microbial fungi detected was 45.15%, and the representative contribution of PC2 was 24.99% (Figure 2b). The structures of the fungal communities in leaves and stems were similar between the samples from Hubei and Fujian. The structures of the fungal communities in the leaf and stem were clustered separately from those of the fungal communities in the roots and rhizosphere soil. In the cluster analysis (Figure 2c), the root, stem, and leaf samples from Hubei clustered together; however, the root samples were not an ingroup of the leaf + stem cluster, but a sister group of it. In the samples from Fujian (Figure 2d), the root and rhizosphere samples clustered together, and the stem and leaf samples clustered together.
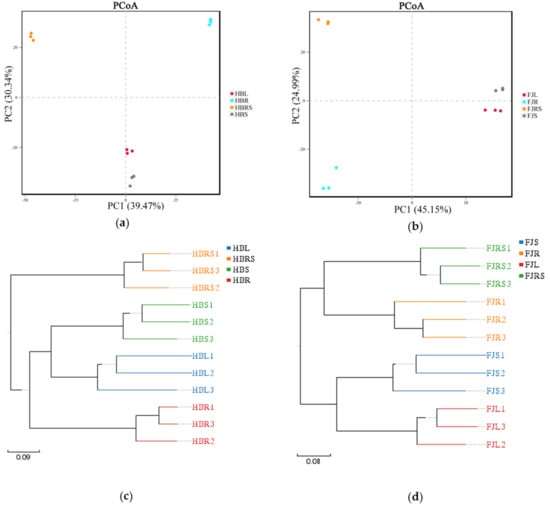
Figure 2.
Principal coordinates analysis (a) Sample of Hubei Province. HBL: H. serrata leaf, HBR: H. serrata root, HBS: H. serrata stem, HBRS: rhizosphere soil; (b) Sample of Fujian Province. FJL: H. serrata leaf, FJR: H. serrata root, FJS: H. serrata stem, FJRS: rhizosphere soil) and cluster analysis (c) Sample of Hubei Province. HBL: H. serrata leaf, HBR: H. serrata root, HBS: H. serrata stem, HBRS: rhizosphere soil; (d) Sample of Fujian Province. FJL: H. serrata leaf, FJR: H. serrata root, FJS: H. serrata stem, FJRS: rhizosphere soil) of endophytic fungi in Huperzia serrata. The numbers 1, 2, and 3 in the names are used to indicate that the same tissue sample is repeated three times.
3.1.3. Composition of Endophytic and Rhizosphere Soil Fungi
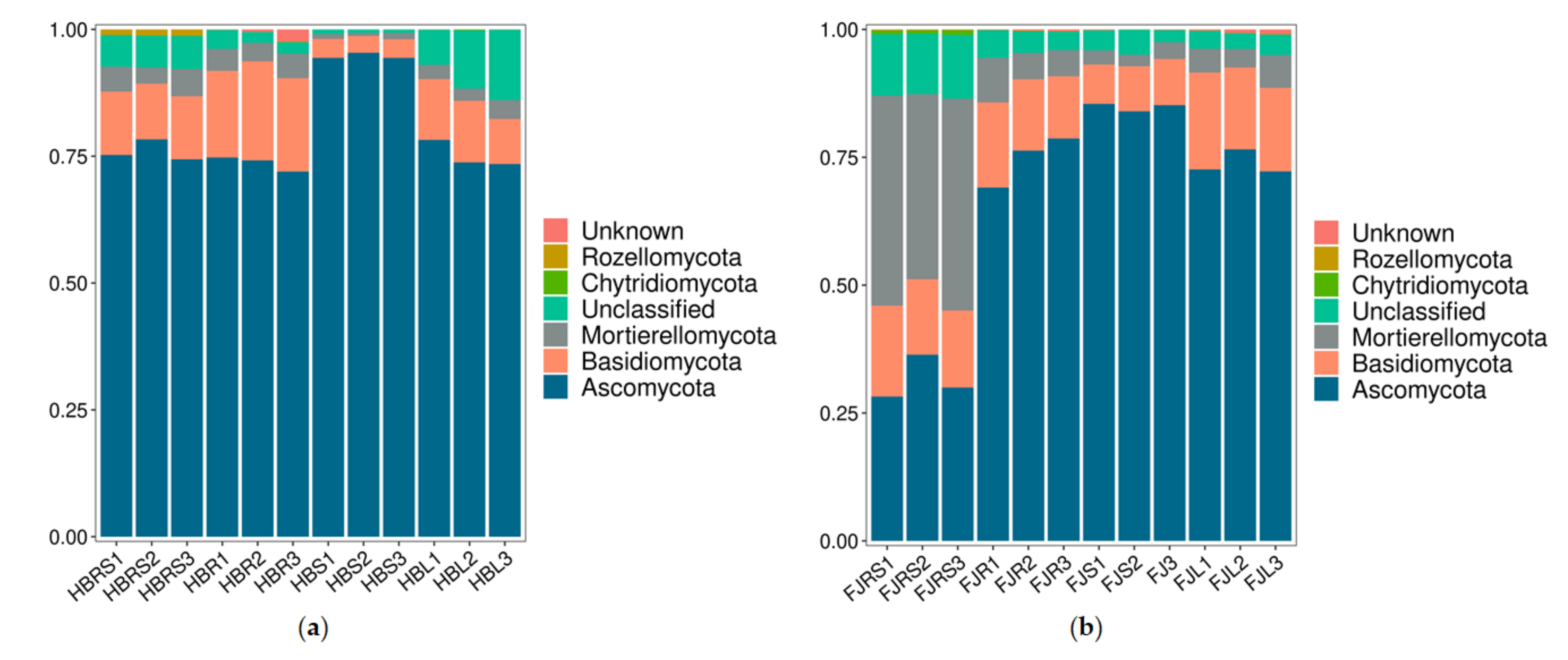
At the phylum level (Figure 3), Ascomycota was the dominant phylum in the Hubei samples, with relative abundances of 74.40–78.30%, 71.95–74.18%, 94.36–95.36% and 73.45–78.22%, respectively, followed by Basidiomycota and Mortierellomycota. Ascomycota was also the dominant phylum in the Fujian samples. However, the relative abundances of the dominant flora in the Fujian rhizosphere soil samples differed significantly from those in the tissue samples. The relative abundances of Ascomycota, Mortierellomycota, and Basidiomycota were 31.63%, 39.55%, and 15.73%, respectively, in the rhizosphere soil; about 75%, 5%, and 15%, respectively, in the roots and leaves; and about 84.83%, 2.75%, and 8.53%, respectively, in the stem samples.
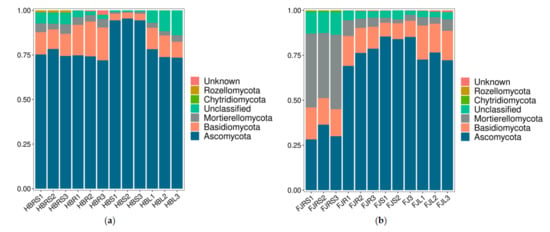
Figure 3.
Fungal community composition at the phylum level. (a) Sample of Hubei Province. HBL: H. serrata leaf, HBR: H. serrata root, HBS: H. serrata stem, HBRS: rhizosphere soil; (b) Sample of Fujian Province. FJL: H. serrata leaf, FJR: H. serrata root, FJS: H. serrata stem, FJRS: rhizosphere soil). The numbers 1, 2, and 3 in the names are used to indicate that the same tissue sample is repeated three times.
3.2. Analysis of Endophytic Fungi Community Structure in H. serrata with Traditional Culture Methods
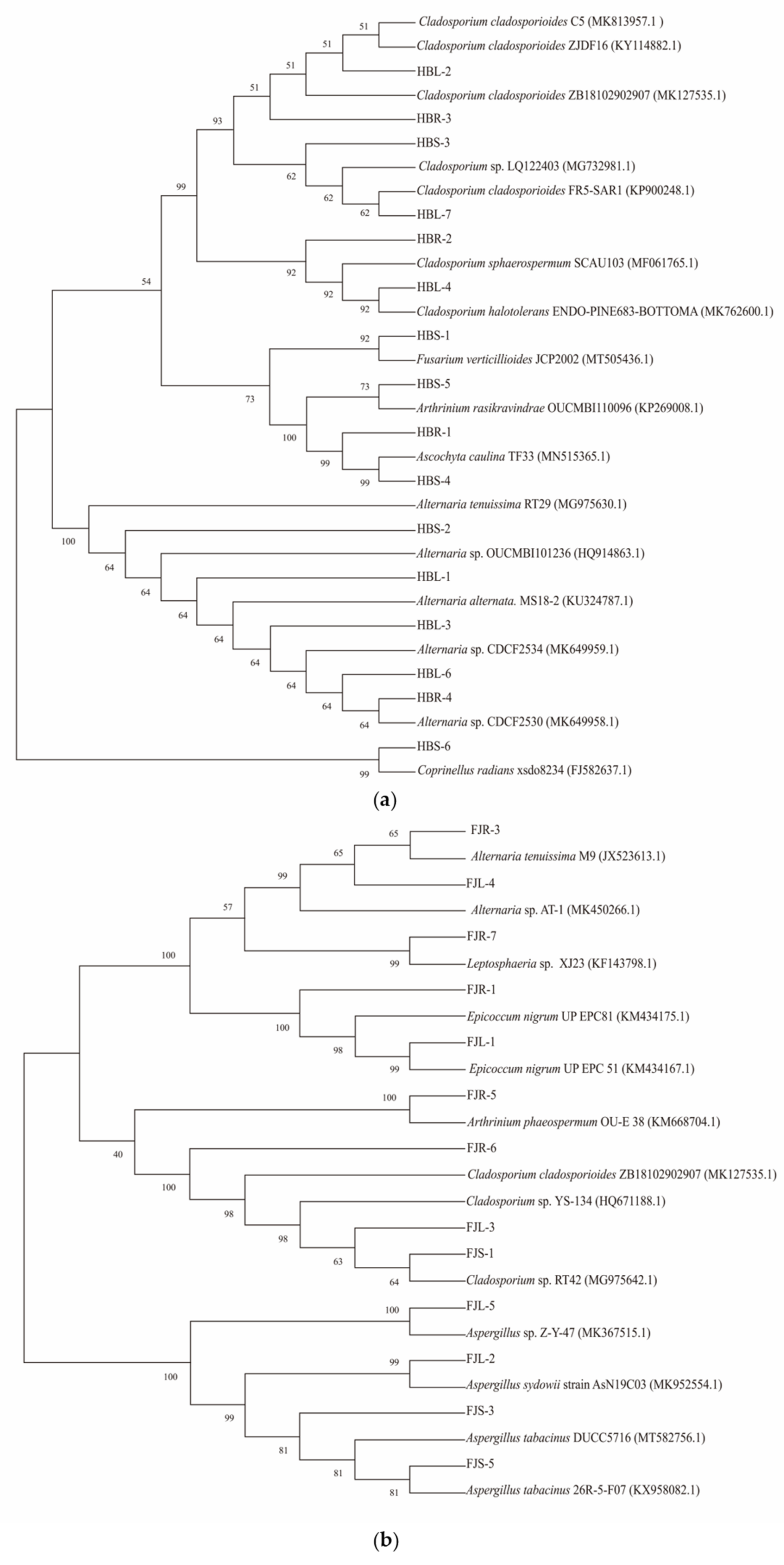
In total, 15 and 13 strains were cultured from the root, stem, and leaf tissues of plants from Hubei and Fujian Provinces, respectively. The ITS rRNA gene sequence of strains was compared with the sequences deposited in the GenBank database (https://blast.ncbi.nlm.nih.gov/Blast.cgi/ (accessed on 17 July 2020)). The species with the highest similarity was obtained and the results are shown in Table 1. The phylogenetic tree was constructed by the neighbor-joining (NJ) method using the endophytic fungi isolated from Hubei and Fujian, and the species of these with the highest similarity. The preliminary identification of the endophytic fungi is given in Figure 4.

Table 1.
Analysis of endophytic fungi isolated with traditional culture methods.
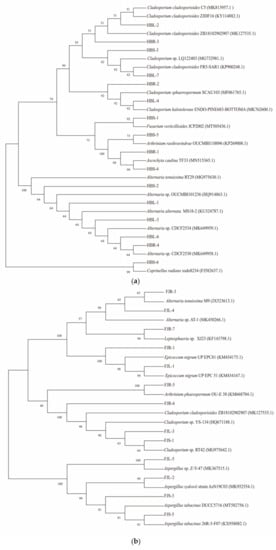
Figure 4.
Neighbor-joining tree of the ITS sequences of endophytic fungi from Huperzia serrata. (a) phylogenetic tree was constructed based on fungi isolated from Huperzia serrata in Hubei province; (b) phylogenetic tree was constructed based on fungi isolated from Huperzia serrata in Fujian province).
3.3. Differences in Endophytic Fungi Detected with Traditional Culture Methods and High-Throughput DNA Sequencing
3.3.1. Differences in Endophytic Fungal Community Structures Detected
When the endophytic fungi were isolated with traditional culture methods, two phyla, three classes, five orders, six families, and six genera were detected in the samples from Hubei, and one phylum, three classes, five orders, five families, and six genera were detected in the samples from Fujian. In contrast, high-throughput sequencing detected five phyla, 22 classes, 55 orders, 120 families, and 178 genera in the samples from Hubei and five phyla, 22 classes, 54 orders, 124 families, and 196 genera in the samples from Fujian (Table 2). These results indicate that traditional culture methods seriously underestimated the species composition of the endophytic fungi in H. serrata. More microbial species were detected with the high-throughput sequencing method than with the traditional culture method. High-throughput sequencing has obvious advantages over the pure culture method, and these advantages were greatest at these levels.

Table 2.
Community structures of endophytic bacteria detected with traditional culture methods and high-throughput sequencing at each classification level.
3.3.2. Differences in Endophytic Fungal Diversity
The microbial diversity indices of the endophytic fungi in H. serrate were analyzed. The Shannon indices for the endophytic fungi of H. serrata isolated with traditional culture methods were 2.099 in Hubei and 2.416 Fujian, whereas the Simpson indices were 0.711 and 0.793, respectively. However, the corresponding Shannon indices calculated from high-throughput sequencing data were 3.426 and 3.939, respectively, and the corresponding Simpson indices were 0.828 and 0.936, respectively. These findings show that the diversity of endophytic fungi detected with high-throughput sequencing was higher than that detected with traditional culture methods. They also show that the endophytic fungus community’s structure in H. serrata is complex and stable.
3.4. Acetylcholine-Inhibiting Activity of Endophytic Fungi Isolated with Traditional Culture Methods
The in vitro AChE inhibitory activity of endophytic fungal extracts from two Chinese provinces were tested (Table 1). Among the strains isolated with traditional culture methods, those with high AChE inhibitory activity (>50% inhibition) accounted for 33.33% and 23.08% in the samples from Hubei and Fujian, respectively; those with moderate activity (30–50% inhibition) for 40% and 38.46%, respectively; and those with low activity (<30% inhibition) for 26.67% and 38.46%, respectively. The endophytic fungal sample HBR-1 from Hubei and sample FJL-5 from Fujian had the strongest inhibitory effects on AChE, with inhibition rates of 72.34% and 60.54%, respectively.
4. Discussion
In this study, traditional culture methods and high-throughput sequencing were used to study the diversity of endophytic fungi in H. serrata from Hubei and Fujian. Both methods showed that these endophytic fungi were highly diverse. Compared with high-throughput sequencing, the traditional culture methods identified relatively fewer cultures, which may be attributable to the influence of the culture conditions, which limited the numbers of microorganisms obtained.
When the different tissues of the plants from Hubei and Fujian were processed with traditional culture methods, 15 and 13 endophytic fungi were isolated, respectively (Table 1). The endophytes in both places were predominantly from the phylum Ascomycota, except a unique strain from Basidiomycota in the Hubei sample, but the distribution was different at the genus level. Both the Hubei and Fujian samples contained Alternaria, Arthrinium, and Cladosporium. Ascochyta, Fusarium and Coprinellus were only isolated from the Hubei sample and Epicoccum, Leptosphaeria, and Aspergillus from the Fujian sample. These genera have also been isolated in previous studies [21,30,31,32,33]. The results show some differences in the diversity of endophytic fungi at different geographic locations. Wang et al. studied the composition of endophytic fungi of H. serrata from Lushan Mountain and Jinggang Mountain, and also found that there were many differences, indicating that geographical environment plays a more important role in affecting the diversity of endophytic fungi, even if from the same plant [21,33]. Plants in different habitats provide different environmental and nutritional conditions for the microorganisms within them, and these conditions affect the complex symbiotic relationships between the microorganisms and plants, generating different endophytic flora. When isolating and culturing the endophytic fungi of H. serrata, the natural ecological environment in which the plant grows must be simulated as closely as possible to maximize the endophytic fungal resources available and better reflect the natural endophytic fungal diversity of H. serrata. Because we cannot completely simulate this natural environment, fewer endophytic fungi were isolated in the laboratory. During the process of isolation and culturing in the laboratory, the first thing to be considered is the interaction between endophytes and hosts. The second may be the interconnection among endophytes [34]. We realize that the strategies of culturing “unculturable microorganisms” have attracted widespread attention. Mu et al. concluded that simulating the natural environment is an effective strategy to isolate active bacteria [35,36].
High-throughput sequencing detected Ascomycota, Basidiomycota, Mortierellomycota, Chytridiomycota, and Rozellomycota in the H. serrata plants from both Hubei and Fujian Provinces. The dominant phylum was Ascomycota, followed by Basidiomycota and Mortierellomycota; therefore, the composition of endophytic fungi at the phylum level was similar, to a certain extent, in plants under different growth and environmental conditions, but the relative abundances of the various phyla differed in response to the different environments for plant growth. At the genus level, the traditional separation methods detected only six different strains, far fewer than were detected with high-throughput sequencing. All the genera detected with traditional culture methods were also detected with high-throughput sequencing (Supplementary Materials Tables S1 and S2), except Ascochyta. This exception may be attributable to its missed detection with high-throughput sequencing. These results combined with the α diversity indices analysis show that high-throughput sequencing better reflects the distribution of endophytic fungi in H. serrate than traditional culture methods. The abundance, uniformity, and diversity of OTUs in the roots, stems, leaves, and rhizosphere soil were evaluated in terms of their alpha diversity. The OTU richness in the rhizosphere clearly differed from that in the plant roots, stems, and leaves, and the rhizosphere microbial richness in the two provinces was significantly higher than the richness of the plant endophytic flora. However, there were no significant differences in the OTU abundances among the different tissues. Our results differ from the results of Beckers et al. [37]. Their research showed that the endophytic bacteria in the rhizosphere soil, root, and stem had clearly distinguishable OTU abundances. This may be related to the different plant species and microflora types studied. To further compare the structures of fungal flora in the plant rhizosphere soil and in different plant tissues, all the samples were clustered with PCoA and cluster analysis. All the fungal samples were clustered according to the different plant tissues, and the result indicated that endophytic fungi differ across different tissues. The cluster analysis showed that the endophytic fungi in Fujian showed similar clustering to that of bacteria [37]. The endophytic fungi of Fujian samples in the rhizosphere soil and root tissues clustered together, and the stem and leaf tissues clustered together. These findings support the idea that rhizosphere sediments and the root exudates of the host plant enhance the richness of the microbial community in the rhizosphere soil [38,39]. The microorganisms in the rhizosphere soil travel through the endoderm and root stele to the xylem vessels, ultimately colonizing the whole plant [40]. However, the clustering of the endophytic fungi in the rhizosphere soil and the plant tissues in the Hubei region, where the endophytic fungi from the roots, stems, and leaves clustered together, differed from that in Fujian Province. This difference may be related to the different environments of plant growth in these two places. The soil in which the plants grew in Fujian was relatively moist, which may have promoted the movement of the rhizosphere soil fungi into the plants to colonize them. Several studies have shown that the structures of fungal communities can vary according to the soil type, plant compartment, plant species, and season [41,42,43].
The endophytic fungi of H. serrata in the Hubei and Fujian Provinces were separated and purified with traditional culture methods. The endophytic fungi HBR-1 and FJL-5 from Hubei and Fujian, respectively, showed the best inhibitory effects on AChE (inhibition rate > 50%), with inhibition rates of 72.34% and 60.54%, respectively. This indicates that the production and development of AchE inhibitors by endophytic fungi have certain potential. A statistical analysis of the fungi with higher AChE inhibitory activity (inhibition rates > 50%) indicated that most of the strains with high AChE inhibitory activity were isolated from H. serrata leaf tissue, which accounted for 80% and 66.67% of the high enzyme producers from Hubei and Fujian, respectively. Cui et al. analyzed the content of HupA in the endophytic fungi community and the species of HupA, and found that most of the endophytic fungi that showed a significant positive correlation with the acetylcholinesterase inhibitor HupA were isolated from leaves [44]. This phenomenon may be related to the higher content of the AChE inhibitor in the leaves of H. serrata plants than in the stems or roots [45]. It also supports the theory of Young et al. that, through evolution, symbiotic endophytes have developed mechanisms for the biosynthesis and tolerance of high levels of secondary metabolites, so as to better compete with and survive in the tissues of medicinal plants [46].
The results of the high-throughput sequencing method more comprehensively reflect the diversity of endophytic fungi in H. serrata than those of the traditional culture methods, which has major significance for research into endophytic fungi. The isolation of endophytic fungal resources with traditional culture methods also has valuable applications, especially because some of these fungi have antibacterial and growth-promoting activities. A variety of metabolites can be isolated from these endophytic fungi, which have broad potential utility in the development and utilization of natural products. The combination of high-throughput sequencing and traditional pure culture methods reflects the diversity of the endophytic fungi of H. serrata most comprehensively, and also allows their targeted isolation and purification, the isolation of greater numbers of endophytic fungi, the enrichment of the endophytic fungal resource pool, and the provision of materials for research. However, further research is needed. Furthermore, the use of degenerated primer pairs for genes involved in the biosynthesis of the AchE inhibitor, in order to assess the diversity of fungi able to produce this compound, is still worth exploring [47,48].
Supplementary Materials
The following are available online at https://www.mdpi.com/article/10.3390/su132112073/s1, Table S1. Community structure of endophytic bacteria in Hubei at the genus level, detected with high-throughput sequencing. Table S2. Community structure of endophytic bacteria in Fujian at the genus level, detected with high-throughput sequencing.
Author Contributions
Conceptualization, project administration, resources, Funding acquisition, Supervision H.Y.; writing—original draft preparation, Z.L.; writing—review and editing, Y.M.; supervision, D.Z.; visualization, L.X.; investigation, Methodology, Z.L. and Y.M. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Natural Science Foundation of Jiangxi Province, grant number 20181BAB214003, and funded by Project of Graduate Innovation Foundation from Education Department of Jiangxi Province, grant number YC2020-S184.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are openly available in National Center for Bio-technology Information (NCBI) database, BioProject accession number PRJNA655213 and Sequence Read Archive (SRA) accession numbers SRR12401455–SRR12401478.
Conflicts of Interest
The authors declare no conflict of interest.
References
- Prieto, J.A.F.; Aguiar, C.; Dias, E.; Casado, M.D.L.A.F.; Homet, J. The genus Huperzia (Lycopodiaceae) in the Azores and Madeira. Bot. J. Linn. Soc. 2008, 158, 522–533. [Google Scholar] [CrossRef][Green Version]
- Yang, Q.P.; Kou, L.X.; Fugal, K.B.; McLaughlin, J.L. Determination of huperzine A in formulated products by reversed-phase-liquid chromatography using diode array and electrospray ionization mass spectrometric detection. Phytomedicine 2003, 10, 200–205. [Google Scholar] [CrossRef]
- Chen, X.Y.; Qi, Y.D.; Wei, J.H.; Zhang, Z.; Wang, D.L.; Feng, J.D.; Gan, B.C. Molecular identification of endophytic fungi from medicinal plant Huperzia serrata based on rDNA ITS analysis. World J. Microbiol. Biotechnol. 2011, 27, 495–503. [Google Scholar] [CrossRef]
- Ma, X.Q.; Tan, C.H.; Zhu, D.Y.; Gang, D.R.; Xiao, P.G. Huperzine A from Huperzia species—An ethnopharmacolgical review. J. Ethnopharmacol. 2007, 113, 15–34. [Google Scholar] [CrossRef] [PubMed]
- Liu, Y.C.; Zhang, Z.J.; Su, J.; Peng, L.Y.; Pan, L.T.; Wu, X.D.; Zhao, Q.S. Lycodine-Type Lycopodium Alkaloids from the Whole Plants of Huperzia serrata. Nat. Prod. Bioprospect. 2017, 7, 405–411. [Google Scholar] [CrossRef]
- Wu, J.G.; Wang, Y.Y.; Zhang, Z.L.; Yu, B. Herbal medicine in the treatment of Alzheimer’s disease. Chin. J. Integr. Med. 2015, 2, 102–107. [Google Scholar] [CrossRef] [PubMed]
- Rasic, J.S.; Ivanovic, N.D.; Andjelkovic, M.S.; Nedeljkovic, I.P.; Nikolic, I.R.; Stojanovic, S.D.; Ristic-Medic, D.K.; Takic, M.M.; Djordjevic, B.I.; Dikic, N.V. Influence of Higenamine on Exercise Performance of Recreational Female Athletes: A Randomized Double-Blinded Placebo-Controlled Trial. Front. Psychol. 2021, 12. [Google Scholar] [CrossRef]
- Petrini, O. Fungal Endophytes of Tree Leaves. In Microbial Ecology of Leaves; Andrews, J.H., Hirano, S.S., Eds.; Springer: New York, NY, USA, 1991; pp. 179–197. [Google Scholar]
- Saikkonen, K.; Faeth, S.H.; Helander, M.; Sullivan, T.J. Fungal endophytes: A continuum of interactions with host plants. Annu. Rev. Ecol. Syst. 1998, 29, 319–343. [Google Scholar] [CrossRef]
- Li, J.; Zhao, J.L.; Xu, L.J.; Zhou, L.G.; Li, X.; Wang, J.G. Endophytic fungi from rhizomes of Paris polyphylla var. yunnanensis. World J. Microbiol. Biotechnol. 2008, 24, 733–737. [Google Scholar] [CrossRef]
- Reiter, B.; Sessitsch, A. Bacterial endophytes of the wildflower Crocus albiflorus analyzed by characterization of isolates and by a cultivation-independent approach. Can. J. Microbiol. 2006, 52, 140–149. [Google Scholar] [CrossRef]
- Compant, S.; Duffy, B.; Nowak, J.; Clément, C.; Barka, E.A. Use of Plant Growth-Promoting Bacteria for Biocontrol of Plant Diseases: Principles, Mechanisms of Action, and Future Prospects. Appl. Environ. Microbiol. 2005, 71, 4951–4959. [Google Scholar] [CrossRef] [PubMed]
- Frankowski, J.; Lorito, M.; Scala, F.; Schmid, R.; Berg, G.; Bahl, H. Purification and properties of two chitinolytic enzymes of Serratia plymuthica HRO-C48. Arch. Microbiol. 2001, 176, 421–426. [Google Scholar] [CrossRef] [PubMed]
- Ezra, D.; Castillo, U.F.; Strobel, G.A.; Hess, W.M.; Porter, H.; Jensen, J.B.; Condron, M.A.M.; Teplow, D.; Sears, J.; Maranta, M.; et al. Coronamycins, peptide antibiotics produced by a verticillate Streptomyces sp. (MSU—2110) endophytic on Monstera sp. Microbiology 2004, 150, 785–793. [Google Scholar] [CrossRef]
- Aken, B.V.; Peres, C.M.; Doty, S.L.; Yoon, J.M.; Schnoor, J.L. Methylobacterium populi sp. nov. a novel aerobic, pink-pigmented, facultatively methylotrophic, methane-utilizing bacterium isolated from poplar trees (Populus deltoidesxnigra DN34). Int. J. Syst. Evol. Microbiol. 2004, 54, 1191–1196. [Google Scholar] [CrossRef] [PubMed]
- Aly, A.H.; Debbab, A.; Kjer, J.; Proksch, P. Fungal endophytes from higher plants: A prolific source of phytochemicals and other bioactive natural products. Fungal Divers 2010, 41, 1–16. [Google Scholar] [CrossRef]
- Stierle, A.; Strobel, G.; Stierle, D. Taxol and taxane production by Taxomyces andreanae, an endophytic fungus of Pacific yew. Science 1993, 260, 214. [Google Scholar] [CrossRef] [PubMed]
- Strobel, G.; Daisy, B.; Castillo, U.; Harper, J. Natural products from endophytic microorganisms. J. Nat. Prod. 2004, 67, 257–268. [Google Scholar] [CrossRef]
- Strobel, G.; Daisy, B. Bioprospecting for microbial endophytes and their natural products. Microbiol. Mol. Biol. Rev. 2003, 67, 491–502. [Google Scholar] [CrossRef]
- Han, W.X.; Han, Z.W.; Jia, M.; Zhang, H.; Li, W.Z.; Yang, L.B.; Liang, F.; Han, L. Zhao, N.; Li, X.F. Five novel and highly efficient endophytic fungi isolated from Huperzia serrata expressing huperzine A for the treatment of Alzheimer’s disease. Appl. Microbiol. Biotechnol. 2020, 104, 9159–9177. [Google Scholar] [CrossRef]
- Wang, Y.; Zeng, G.Q.; Zhang, Z.B.; Yan, R.M.; Wang, L.Y.; Zhu, D. Isolation and characterization of endophytic huperzine A-producing fungi from Huperzia serrata. J. Ind. Microbiol. Biotechnol. 2011, 38, 1267–1278. [Google Scholar] [CrossRef]
- Beckers, B.; Michiel, O.D.B.; Weyens, N.; Acker, R.V.; Montagu, M.V.; Boerjan, W.; Vangronsveld, J. Lignin engineering in field-grown poplar trees affects the endosphere bacterial microbiome. Proc. Natl. Acad. Sci. USA 2016, 113, 2312. [Google Scholar] [CrossRef] [PubMed]
- Mcinroy, J.A.; Kloepper, J.W. Survey of indigenous bacterial endophytes from cotton and sweet corn. Plant Soil 1995, 173, 337–342. [Google Scholar] [CrossRef]
- Bokulich, N.A.; Mills, D.A. Improved Selection of Internal Transcribed Spacer-Specific Primers Enables Quantitative, Ultra-High-Throughput Profiling of Fungal Communities. Appl. Environ. Microbiol. 2013, 79, 2519–2526. [Google Scholar] [CrossRef] [PubMed]
- Edgar, R.C. Search and clustering orders of magnitude faster than BLAST. Bioinformatics 2010, 26, 2460–2461. [Google Scholar] [CrossRef] [PubMed]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Peña, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 2010, 7, 335–336. [Google Scholar] [CrossRef]
- Tamura, K.; Stecher, G.; Peterson, D.; Filipski, A.; Kumar, S. MEGA6: Molecular Evolutionary Genetics Analysis Version 6.0. Mol. Biol. Evol. 2013, 30, 2725–2729. [Google Scholar] [CrossRef] [PubMed]
- SzypulA, W.; Pietrosiuk, A.; Suchocki, P.; Olszowska, O.; Furmanowa, M.; Kazimierska, O. Somatic embryogenesis and in vitro culture of Huperzia selago shoots as a potential source of huperzine A. Plant Sci. 2005, 168, 1443–1452. [Google Scholar] [CrossRef]
- Rahman, A.U.; Choudhary, M.I.; Thomsen, W.J. Bioassay Techniques for Drug Development, 1st ed.; CRC Press: Florida, FL, USA, 2001. [Google Scholar]
- Le, T.T.M.; Hoang, A.T.H.; Le, T.T.B.; Vo, T.T.B.; Quyen, D.V.; Chu, H.H. Isolation of endophytic fungi and screening of Huperzine A–producing fungus from Huperzia serrata in Vietnam. Sci. Rep. 2019, 9, 16152. [Google Scholar] [CrossRef]
- Yang, H.Y.; Qi, B.W.; Ding, N.; Jiang, F.F.; Jia, F.F.; Luo, Y.; Xu, X.P.; Wang, L.L.; Zhu, Z.X.; Liu, X.; et al. Polyketides from Alternaria alternata MT-47, an endophytic fungus isolated from Huperzia serrata. Fitoterapia 2019, 137, 104282. [Google Scholar] [CrossRef]
- Zhang, Z.B.; Zeng, Q.G.; Yan, R.M.; Wang, Y.; Zou, Z.R.; Zhu, D. Endophytic fungus Cladosporium cladosporioides LF70 from Huperzia serrata produces Huperzine A. World J. Microbiol. Biotechnol. 2011, 27, 479–486. [Google Scholar] [CrossRef]
- Wang, Y.; Lai, Z.; Li, X.X.; Yan, R.M.; Zhang, Z.B.; Yang, H.L.; Zhu, D. Isolation, diversity and acetylcholinesterase inhibitory activity of the culturable endophytic fungi harboured in Huperzia serrata from Jinggang Mountain, China. World J. Microb. Biot. 2016, 32. [Google Scholar] [CrossRef] [PubMed]
- Peng, L.L.; Wang, Q.; Xin, M.X. Advanced in uncultivable microorgnisms in nature. J. Microbiol. 2011, 31, 75–79. [Google Scholar] [CrossRef]
- Mu, D.S.; Ouyang, Y.; Chen, G.J.; Du, Z.J. Strategies for culturing active/dormant marine microbes. Marine Life Sci. Technol. 2020, 3, 121–131. [Google Scholar] [CrossRef]
- Kaeberlein, T.; Lewis, K.; Epstein, S.S. Isolating “Uncultivable” Microorganisms in Pure Culture in a Simulated Natural Environment. Science 2002, 296, 1127–1129. [Google Scholar] [CrossRef]
- Beckers, B.; Beeck, M.O.D.; Weyens, N.; Boerjan, W.; Vangronsveld, J. Structural variability and niche differentiation in the rhizosphere and endosphere bacterial microbiome of field-grown poplar trees. Microbiome 2017, 5, 25. [Google Scholar] [CrossRef] [PubMed]
- Lugtenberg, B.; Kamilova, F. Plant-growth-promoting rhizobacteria. Annu. Rev. Microbiol. 2009, 63, 541–556. [Google Scholar] [CrossRef]
- Bais, H.P.; Weir, T.L.; Perry, L.G.; Gilroy, S.; Vivanco, J.M. The role of root exudates in rhizosphere interactions with plants and other organisms. Annu. Rev. Plant Biol. 2006, 57, 233–266. [Google Scholar] [CrossRef] [PubMed]
- Compant, S.; Clément, C.; Sessitsch, A. Plant growth-promoting bacteria in the rhizo- and endosphere of plants: Their role, colonization, mechanisms involved and prospects for utilization. Soil Biol. Biochem. 2009, 42, 669–678. [Google Scholar] [CrossRef]
- Devin, C.D.; Damaris, D.; Citlali, F.G.; Stephen, G.; Scott, C.; Tanja, W.; North, G.; Visel, A.; Partida-Martinez, L.P.; Tringe, S.G. Plant compartment and biogeography affect microbiome composition in cultivated and native Agave species. New Phytol. 2016, 209, 798–811. [Google Scholar] [CrossRef]
- Aurore, C.; Tristan, C.; Lengellé, J.; Defossez, E.; Vacher, C.; Robin, C.; Buée, M.; Marcais, B. Leaf and Root-Associated Fungal Assemblages Do Not Follow Similar Elevational Diversity Patterns. PLoS ONE 2014, 9, e100668. [Google Scholar] [CrossRef]
- Shakya, M.; Gottel, N.; Castro, H.F.; Yang, Z.K.; Gunter, L.; Labbé, J.; Muchero, W.; Bonito, G.M.; Vilgalys, R.; Tuskan, G.; et al. A Multifactor Analysis of Fungal and Bacterial Community Structure in the Root Microbiome of Mature Populus deltoides Trees. PLoS ONE 2013, 8, e76382. [Google Scholar] [CrossRef] [PubMed]
- Cui, L.L.; Noushahi, H.A.; Zhang, Y.P.; Liu, J.X.; Cosoveanu, A.; Liu, Y.; Yan, L.; Zhang, J.; Shu, S.H. Endophytic Fungal Community of Huperzia serrata: Diversity and Relevance to the Production of Huperzine A by the Plant Host. Molecules 2021, 26, 892. [Google Scholar] [CrossRef] [PubMed]
- Yuan, J.Q.; Zhou, L.X.; Wang, S.; Yan, Z.G.; Feng, S.X.; Jiang, J.M.; Zhu, D.Y.; Ma, X.J. Simultaneous determination of huperzine A and huperzine B in different parts of Huperzia serrata from different habitats by HPLC. J. Pharm. Anal. 2012, 32, 1541–1544. [Google Scholar] [CrossRef]
- Young, D.H.; Michelotti, E.L.; Swindell, C.S.; Krauss, N.E. Antifungal properties of taxol and various analogues. Experientia 1992, 48, 882–885. [Google Scholar] [CrossRef] [PubMed]
- Yang, M.Q.; Wu, S.W.; You, W.J.; Jaisi, A.; Xiao, Y.L. Selection of Reference Genes for Expression Analysis in Chinese Medicinal Herb Huperzia serrata. Front. Pharmacol. 2019, 10, 44. [Google Scholar] [CrossRef] [PubMed]
- Yang, G.D.; Li, P.; Meng, L.F.; Xv, K.Y.; Dong, F.M.; Qiu, Y.; He, L.; Lin, L. Diversity and communities of culturable endophytic fungi from different tree peonies (geoherbs and non-geoherbs), and their biosynthetic potential analysis. Braz. J. Microbiol. 2018, 49, 47–58. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).