Abstract
Balkan Livestock Guardian Dogs (LGD) were bred to help protect sheep flocks in sparsely populated, remote mountainous areas in the Balkans. The aim of this study was genomic characterization (107,403 autosomal SNPs) of the three LGD breeds from the Balkans (Karst Shepherd, Sharplanina Dog, and Tornjak). Our analyses were performed on 44 dogs representing three Balkan LGD breeds, as well as on 79 publicly available genotypes representing eight other LGD breeds, 70 individuals representing seven popular breeds, and 18 gray wolves. The results of multivariate, phylogenetic, clustering (STRUCTURE), and FST differentiation analyses showed that the three Balkan LGD breeds are genetically distinct populations. While the Sharplanina Dog and Tornjak are closely related to other LGD breeds, the Karst Shepherd is a slightly genetically distinct population with estimated influence from German Shepard (Treemix analysis). Estimated genomic diversity was high with low inbreeding in Sharplanina Dog (Ho = 0.315, He = 0.315, and FROH>2Mb = 0.020) and Tornjak (Ho = 0.301, He = 0.301, and FROH>2Mb = 0.033) breeds. Low diversity and high inbreeding were estimated in Karst Shepherds (Ho = 0.241, He = 0.222, and FROH>2Mb = 0.087), indicating the need for proper diversity management. The obtained results will help in the conservation management of Balkan LGD dogs as an essential part of the specific grazing biocultural system and its sustainable maintenance.
1. Introduction
Livestock Guardian Dogs (LGDs, Canis familiaris) are a specialized type of dog breeds kept with grazing livestock as they are able to protect them from large predators, including gray wolves, brown bears, foxes, lynxes, and others. Together with the livestock, mostly sheep flocks, LGDs typically live under harsh mountain environments in the open and free-range grazing production systems. They are large-bodied dogs that are locally adapted and selected to demonstrate intelligence, trustworthiness, attentiveness, and protectiveness [1]. While most LGD breeds are common in Europe and Asia, they can be found all over the world. Today, LGDs are a very important component in the sustainable maintenance of the specific grazing biocultural systems. Despite several hypotheses that can be traced from the Tibetan Mastiff to the Molossers given to Alexander the Great by an Indian king, little is known about their precise origin. The oldest link between dogs and sheep can only be traced back to 5600 BP [2]. However, it would not be surprising if their functional role and interaction with humans date back to the beginnings of the Neolithization process.
Karst Shepherd (KRA), Sharplanina Dog (SAR), and Tornjak (TOR) are breeds recognized by the Fédération Cynologique Internationale (FCI), belonging to the LGD subgroup, geographically linked to the Balkans and considered molosser-like mountain dogs. Originally the Karst Sheperd, formerly also called the Illyrian Shepherd, was native to the Slovenian region of the Karst, and the Sharplanina Dog comes from Sharplanina Massif, while the name Tornjak comes from the Croatian word “tor”, which means a fenced area for keeping livestock in the mountainous part of Croatia and Bosnia and Herzegovina. The ancestors of KRA and SAR were listed as subtypes of the same breed, but in 1968 they were internationally recognized as separate breeds. By then the number of KRAs had dropped to the critical point, whereupon two Newfoundland males and some German Shepherds were added to the breed. SAR became a rare breed after World War II, probably mixed with several breeds, including Caucasian Shepherd dogs, but still representing a native population of herding dogs. In 2007, Tornjak, the Bosnian and Herzegovinian-Croatian Shepherd dog was provisionally recognized by the FCI. During the recognition process, the breed was listed in two subtypes (Bosnian-Herzegovinian subtype and Croatian subtype), a practice that was later abandoned. These breeds are large-framed, often around 70 cm at the withers and more than 45 kg body weight. They were formed to protect livestock from wild predators in the absence of a human shepherd, as shown in Figure 1.

Figure 1.
Tornjak dogs guarding sheep on a mountain pasture. The photo was provided by Ljuban Gudelj and Ilija Smiljanić (Tomislavgrad, Bosnia & Herzegovina).
The dog was the first domesticated animal [3,4], and currently, there are 353 dog breeds recognized by the FCI (http://www.fci.be/en/Presentation-of-our-organisation-4.html (accessed on 19 February 2021)). The concept of dog breeds dates back to the 19th century [5] and refers to the specific group of animals with homogenous morphological appearance, behavior, and other phenotypic characteristics that are uniformly transmitted to the next generation. Breeds are formed by the combined forces of genetic drift, natural selection for adaptive traits to the local environment, and artificial selection application [6,7,8].
The first comprehensive genetic variability was studied by Irion et al. [6], who used 100 autosomal microsatellite markers in the study of genotypic variation within and between globally representative samples of 28 breeds. As the authors noted, “While the set of autosomal microsatellites was useful in describing genetic variation within breeds, establishing the genetic relatedness between breeds was less conclusive”. In addition, the use of microsatellites was limited because it was difficult to link genotypes obtained in different laboratories/machines. For this response, results from a number of studies focusing on small native breeds were underutilized. The genetic diversity of LGD by microsatellites has been analyzed in several studies. For example, genetic diversity and relatedness of Balkan LGD breeds were analyzed by Ceh and Dovc (2014) [9]. Dimitrijević et al. (2020) [10] analyzed genetic diversity and population structure within SAR sampled in Serbia, while the genetic diversity and relatedness of Italian LGD breeds have been analyzed in several studies [11,12]. Recent advances in next-generation sequencing provide affordable access to high-throughput genotypic information and enable more comprehensive estimates of genetic diversity, population structure, genetic admixture, inbreeding levels, and effective population size at global scales [13,14]. At the same time, developments in the isolation and analysis of ancient DNA have led to a better understanding of the origin and domestication process [15] and the interrelationship between dogs and wolves [16]. In general, the genetic diversity of popular dog breeds (PDB) has been frequently analyzed [17,18,19], although there are several studies that have investigated the genomic diversity of local native breeds [20]. At the same time, the number of studies analyzing the genetic variability of LGDs is far from its matching their importance [9,11], especially those using high-throughput molecular information [21].
The aim of this study was to provide information relevant to the genomic characterization and conservation of the three Balkan LGD breeds (KRA, SAR, and TOR) by hypothesizing the following: (i) serious depletion of genetic diversity and high inbreeding is present in KRA, SAR and TOR; (ii) using genomic arrays, we can classify KRA, SAR, and TOR with other LGD breeds; (iii) KRA, SAR, and TOR are genetically pure breeds with little admixture with other LGD breeds; and (iv) we can detect genomic introgression of gray wolves within KRA, SAR, and TOR. To answer these hypotheses, we performed several high-resolution population genomic analyses, such as principal component discriminant analyses, phylogenetic network graph reconstruction, unsupervised clustering, estimation of migrations, and estimation of conservation genetics parameters (diversity and genomic inbreeding). In our analyses, we made the comparisons of Balkan LGDs with other eight LGD breeds, seven popular dog breeds, and a gray wolf as a wild reference population.
2. Materials and Methods
2.1. Sampling, DNA Extraction, Breed Origin, and Genomic Information
2.1.1. Sampling, DNA Extraction, and Genotyping
Overall, 52 dogs registered to the three Balkan LGD breeds (KRA, 14; SAR, 14; and TOR, 24) were sampled as buccal swabs or/and hair follicles at dog shows or were obtained from breeders and owners. Geographical origin and distribution, together with illustrated physical appearance (photo), of the sampled dogs, is shown in Figure 2. An additional four gray wolves representing wild reference populations from Croatia were provided (tissue) by Ana Galov and Haidi Arbanasić (the University of Zagreb, Faculty of Science). DNA from buccal swabs or hair follicles was extracted using the standard protocol of the DNeasy Blood & Tissue kit reagents (Qiagen, Germany). The quantity of DNA was evaluated using NanoVue (GE Healthcare Life Sciences, USA). All 56 DNA samples were genotyped using Illumina CanineHD BeadChip (Illumina Inc., San Diego, CA, USA) containing 172,115 SNPs. We performed quality control using SNP & Variation Suite v8.7.0 (Golden Helix, Inc., Bozeman, MT, USA www.goldenhelix.com, (accessed on 19 February 2021)), and after filtering (SNP call rate <95%) and excluding nonautosomal and unassigned markers, a total of 137,725 markers remained. We also excluded four KRA and four TOR with an individual call rate < 90%.
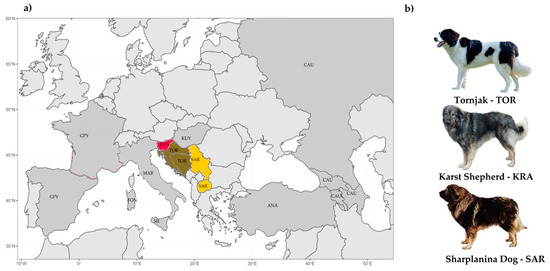
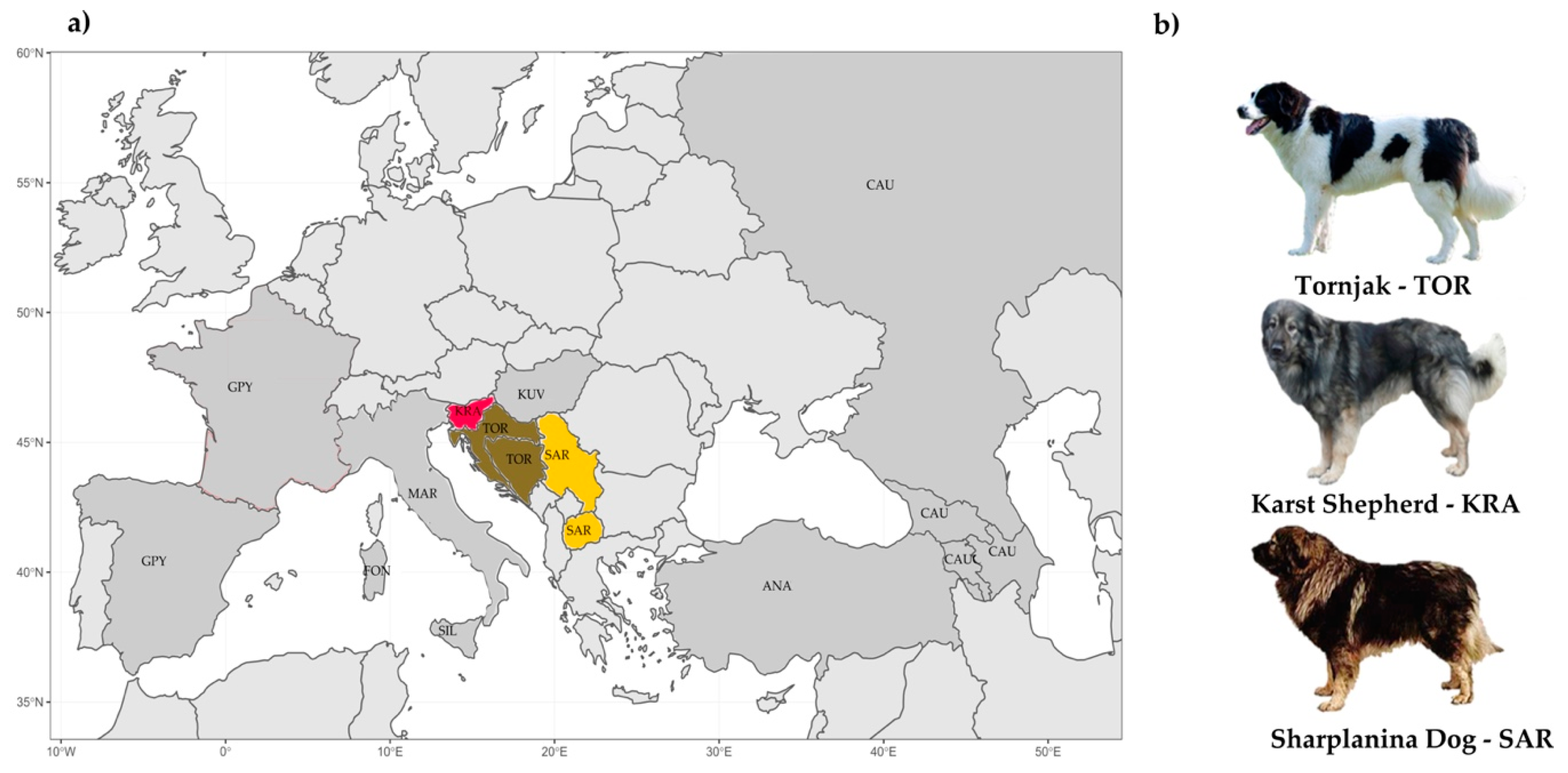
Figure 2.
Geographic origin and distribution of 10 livestock guardian dog (LGD) breeds: (a) Origin and distribution of 10 LGD breeds and (b) Illustration of the physical appearance of the three Balkan LGD breeds. The breeds shown are the Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), and Tornjak (TOR). Information for the Tibetan Mastiff not presented (geographical distance).
2.1.2. Publicly Available Genomic Information
For this study, we obtained publicly available SNP data [8,16], from which we selected a representative sample of LGD, PDB, and gray wolves (WOL). Publicly available SNP data were merged with the Balkan LGD dataset described above. The final dataset contained 107,403 autosomal SNPs that were common to both previously described datasets and 211 dogs representing 18 breeds with a population of 18 gray wolves (Table 1).

Table 1.
Genomic diversity and inbreeding in the dog breeds and gray wolves (reference wild population).
2.2. Genomic Diversity, Inbreeding, and Molecular Coancestry
2.2.1. Genomic Diversity and Inbreeding
Heterozygosity (observed and expected) and genomic individual inbreeding coefficient (FIS), were calculated in SNP & Variation Suite v8.7.0 (Golden Helix, Inc., Bozeman, MT, USA, www.goldenhelix.com, (accessed on 19 February 2021)) using the “inbreeding coefficient” option on autosomal markers of the respective population. The FIS indicates the deviation from Hardy-Weinberg heterozygosity equilibrium and can range from −1 to +1, indicating mating randomness. While negative values usually indicate outbreeding, positive values result from inbreeding [22].
2.2.2. Runs of Homozygosity Inbreeding
ROH segments were detected using SNP & Variation Suite v8.7.0 (Golden Helix, Inc., Bozeman, MT, USA www.goldenhelix.com (accessed on 19 February 2021)). According to the recommendations in Ferenčaković et al. (2013) [23], ROH segments with different length categories (from 2 to 5 Mb, from 5 to 10 Mb, and longer than 10 Mb) were identified based on different criteria. Thus, ROH segments were named in the category from 2 to 5 Mb if there were at least 15 consecutive homozygous SNPs covering more than 2 Mb, with one missing SNP allowed. In the 5 to 10 Mb category, two missing SNPs and one heterozygous SNP were allowed. For segments greater than 10 Mb, four missing and two heterozygous SNPs were allowed. Where more than one missing or heterozygous call was allowed, they were not allowed to be consecutive. The maximum gap between the two SNPs was set at 1 Mb, and the minimum required density was 1 SNP per 75 kb. The number of missing and heterozygous calls allowed was set following Ferenčaković et al. (2013) [23]. Next, the genomic inbreeding coefficient FROH was calculated following McQuillan et al. (2008) [24] and Curik et al. (2014) [25], where FROH is defined as genome length in ROH/autosomal genome length covered by the SNP chip (here 2,200 Mb distributed over 38 chromosomes). Inbreeding coefficients with different ROH lengths (FROH>2Mb, FROH2-5Mb, FROH5-510Mb, and FROH>10Mb,) were calculated as they correspond to a different base population and allow decomposition of the FROH>2Mb into its remote (FROH2-5Mb), intermediate (FROH2-5Mb), or recent (FROH>10Mb) origin. Boxplots and stacked-bar-plots were created using the ggplot2 package for R software [26].
2.2.3. Molecular Coancestry
Molecular (marker-by-marker) coancestry coefficients were estimated for each population using the “Relationship Matrix” option in JMP Genomics and IBS statement (JMP® Genomics, Version 9.1. SAS Institute Inc., Cary, NC, 1989-2019). Pairwise molecular coancestry coefficients, all unique (lower) off-diagonal elements from the coancestry matrix, were further analyzed within each breed/population, with mean values presenting the probability that two randomly sampled alleles, each from one individual, are the same copy of an allele. Note that molecular coancestry refers to alleles that are identical-by-state, in contrast to genealogical coancestry coefficients which refer to identical-by-descent alleles inherited from the same ancestor [27].
2.3. Population Structure and Gene Flow
2.3.1. Discriminant Analysis of Principal Components
We performed a discriminant analysis of principal components (DAPC) to identify and visualize clusters of genetically related individuals [28]. All calculations and visualizations required for DAPC were performed in the R package adegenet [29].
2.3.2. Phylogenetic Analyses and Gene Flow
Inference of the phylogenetic relationship between 18 dog breeds and WOL was represented as the NeighborNet network inferred from pairwise Nei’s genetic distances [30]. Nei’s distances were calculated using the R package StAMPP R [31]. A NeighborNet network was created and represented using SplitsTree5 software [32].
In addition, patterns of splits and mixtures of a subset of 18 dog breeds, and the gray wolf population as a rooted outgroup, were detected by the maximum-likelihood algorithm implemented in the TreeMix program [33]. First, we constructed a maximum-likelihood tree of the populations with no migration events considered. We then constructed a phylogenetic network for all selected populations, sequentially increasing migration events up to 12 migrations. Residuals from fitting the model to the data were visualized using the R script implemented in TreeMix.
2.3.3. Unsupervised Clustering with STRUCTURE Analyses
Population genetic structure was inferred by the algorithm implemented in the STRUCTURE program [34,35]. We had to reduce the number of SNP genotypes in the dataset to 22,220 to enable the complex calculations that would not otherwise have been possible with our computational resources. The reduction of the dataset (pruning) was based on the threshold of linkage disequilibrium between SNPs, here defined as the pairwise r2, which was equal to 0.1, with the window size equal to 100 SNPs and a step size equal to 20. A model assuming admixture and correlated allele frequencies was used for all STRUCTURE runs, with a burn-in of 105 iterations followed by 104 MCMC iterations. Runs were repeated ten times for each assumed K, starting with K = 1 to K = 24. The resulting individual genotype membership coefficients were displayed using DISTRUCT [36].
To gain additional insight into population structure and quantify genetic differentiation among the analyzed populations, we also estimated pairwise Wright’s FST fixation indices [37] using the R package StAMPP [31].
3. Results
3.1. Genetic Diversity, Inbreeding, and Molecular Coancestry
3.1.1. Genomic Diversity and Inbreeding
The estimated observed (Ho) and expected (He) heterozygosity, FIS, and FROH>2Mb means in 18 dog breeds and gray wolves are shown in Table 1. Compared to PDB and gray wolves, significantly higher heterozygosity (Ho and He) and significantly lower FROH>2Mb were observed in LGD breeds. For example, the estimated 95% confidence intervals for Ho ranged from 0.284 to 0.296 for LGD, from 0.200 to 0.216 for PDB, and from 0.155 to 0.196 for WOL. Because the confidence intervals did not overlap, the estimated Ho were significantly higher in LGD than in PDB and/or WOL. Considerably lower FIS, although the difference was not significant, was observed in LGD than in PDB. The values observed in KRA, with exception of FIS, were within the PDB estimates (see Table 1). Thus, in LGD breeds with exception of KRA, estimates of observed (expected) heterozygosity ranged from 0.277 in Great Pyrenees (GPY) to 0.315 in SAR (from 0.251 in Caucasian Shepherd (CAU) to 0.329 in Anatolian Shepherd (ANA)) while FROH>2Mb ranged from 0.018 in Tibetan Mastiff (TIB) to 0.045 in Kuvazs (KUV). Interestingly, almost the lowest negative FIS value (−0.081) was observed in KRA, indicating strong avoidance of inbreeding among recent genealogical relatives. The contrast between high FROH>2Mb and low FIS suggests that low diversity and relatively high ROH inbreeding in KRA resulted from small population size and genetic drift. Very high heterozygosity and low ROH inbreeding, even among LGD, were observed in SAR and TOR, indicating importance for genetic diversity. Zero FIS values observed in these two breeds show complete randomness in mating which could be improved by pedigree management and mating avoidance of genealogical relatives. At the same time, the lowest heterozygosity and the highest inbreeding values (FIS) were observed in WOL.
Ho is observed heterozygosity, He is expected heterozygosity (gene diversity), FIS is Wright’s inbreeding coefficient, and FROH>2Mb is ROH inbreeding coefficient with the assumption that 2Mb long ROHs are autozygous, SE is the standard error.
3.1.2. Runs of Homozygosity, Inbreeding, and Molecular Coancestry
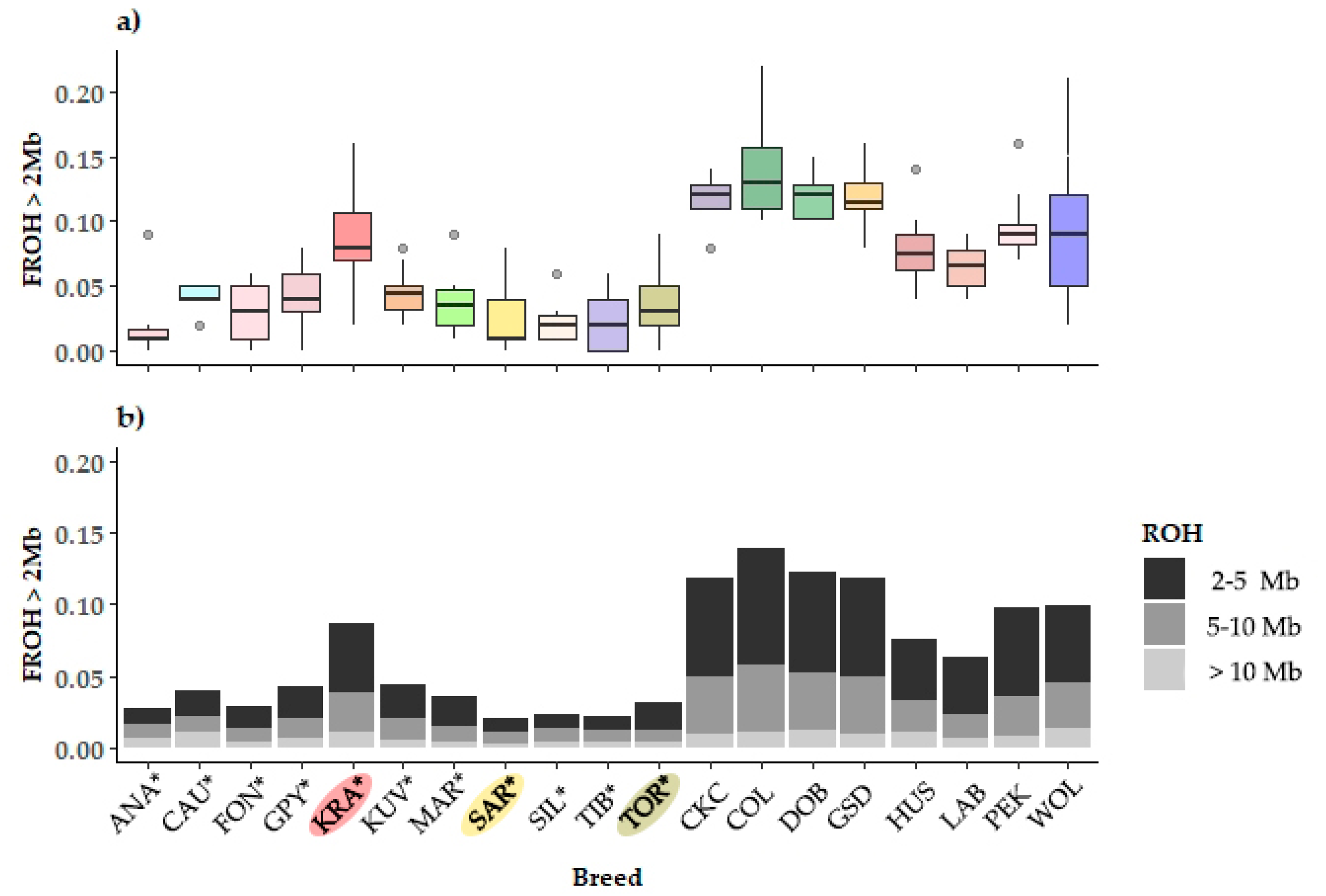
The distribution of FROH>2Mb and its partitioning with respect to the origin of autozygosity (remote, intermediate, and recent) is shown in Figure 3. While LGD breeds have a much lower inbreeding in several breeds (ANA, KUV, Maremmano-Abruzzese Sheepdog (MAR), and Pastore della Sila (SIL)), outlining dogs with much higher inbreeding has been observed, a similar pattern was also observed in PGD breeds (Husky (HUS) and Pekingese (PEK)), see Figure 3a. This points to the lack of pedigree inbreeding management or intentional mating with respect to the desired ancestor.
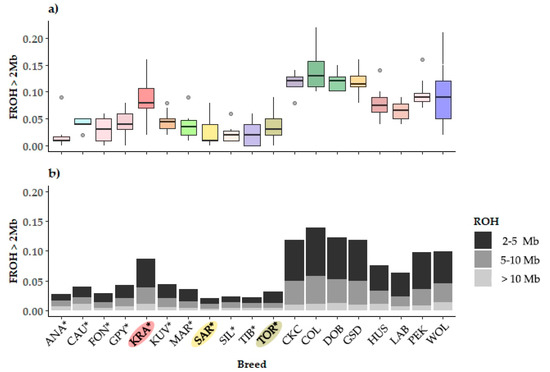
Figure 3.
Distribution of genomic inbreeding coefficients in 18 dog breeds and gray wolf: (a) boxplot presentation of the runs of homozygosity inbreeding coefficients (FROH>2Mb); (b) stacked-bar presented partition of FROH>2Mb to the remote (FROH2-5Mb), intermediate (FROH5-10Mb), and recent (FROH>10Mb) autozygosity origin. The breeds shown are Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), Tibetan Mastiff (TIB), and Tornjak (TOR) as livestock guarding dogs, Cavalier King Charles Spaniel (CKC), Rough Collie (COL), Doberman Pinscher (DOB), German Shepherd Dog (GSD), Husky (HUS), Labrador Retriever (LAB), and Pekingese (PEK) as popular dog breeds and gray wolves (WOL) as a reference wild population.
Assuming that the expected length of an IBD haplotype segregating in a population follows an exponential distribution, we can infer the number of generations from the inbred individual to the common ancestor. It is generally assumed that if 1 cM is approximated by the length of 1 Mb, we would expect ROH that are 16.6, 10.0, or 5.0 Mb in length to be derived from a common ancestor 3 generations back (6 meioses), 5 generations back (10 meioses), or 10 generations back (20 meioses), respectively. Further details can be found in Curik et al. (2014) [25]. Thus, with the exception of KRA, which exhibits the PDB pattern, the differences in the degree of inbreeding between LGD and PDB are mainly the result of distant and intermediate inbreeding, originating from 25 to 10 (FROH2-5Mb) and from 10 to 5 (FROH5-10Mb) generations distant ancestors, respectively (Figure 3b).
Mean molecular coancestry in a breed is functionally related to expected heterozygosity, which is also defined as the probability that two randomly sampled alleles are not identical-by-state. It is evident that much higher molecular coancestry was observed in PDB and WOL compared to LGD (Table 2). However, we also analyzed the range of all pairwise molecular coancestry coefficients and found that the mean range (the difference between the minimum and maximum molecular coancestry averaged over LGD or PDB) in LGD was 0.088 compared to 0.043 in PDB. By comparing the mean range of molecular coancestry, we were able to analyze the genetic heterogeneity (or compactness) of each breed. WOL had a fairly large molecular coancestry range (0.172), but this was a sampling effect as gray wolves were sampled from several different populations.

Table 2.
Population differentiation among 18 dog breeds and gray wolves (wild reference population) based on genome-wide FST estimates.
3.2. Population Structure and Gene Flow
Discriminant Analysis of Principal Components and Phylogenetic Analyses with Gene Flow
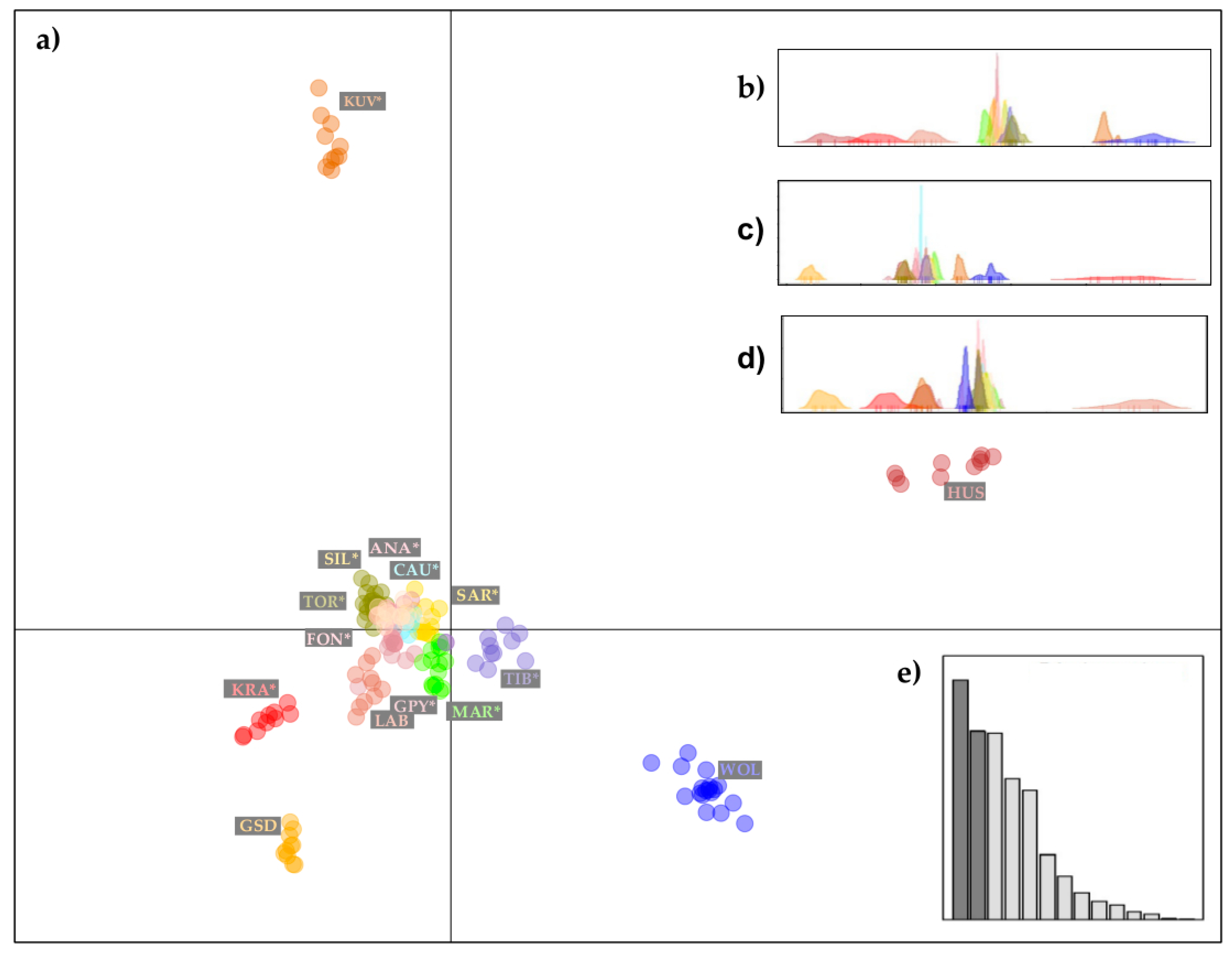
Presentation of the DAPC analysis, variation in five principal components of the DAPC method performed on 14 dog breeds and a gray wolf population, is shown in Figure 4. We have omitted Cavalier King Charles Spaniel (CKC), Rough Collie (COL), Doberman Pinscher (DOP), and PEK from this illustration (Figure 4) as these four breeds are distant from the LGD breeds and did not help to clarify their genetic relationship. From the first two principal components of DAPC (Figure 4a), it was clear that LGDs form a distinct group of dogs, although a slight separation was observed for KRA, while KUV was quite distinct. The first two principal components of DAPC explained 40.1% of the variation, while most of the variation remained in the third, fourth, fifth, and further principal components of DAPC (Figure 4b–e).
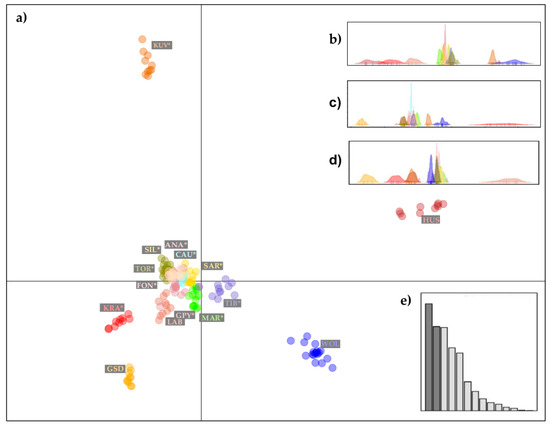
Figure 4.
Graphical illustrations of the population genetic structure of 14 dog breeds and gray wolves, generated by the “supervised” DAPC algorithm: (a) scatter-plot presenting a variation of the first two principal components of the DAPC; (b) distribution of the third principal components of the DAPC; (c) distribution of the fourth principal components of the DAPC; (d) distribution of the fifth principal components of the DAPC and (e) Barr plot visualization of the contribution of discriminant functions explaining sample variation. The breeds shown are Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), Tibetan Mastiff (TIB), and Tornjak (TOR) as livestock guarding dogs, German Shepherd Dog (GSD), Husky (HUS), Labrador Retriever (LAB) as popular dog breeds, and gray wolves (WOL) as a reference wild population.
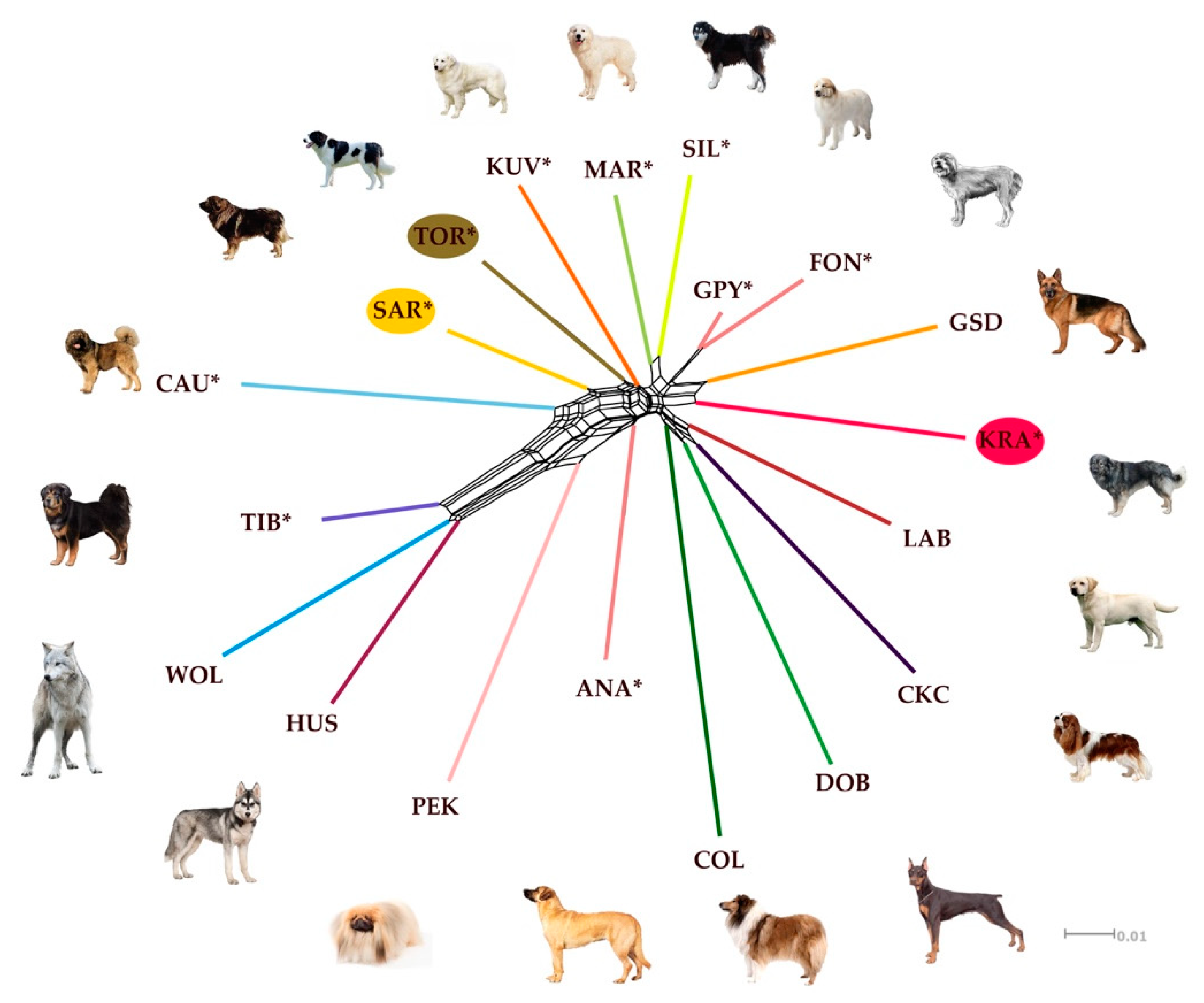
However, the NeighborNet network inferred from pairwise Nei’s genetic distances was able to complement the DAPC approach in presenting genetic relationships between the analyzed breeds and gray wolves. According to the NeighborNet network (Figure 5) CAU, SAR, TOR, and KUV form a closely related cluster within the main reticulation. TIB slightly separated towards WOL. MAR and SIL are clustered on a separate reticulation, in a similar way as GPY and Fonni’s Dog (FON). This clustering pattern can be explained to some extent by the geographical origin of LGD. KUV, SAR, and TOR are geographically very close to each other, while CAU is further away but in the same direction on the continent. However, the genetic relatedness of KUV, SAR, and TOR with CAU could be explained by human migrations, but this aspect is not considered in our analysis. The clustering of MAR and SIL is logical since both are Italian LGD breeds. Since there is a historical link between Sardinia and Spain, we were not surprised that FON and GPY share the same cluster. In contrast, ANA was separated from other LGD breeds while KRA was clustered with German Shepherd Dog (GSD) indicating the documented influence of GSD on the breeding history of KRA.
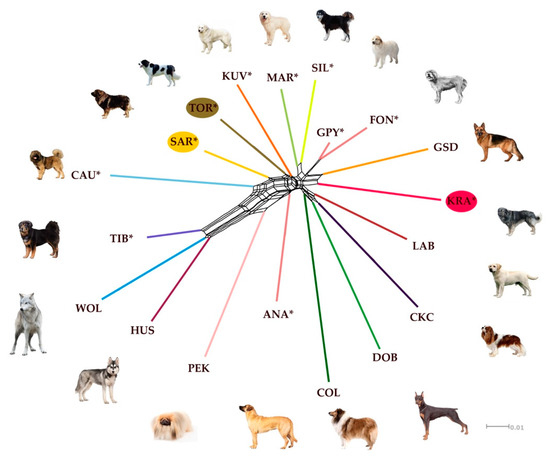
Figure 5.
NeighborNet network inferred from pairwise Nei’s genetic distances, estimated from the CanineHD BeadChip information, among in 10 livestock guarding dog breeds, 8 popular dog breeds, and gray wolves (wild reference population). Reticulations on the graph indicate past hybridization events among populations. The breeds shown are Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), Tibetan Mastiff (TIB), and Tornjak (TOR) as livestock guarding dogs, Cavalier King Charles Spaniel (CKC), Rough Collie (COL), Doberman Pinscher (DOB), German Shepherd Dog (GSD), Husky (HUS), Labrador Retriever (LAB), and Pekingese (PEK) as popular dog breeds, and gray wolf (WOL) as a reference wild population.
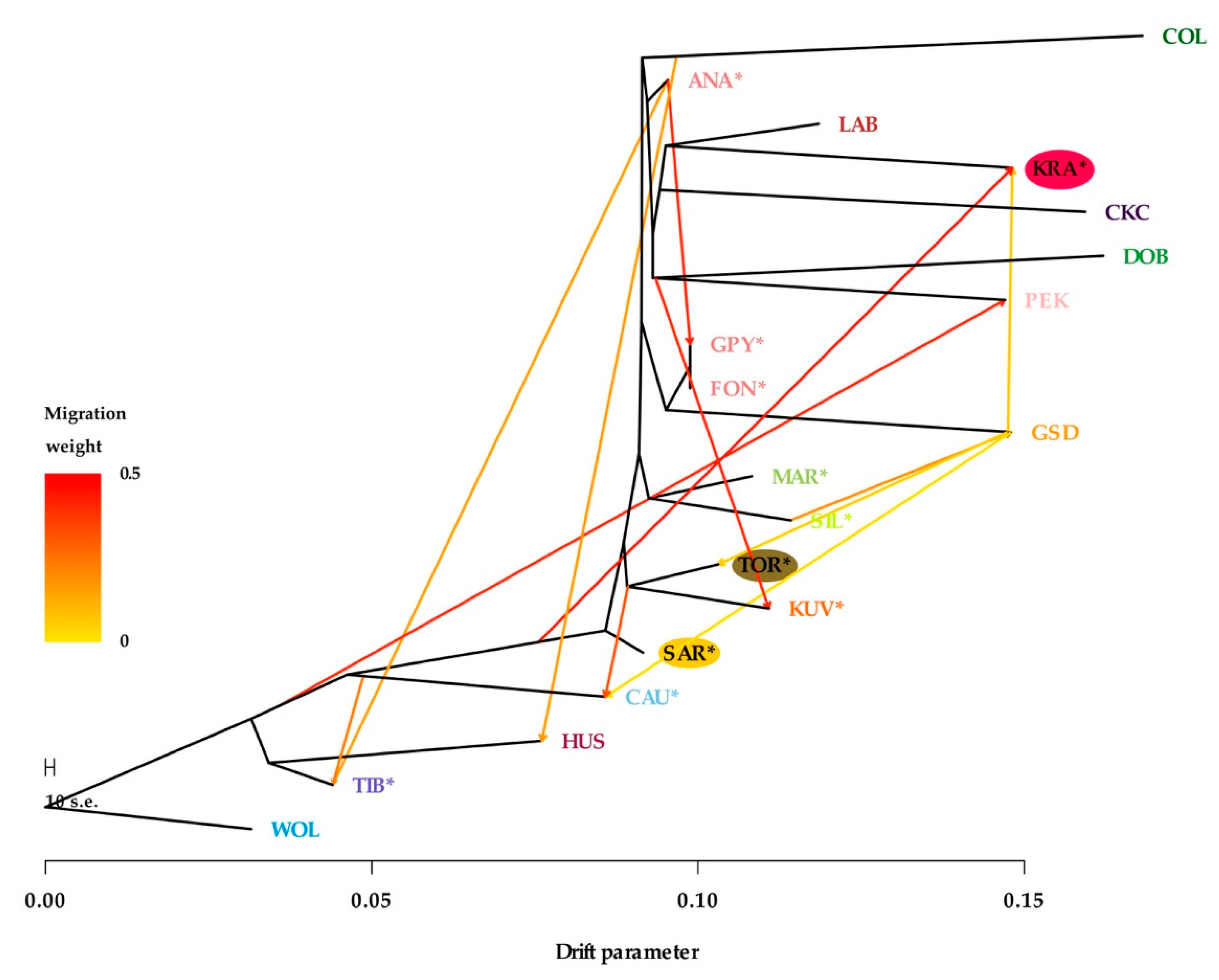
Genetic relatedness and migration patterns, allowing 12 migration events (that best fit the model), were observed using TreeMix analysis (Figure 6). While some differences from the NeighborNet network analysis were observed, much of the relatedness was logical and in agreement with other analyses. For example, KRA was clustered slightly differently, but the migration influence of GSD and the root of LGD breeds was clearly identified (Figure 5). Interestingly, the migration pattern of TOR and KUV root towards CAU may explain why CAU was clustered with KUV, SAR, and TOR in NeighborNet network analysis (Figure 5). Analyses implemented in the TreeMix algorithm revealed no migration flow between dog breeds and gray wolves.
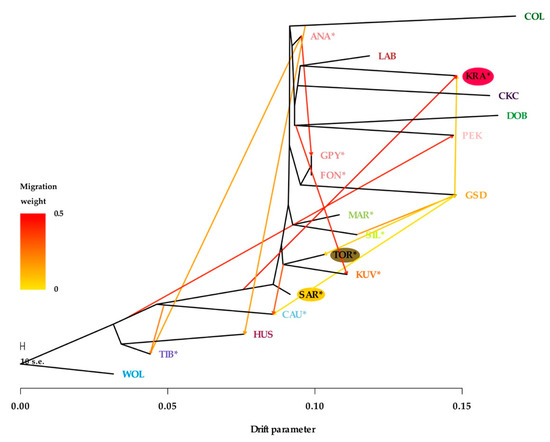
Figure 6.
TreeMix graph visualizing the phylogenetic relationship among 18 dog breeds and gray wolf as a root, in a model allowing 12 migration events. Migration arrows are colored according to their weights, while branch lengths are proportional to the estimated genetic drift. The breeds shown are Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), Tibetan Mastiff (TIB), and Tornjak (TOR) as livestock guarding dogs, Cavalier King Charles Spaniel (CKC), Rough Collie (COL), Doberman Pinscher (DOB), German Shepherd Dog (GSD), Husky (HUS), Labrador Retriever (LAB), and Pekingese (PEK) as popular dog breeds and gray wolves (WOL) as a reference wild population.
3.3. Population Structure and Admixture
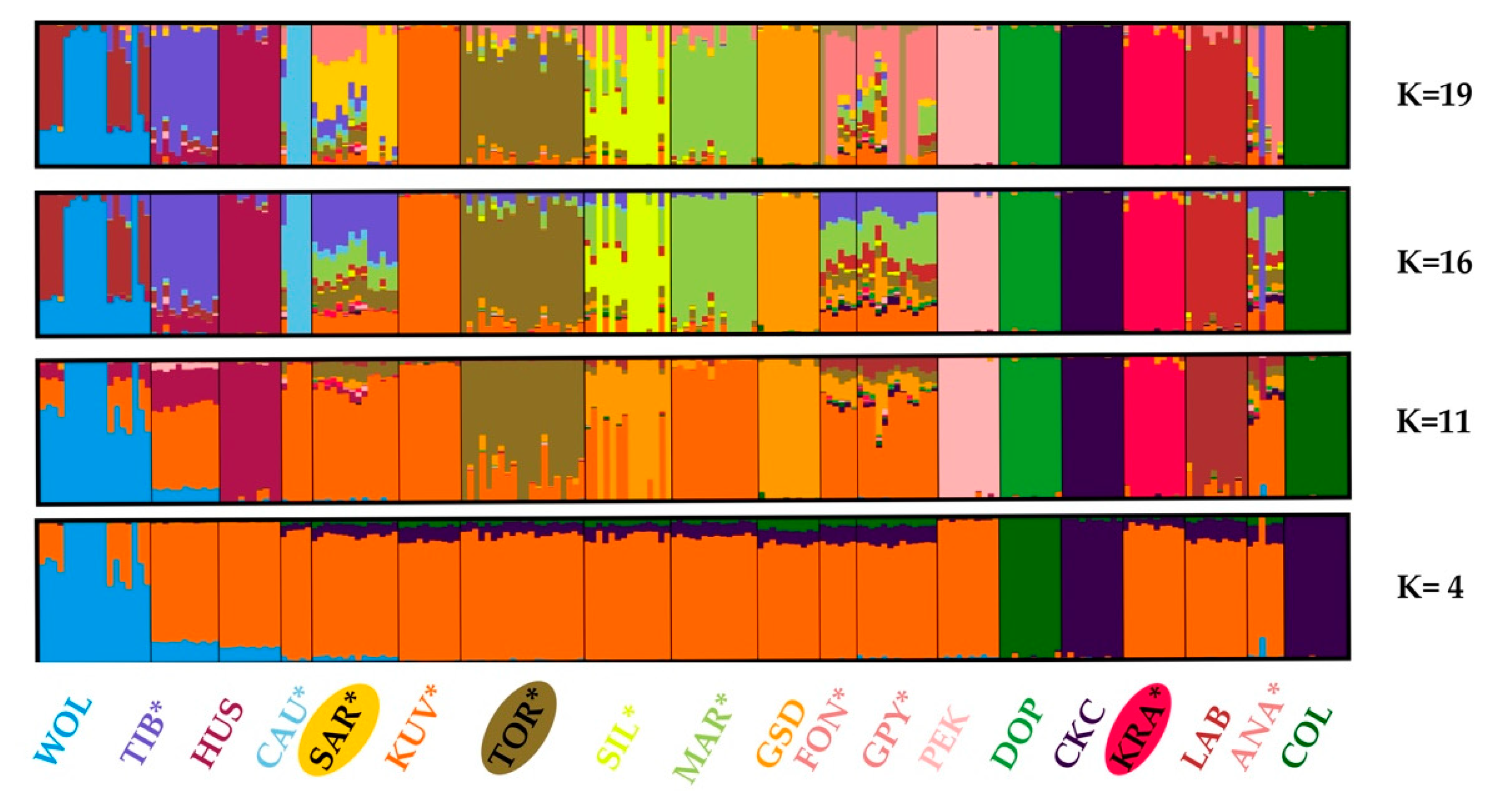
We estimated population structure and admixture by the algorithm implemented in the STRUCTURE software. The STRUCTURE can reveal “hidden structure” without a priori assigning individual breed or population membership. According to recommendations [34,35], the most likely K is the one where Ln Pr(G|K) is maximized or where the smallest value of K captures the main structure in the data. In our analyses, it was challenging to decide which was the optimal number of clusters in the dataset (the most likely parameter K), as the Ln Pr(G|K) values increased continuously with the number of clusters (Ks) used in the model (Figure S1A in the Supplement). We further analyzed the rate of change in the Ln Pr(G|K) between successive K values as suggested by Evanno et al. (2005) [38]. In the presence of a hierarchical island population structure, this approach could reveal a more subtle structure within populations. In our dataset, see Figure S1B in the Supplement, several peaks were observed at K = 4, K = 11, K = 16, and K = 19, indicating the presence of a complex hierarchical structure. Therefore, we decided to present from the STRUCTURE analyses (Figure 7), only the results that are in concordance with the observed divergent change in ΔK [38].
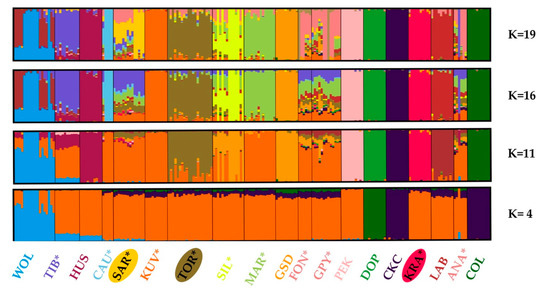
Figure 7.
Graphical presentation of the Bayesian unsupervised clustering of individual 193 dogs and 18 gray wolves performed with the STRUCTURE algorithm. The selection of the meaningful and relevant clusters (K = 19, K = 16, K = 11, and K = 4) is concordant with the procedure suggested by Evanno et al. (2005) [38]. The breeds shown are Anatolian Shepherd (ANA), Caucasian Shepherd (CAU), Fonni’s Dog (FON), Great Pyrenees (GPY), Karst Shepherd (KRA), Kuvazs (KUV), Maremmano-Abruzzese Sheepdog (MAR), Sharplanina Dog (SAR), Pastore della Sila (SIL), Tibetan Mastiff (TIB), and Tornjak (TOR) as livestock guarding dogs (*), Cavalier King Charles Spaniel (CKC), Rough Collie (COL), Doberman Pinscher (DOB), German Shepherd Dog (GSD), Husky (HUS), Labrador Retriever (LAB), and Pekingese (PEK) as popular dog breeds, and gray wolves (WOL) as a reference wild population.
At K = 19, the STRUCTURE algorithm distinguished three Balkan LGD breeds, although considerable admixture was observed within SAR and TOR. Notable admixture patterns were observed in a majority of LGD breeds (GPY, FON, KUV, MAR, SAR, SIL, and TOR). The presence of admixture patterns in LGD breeds is undoubtedly consistent with the explanations for the estimated higher heterozygosity and lower ROH inbreeding values, especially in those of intermediate and remote ancestry. Gene exchange, in both directions, was observed in all STRUCTURE results presented but was most pronounced in K = 4. Very strong introgression of dog genes was observed in some gray wolves, while the migration signal was not specific to a particular dog breed (Figure 7). We also recorded the introgression of gray wolf genes into some dog breeds. The strongest WOL introgression was observed in HUS and TIB, weak in SAR and ANA, and barely visible in GPY, PEK, and TOR.
Using pairwise Wright’s coefficients, we also estimated population differentiation between the analyzed breeds and the WOL, as well as average genetic relatedness to other dog breeds in the dataset (Table 2). According to the mean FST values across all other dog breeds, SAR (MFST = 0.13) and TOR (MFST = 0.15) were in the group of LGD breeds (GPY, MFST = 0.11; ANA, MFST = 0.12, and FON, MFST = 0.14) with the lowest mean FST values, occupying a central position among the analyzed dogs. In contrast, KRA (MFST = 0.27) was the most pronounced among the other LGD breeds with MFST values ranging from 0.11 to 0.23, which could be the consequence of the strong influence of GSD (MFST = 0.27) migrations.
4. Discussion
Although they do not achieve the popularity of some other dog breeds, LGD breeds have specific functional roles and behavioral abilities that were historically selected by humans during the domestication process. Today, their ability to protect sheep from large wild predators, usually wolves, ensures the livelihood of people in remote rural areas with harsh environmental conditions. Thus, LGD breeds, together with sheep, are important participants in the sustainable maintenance of specific biocultural grazing systems [39].
Three well-known Balkan LGD breeds that are used in mountainous areas of Bosnia and Herzegovina (TOR), Croatia (TOR), North Macedonia (SAR), Serbia (SAR), and Slovenia (KRA) were analyzed for the first time using high-throughput genomic information. The availability and use of high-density genome-wide arrays and the development of some population genomic methods, e.g., estimation of ROH-based inbreeding, enabled a more precise estimation of genetic conservation parameters compared to microsatellite-based studies [9,10]. More so, the open data sharing policy (samples were taken from Parker et al. (2017) [8] and Talenti et al. (2018) [21]) allowed comparison, with the same methodology used, of the targeted breeds (KRA, SAR, and TOR) with representative samples of LGDs and other popular dog breeds, as well as with gray wolves.
Sample sizes (from 5 to 20 in TOR) were very small for some breeds (ANA, CAU, and FON), but still comparable to other similar analyses [8,13,21]. However, we stand on the conclusions made because in many methods used (DAPC, NeighborNet, TreeMix analyses, STRUCTURE) the high number of informative SNPs can compensate for the small sample size since the mosaicism of the segregating segments is high. For example, looking back 5 or 10 generations, each individual inherits haplotypes from 32 or 1024 parents, respectively. When we have presented estimates related to genetic diversity and the degree of inbreeding, we have provided standard errors to allow the calculation of confidence intervals. For example, the same order of genetic diversity, SAR (highest), TOR (second highest), and KAR (lowest), was estimated in Ceh and Dovc (2014) [9] in 471 dogs (KRA = 326, SAR = 109, and TOR = 36) using 18 microsatellite loci.
On the other hand, we were not confident in estimating effective population size or selection signatures because these estimates are very sensitive to sample size [40]. A small sample size may be the reason for not detecting a subtle subdivision of the population. Dimitrijević et al. (2020) [10] detected the presence of two clusters when analyzing 94 SAR dogs based on 10 nuclear microsatellites. Therefore, it could be possible that we only sampled individuals from a single SAR cluster.
In our analysis, SAR dogs, like the majority of LGD breeds, showed some degree of admixture that was not detected by Dimitrijević et al. (2020) [10] using microsatellites. A large proportion of SAR dogs was assigned to the CAU breed in Ceh and Dovc (2014) [9]. The same authors (Ceh and Dovc, 2014 [9]) were able to show the presence of two subpopulations within the breed TOR when they performed a separate STRUCTURE analysis only for TOR individuals. We could not detect this subtle substructure since we did not perform a separate STRUCTURE analysis. TOR dogs are bred in two countries separated by the border (non-EU member Bosnia and Herzegovina versus EU member Croatia), which is a strong factor for modern breed differentiation (a good example in Druml et al. 2007 [41]). Therefore, we would not be surprised if the small level of subtle population structure is present in TOR. However, we do not believe that supporting breeding to separate populations is the sensible option, as these are dogs kept under strong natural selection pressure, and reduced genetic diversity would certainly reduce their ability to respond to natural selection. Identification of adaptive genes in Balkan LGDs is desirable but requires a large sample size.
Dreger et al. (2016) [42] found that the FON is present in the genetic profiles of other Mediterranean breeds and that the breed originated from the interbreeding of sighthound and mastiff breeds. It was further speculated that the relationship of FON with breeds from the eastern (Komondor) and southern (Saluki) Mediterranean parallels human migration to Sardinia [14,21,43]. We observed that the predominant cluster present in GPY and FON is also present in varying degrees of admixture in KUV, MAR, SAR, SIL, and TOR. It would be interesting to investigate whether the common origin present within LGD breeds is related to migrations in the Neolithization process. For example, the existence of hunting dogs similar to FON dates back to the Bronze Age [43]. Although this is speculation, it may be worthwhile to conduct further research on a large dataset, supplemented by additional archaeological evidence and evidence observed in analyses of ancient human and sheep DNA.
5. Conclusions
Using high-throughput genomic information and a series of analyses, we were able to answer all four hypotheses posed in this study and reached the following conclusions: (i) higher genetic diversity and low inbreeding, following the pattern observed in other LGD breeds, was observed in SAR (95% confidence interval; for Ho from 0.305 to 0.325, for He from 0.313 to 0.317, for FIS from −0.033 to 0.033, and for FROH>2Mb from 0.006 to 0.034) and TOR (95% confidence interval; for Ho from 0.295 to 0.307, for He from 0.299 to 0.302, for FIS from −0.020 to 0.020, and for FROH>2Mb from 0.023 to 0.043). In contrast, much lower genetic diversity and higher inbreeding, following the pattern observed in PDB, was observed in KRA (95% confidence interval; for Ho from 0.235 to 0.247, for He from 0.220 to 0.224, for FIS from −0.108 to −0.054, and for FROH>2Mb from 0.063 to 0.111). (ii) Population genetic structure analyses (multivariate, phylogenetic, and unsupervised clustering) showed that SAR and TOR are closely related to other LGD breeds, whereas KRA is a slightly genetically divergent population with estimated influence from GSD. (iii) Analysis of STRUCTURE revealed the presence of admixture in SAR and TOR, which seems to be characteristic of most LGD breeds, while genetic purity was observed in KRA. (iv) Traces of admixture from dogs to gray wolves and from gray wolves to dogs were most pronounced at K = 4 in the STRUCTURE analysis. The strongest introgression of gray wolf genes was observed in HUS and TIM, while signals were much lower in SAR and ANA and negligible in GPY, PEK, and TOR.
Supplementary Materials
The following are available online at https://www.mdpi.com/2071-1050/13/4/2289/s1, Figure S1. The choice of the representative number of clusters (K): (A) Plot of Ln Pr(G|K) values presented as a function of the number of clusters [34,35], 10 runs presented. (B) Plot of ΔKs presented as a function of the number of clusters [38].
Author Contributions
V.C.-C., I.C., and P.D. conceived and designed the study; M.J., V.C.-C., and P.D. organized sample collection, extraction, and genotyping; M.J., M.Z., and M.F. performed the analyses with the supervision of I.C.; M.J., I.C., and V.C.-C. wrote the manuscript; V.C.-C. and P.D. funding acquisition. All authors have read and agreed to the published version of the manuscript.
Funding
This study was supported by the Croatian Science Foundation (Project ANAGRAMS-IP-2018-01-8708 “Application of NGS in assessment of genomic variability in ruminants”), Slovenian Research Agency (Research programme P4-0220, Comparative genomics and genomic biodiversity), and the support of the Ministry of Science and Education of the Republic of Croatia (bilateral cooperation project between Croatia and Slovenia-BI-HR/14-15-033). This work is part of the Ph.D. dissertation of Mateja Janeš and was additionally supported by CEEPUS Network scholarship Number: CIII-SK-0044-09-1415, CEEPUS IIIFREEMOVER-1516-89203 mobility Grant and ERASMUS + Student Mobility for a traineeship 2016/17.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
The data presented in this study are available on request from the corresponding author.
Acknowledgments
The authors wish to express their gratitude to the dog breeders and owners who provided samples and information about these beautiful livestock guardian dog breeds. In particular, we thank the Croatian Kennel Association (HKS) and Cynological Association Slovenia (KZS) for the organization and sample collection. We thank Ana Galov and Haidi Arbanasić for providing four samples (tissue) of gray wolves from Croatia and Anamarija Jakšić for providing several Tornjak samples. Finally, we thank the two anonymous reviewers whose comments and suggestions helped improve and clarify this manuscript.
Conflicts of Interest
The authors state that the study was conducted without any commercial or financial relationship that could be interpreted as a potential conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- Yilmaz, O.; Coskun, F.; Ertugrul, M. Livestock damage by carnivores and use of livestock guardian dogs for its prevention in Europe—A review. J. Livest. Sci. 2015, 6, 23–35. [Google Scholar]
- Linnell, J.D.C.; Lescureux, N. Livestock Guarding Dogs: Cultural Heritage Icons with a New Relevance for Mitigating Conservation Conflicts; Norwegian Institute for Nature Research: Trondheim, Norway, 2015; p. 76. [Google Scholar]
- Frantz, L.A.F.; Mullin, V.E.; Pionnier-Capitan, M.; Lebrasseur, O.; Ollivier, M.; Perri, A.; Linderholm, A.; Mattiangeli, V.; Teasdale, M.D.; Dimopoulos, E.A.; et al. Genomic and archaeological evidence suggest a dual origin of domestic dogs. Science 2016, 352, 1228–1231. [Google Scholar] [CrossRef]
- Freedman, A.H.; Gronau, I.; Schweizer, R.M.; Ortega-Del Vecchyo, D.; Han, E.; Silva, P.M.; Galaverni, M.; Fan, Z.; Marx, P.; Lorente-Galdos, B.; et al. Genome Sequencing Highlights the Dynamic Early History of Dogs. PLoS Genet. 2014, 10, e1004016. [Google Scholar] [CrossRef]
- Gagliardi, R.; Silvia, L.; García, C.; Arruga, M.V. Microsatellite characterization of Cimarron Uruguayo dogs. Genet. Mol. Biol. 2010, 34, 165–168. [Google Scholar] [CrossRef]
- Irion, D.N. Analysis of Genetic Variation in 28 Dog Breed Populations With 100 Microsatellite Markers. J. Hered. 2003, 94, 81–87. [Google Scholar] [CrossRef]
- Lindblad-Toh, K.; Wade, C.M.; Mikkelsen, T.S.; Karlsson, E.K.; Jaffe, D.B.; Kamal, M.; Clamp, M.; Chang, J.L.; Kulbokas, E.J.; Zody, M.C.; et al. Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 2005, 438, 803–819. [Google Scholar] [CrossRef]
- Parker, H.G.; Dreger, D.L.; Rimbault, M.; Davis, B.W.; Mullen, A.B.; Carpintero-Ramirez, G.; Ostrander, E.A. Genomic Analyses Reveal the Influence of Geographic Origin, Migration, and Hybridization on Modern Dog Breed Development. Cell Rep. 2017, 19, 697–708. [Google Scholar] [CrossRef]
- Ceh, E.; Dovc, P. Population structure and genetic differentiation of livestock guard dog breeds from the Western Balkans. J. Anim. Breed. Genet. 2014, 131, 313–325. [Google Scholar] [CrossRef]
- Dimitrijević, V.; Savić, M.; Tarić, E.; Stanišić, L.; Stanimirović, Z.; Tabaković, A.; Aleksić, M.J. Genetic Characterization of the Yugoslavian Shepherd Dog—Sharplanina, a Livestock Guard Dog from the Western Balkans. Acta Vet. Brno 2020, 70, 329–345. [Google Scholar] [CrossRef]
- Bigi, D.; Marelli, S.P.; Randi, E.; Polli, M. Genetic characterization of four native Italian shepherd dog breeds and analysis of their relationship to cosmopolitan dog breeds using microsatellite markers. Animal 2015, 9, 1921–1928. [Google Scholar] [CrossRef]
- Bigi, D.; Marelli, S.P.; Liotta, L.; Frattini, S.; Talenti, A.; Pagnacco, G.; Polli, M.; Crepaldi, P. Investigating the population structure and genetic differentiation of livestock guard dog breeds. Animal 2018, 12, 2009–2016. [Google Scholar] [CrossRef] [PubMed]
- VonHoldt, B.M.; Pollinger, J.P.; Lohmueller, K.E.; Han, E.; Parker, H.G.; Quignon, P.; Degenhardt, J.D.; Boyko, A.R.; Earl, D.A.; Auton, A.; et al. Genome-wide SNP and haplotype analyses reveal a rich history underlying dog domestication. Nature 2010, 464, 898–902. [Google Scholar] [CrossRef]
- Ostrander, E.A.; Wayne, R.K.; Freedman, A.H.; Davis, B.W. Demographic history, selection and functional diversity of the canine genome. Nat. Rev. Genet. 2017, 18, 705–720. [Google Scholar] [CrossRef] [PubMed]
- Bergström, A.; Frantz, L.; Schmidt, R.; Ersmark, E.; Lebrasseur, O.; Girdland-Flink, L.; Lin, A.T.; Storå, J.; Sjögren, K.-G.; Anthony, D.; et al. Origins and genetic legacy of prehistoric dogs. Science 2020, 370, 557–564. [Google Scholar] [CrossRef]
- Sinding, M.-H.S.; Gopalakrishnan, S.; Ramos-Madrigal, J.; de Manuel, M.; Pitulko, V.V.; Kuderna, L.; Feuerborn, T.R.; Frantz, L.A.F.; Vieira, F.G.; Niemann, J.; et al. Arctic-adapted dogs emerged at the Pleistocene–Holocene transition. Science 2020, 368, 1495–1499. [Google Scholar] [CrossRef]
- Janeš, M.; Zorc, M.; Cubric-Curik, V.; Curik, I.; Dovc, P. Population structure and genetic history of Tibetan Terriers. Genet. Sel. Evol. 2019, 51, 79. [Google Scholar] [CrossRef] [PubMed]
- Lampi, S.; Donner, J.; Anderson, H.; Pohjoismäki, J. Variation in breeding practices and geographic isolation drive subpopulation differentiation, contributing to the loss of genetic diversity within dog breed lineages. Canine Med. Genet. 2020, 7, 5. [Google Scholar] [CrossRef]
- Letko, A.; Minor, K.M.; Jagannathan, V.; Seefried, F.R.; Mickelson, J.R.; Oliehoek, P.; Drögemüller, C. Genomic diversity and population structure of the Leonberger dog breed. Genet. Sel. Evol. 2020, 52, 61. [Google Scholar] [CrossRef]
- Puente, J.M.A.; Barro, Á.L.P.; de la Haba Giraldo, M.R.; Bermejo, J.V.D.; González, F.J.N. Does Functionality Condition the Population Structure and Genetic Diversity of Endangered Dog Breeds under Island Territorial Isolation? Animals 2020, 10, 1893. [Google Scholar] [CrossRef]
- Talenti, A.; Dreger, D.L.; Frattini, S.; Polli, M.; Marelli, S.; Harris, A.C.; Liotta, L.; Cocco, R.; Hogan, A.N.; Bigi, D.; et al. Studies of modern Italian dog populations reveal multiple patterns for domestic breed evolution. Ecol. Evol. 2018, 8, 2911–2925. [Google Scholar] [CrossRef]
- Wright, S. The Interpretation of Population Structure by F-Statistics with Special Regard to Systems of Mating. Evolution 1965, 19, 395. [Google Scholar] [CrossRef]
- Ferenčaković, M.; Sölkner, J.; Curik, I. Estimating autozygosity from high-throughput information: Effects of SNP density and genotyping errors. Genet. Sel. Evol. 2013, 45, 42. [Google Scholar] [CrossRef]
- McQuillan, R.; Leutenegger, A.L.; Abdel-Rahman, R.; Franklin, C.S.; Pericic, M.; Barac-Lauc, L.; Smolej-Narancic, N.; Janicijevic, B.; Polasek, O.; Tenesa, A.; et al. Runs of Homozygosity in European Populations. Am. J. Hum. Genet. 2008, 83, 359–372. [Google Scholar] [CrossRef] [PubMed]
- Curik, I.; Ferenčaković, M.; Sölkner, J. Inbreeding and runs of homozygosity: A possible solution to an old problem. Livest. Sci. 2014, 166, 26–34. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: Berlin/Heidelberg, Germany, 2016; ISBN 3319242776. [Google Scholar]
- Saura, M.; Fernández, A.; Rodríguez, M.C.; Toro, M.A.; Barragán, C.; Fernández, A.I.; Villanueva, B. Genome-Wide Estimates of Coancestry and Inbreeding in a Closed Herd of Ancient Iberian Pigs. PLoS ONE 2013, 8, e78314. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Devillard, S.; Balloux, F. Discriminant analysis of principal components: A new method for the analysis of genetically structured populations. BMC Genet. 2010, 11, 94. [Google Scholar] [CrossRef] [PubMed]
- Jombart, T.; Collins, C. A tutorial for Discriminant Analysis of Principal Components (DAPC) using adegenet. Rvignette 2015. Available online: http://adegenet.r-forge.r-project.org/files/tutorial-dapc.pdf (accessed on 23 December 2020).
- Nei, M. Genetic Distance between Populations. Am. Nat. 1972, 106, 283–292. [Google Scholar] [CrossRef]
- Pembleton, L.W.; Cogan, N.O.I.; Forster, J.W. StAMPP: An R package for calculation of genetic differentiation and structure of mixed-ploidy level populations. Mol. Ecol. Resour. 2013, 13, 946–952. [Google Scholar] [CrossRef]
- Huson, D.H.; Bryant, D. Application of Phylogenetic Networks in Evolutionary Studies. Mol. Biol. Evol. 2006, 23, 254–267. [Google Scholar] [CrossRef]
- Pickrell, J.K.; Pritchard, J.K. Inference of Population Splits and Mixtures from Genome-Wide Allele Frequency Data. PLoS Genet. 2012, 8, e1002967. [Google Scholar] [CrossRef] [PubMed]
- Pritchard, J.K.; Stephens, M.; Donnelly, P. Inference of population structure using multilocus genotype data. Genetics 2000, 155, 945–959. [Google Scholar] [CrossRef] [PubMed]
- Falush, D.; Stephens, M.; Pritchard, J.K. Inference of population structure using multilocus genotype data: Dominant markers and null alleles. Mol. Ecol. Notes 2007, 7, 574–578. [Google Scholar] [CrossRef]
- Rosenberg, N.A. Distruct: A program for the graphical display of population structure. Mol. Ecol. Notes 2003, 4, 137–138. [Google Scholar] [CrossRef]
- Weir, B.S.; Cockerham, C.C. Estimating F -statistics for the analysis of population structure. Evolution 1984, 38, 1358–1370. [Google Scholar] [CrossRef] [PubMed]
- Evanno, G.; Regnaut, S.; Goudet, J. Detecting the number of clusters of individuals using the software structure: A simulation study. Mol. Ecol. 2005, 14, 2611–2620. [Google Scholar] [CrossRef]
- Paraskevopoulou, C.; Theodoridis, A.; Johnson, M.; Ragkos, A.; Arguile, L.; Smith, L.; Vlachos, D.; Arsenos, G. Sustainability Assessment of Goat and Sheep Farms: A Comparison between European Countries. Sustainability 2020, 12, 3099. [Google Scholar] [CrossRef]
- Wiener, P.; Sánchez-Molano, E.; Clements, D.N.; Woolliams, J.A.; Haskell, M.J.; Blott, S.C. Genomic data illuminates demography, genetic structure and selection of a popular dog breed. BMC Genom. 2017, 18, 609. [Google Scholar] [CrossRef]
- Druml, T.; Curik, I.; Baumung, R.; Aberle, K.; Distl, O.; Sölkner, J. Individual-based assessment of population structure and admixture in Austrian, Croatian and German draught horses. Heredity 2007, 98, 114–122. [Google Scholar] [CrossRef]
- Dreger, D.L.; Davis, B.W.; Cocco, R.; Sechi, S.; Di Cerbo, A.; Parker, H.G.; Polli, M.; Marelli, S.P.; Crepaldi, P.; Ostrander, E.A. Commonalities in Development of Pure Breeds and Population Isolates Revealed in the Genome of the Sardinian Fonni’s Dog. Genetics 2016, 204, 737–755. [Google Scholar] [CrossRef]
- Sechi, S.; Polli, M.; Marelli, S.; Talenti, A.; Crepaldi, P.; Fiore, F.; Spissu, N.; Dreger, D.L.; Zedda, M.; Dimauro, C.; et al. Fonni’s dog: Morphological and genetic characteristics for a breed standard definition. Ital. J. Anim. Sci. 2017, 16, 22–30. [Google Scholar] [CrossRef]
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).