Evaluation of Biomonitoring Strategies to Assess Performance of a Bioremediation Bioprocess
Abstract
:1. Introduction
2. Material and Methods
2.1. Chemicals and Growth Media Used in This Study
2.2. Sediment Sampling and Pollutant Spiking
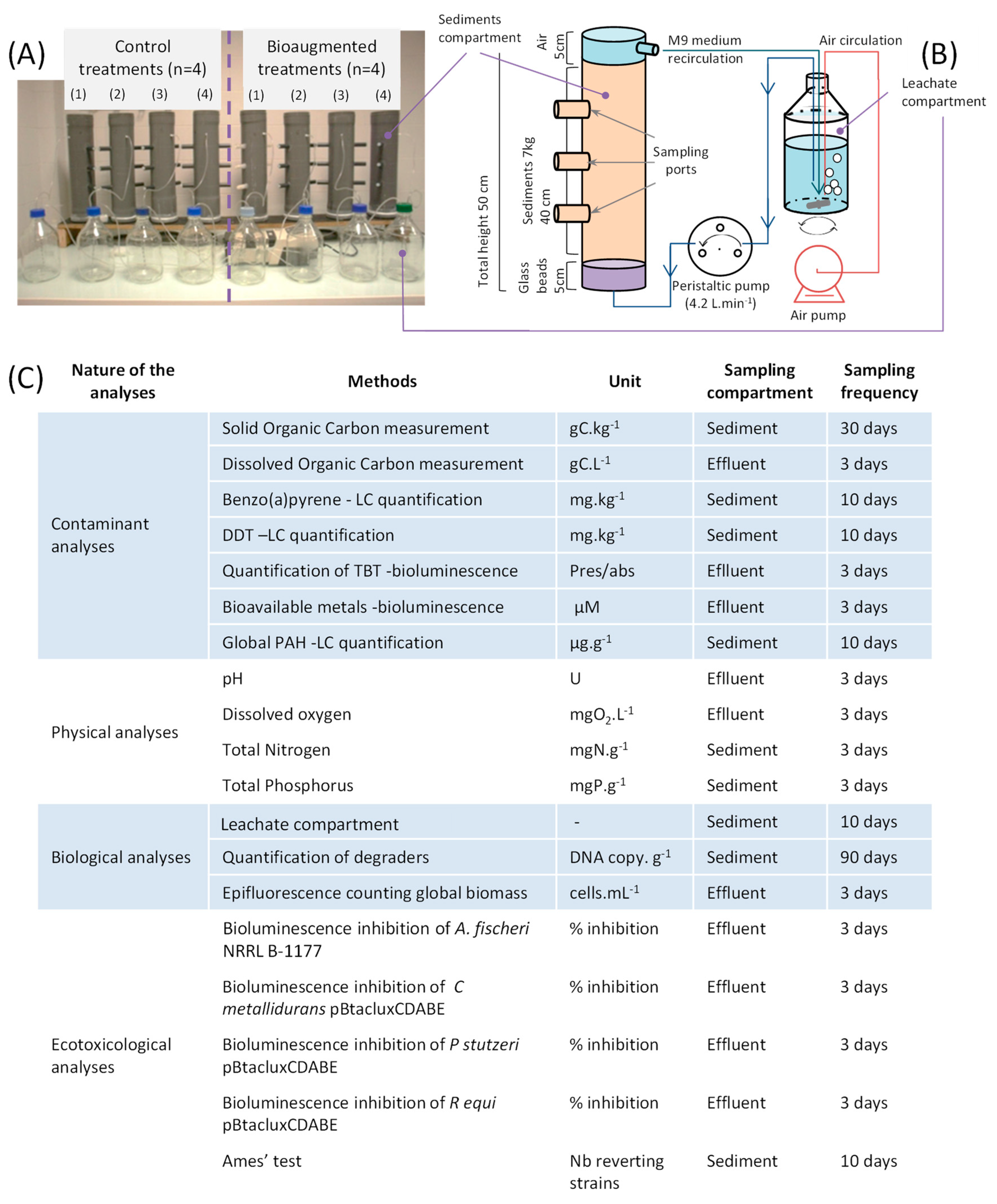
2.3. Bioreactor Configuration and Sampling
2.4. Analysis of Physico-Chemical Parameters
2.4.1. Total Organic Carbon Analysis
2.4.2. Nitrogen and Phosphorus Quantification
2.4.3. Pollutant Extraction and Analysis
2.4.4. Measurement of Bioavailable Metals
2.4.5. Detection of Bioavailable Tributyltin
2.4.6. Leaching Test
2.5. Microbiological Methods
2.5.1. Biodegraders for the Bioremediation
2.5.2. Construction of the Microbial Toxicity Bioreporters
2.5.3. Total Bacterial Counts by Epifluorescence: BacLight Viability Kit
2.5.4. Quantification of Degraders by qPCR Clone Library Construction and Strain Determination
2.6. Ecotoxicity Assessment
2.6.1. Acute Toxicity Assessments
2.6.2. Mutagenicity Assessment with the Ames Test
2.7. Statistical Analysis
3. Results
3.1. Overall Efficiency of the Bioremediation Processes
3.2. Impact of the Bioremediation Processes
4. Discussions
5. Conclusion
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Dangi, A.K.; Sharma, B.; Hill, R.T.; Shukla, P. Bioremediation through microbes: Systems biology and metabolic engineering approach. Crit. Rev. Biotechnol. 2019, 39, 79–98. [Google Scholar] [CrossRef]
- Dua, M.; Singh, A.; Sethunathan, N.; Johri, A. Biotechnology and bioremediation: Successes and limitations. Appl. Microbiol. Biotechnol. 2002, 59, 143–152. [Google Scholar]
- Quintella, C.M.; Mata, A.M.T.; Lima, L.C.P. Overview of bioremediation with technology assessment and emphasis on fungal bioremediation of oil contaminated soil. J. Environ. Manag. 2019, 241, 159–166. [Google Scholar] [CrossRef] [PubMed]
- Margesin, R.; Fonteyne, P.A.; Redl, B. Low-temperature biodegradation of high amounts of phenol by Rhodococcus spp. and basidiomycetous yeasts. Res. Microbiol. 2005, 156, 68–75. [Google Scholar] [CrossRef] [PubMed]
- Teng, Y.; Luo, Y.; Sun, M.; Liu, Z.; Li, Z.; Christie, P. Effect of bioaugmentation by Paracoccus sp. strain HPD-2 on the soil microbial community and removal of polycyclic aromatic hydrocarbons from an aged contaminated soil. Bioresour. Technol. 2010, 101, 3437–3443. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; Da Fonseca, M.M.R.; De Carvalho, C.C.C.R. Bioaugmentation and biostimulation stategies to improve the effectiveness of bioremediation processes. Biodegradation 2011, 22, 231–241. [Google Scholar] [CrossRef]
- Ward, O.; Singh, A.; Van Hamme, J. Accelerated biodegradation of petroleum hydrocarbon waste. J. Ind. Microbiol. Biotechnol. 2003, 30, 260–270. [Google Scholar] [CrossRef]
- Andreoni, V.; Gianfreda, L. Bioremediation and monitoring of aromatic-polluted habitats. Appl. Microbiol. Biotechnol. 2007, 76, 287–308. [Google Scholar] [CrossRef]
- Crisafi, F.; Genovese, M.; Smedile, F.; Russo, D.; Catalfamo, M.; Yakimov, M.; Denaro, R. Bioremediation technologies for polluted seawater sampled after an oil-spill in Taranto Gulf (Italy): A comparison of biostimulation, bioaugmentation and use of a washing agent in microcosm studies. Mar. Pollut. Bull. 2016, 106, 119–126. [Google Scholar] [CrossRef]
- Heitzer, A.; Sayler, G.S. Monitoring the efficacy of bioremediation. Trends Biotechnol. 1993, 11, 334–343. [Google Scholar] [CrossRef]
- Korda, A.; Santas, P.; Tenente, A.; Santas, R. Petroleum hydrocarbon bioremediation: Sampling and analytical techniques, in situ treatments and commercial microorganisms currently used. Appl. Microbiol. Biotechnol. 1997, 48, 677–686. [Google Scholar] [CrossRef] [PubMed]
- Mansouri, A.; Cregut, M.; Abbes, C.; Durand, M.J.; Landoulsi, A.; Thouand, G. The environmental issues of DDT pollution and bioremediation: A multidisciplinary review. Appl. Biochem. Biotechnol. 2017, 181, 309–339. [Google Scholar] [CrossRef]
- Martins, M.V.A.; Zaaboub, N.; Aleya, L.; Frontalini, F.; Pereira, E.; Miranda, P.; Mane, M.; Rocha, F.; Laut, L.; El Bour, M. Environmental quality assessment of Bizerte Lagoon (Tunisia) using living foraminifera assemblages and a multiproxy approach. PLoS ONE 2015, 10, e0137250. [Google Scholar] [CrossRef]
- Phillips, T.M.; Liu, D.; Seech, A.G.; Lee, H.; Trevors, J.T. Monitoring bioremediation in creosote-contaminated soils using chemical analysis and toxicity tests. J. Ind. Microbiol. Biotech. 2000, 24, 132–139. [Google Scholar] [CrossRef]
- Pierro, L.; Matturro, B.; Rossetti, S.; Sagliaschi, M.; Sucato, S.; Alesi, E.; Bartsch, E.; Arjmand, F.; Papini, M.P. Polyhydroxyalkanoate as a slow-release carbon source for in situ bioremediation of contaminated aquifers: From laboratory investigation to pilot-scale testing in the field. New Biotechnol. 2017, 37, 60–68. [Google Scholar] [CrossRef] [PubMed]
- Sabaté, J.; Viñas, M.; Solanas, A.M. Bioavailability assessment and environmental fate of polycyclic aromatic hydrocarbons in biostimulated creosote-contaminated soil. Chemosphere 2006, 63, 1648–1659. [Google Scholar] [CrossRef]
- Van Straalen, N.M. Assessment of soil contamination–a functional perspective. Biodegradation 2002, 13, 41–52. [Google Scholar] [CrossRef]
- D’souza, S.F. Microbial biosensors. Biosens. Bioelectron. 2001, 16, 337–353. [Google Scholar] [CrossRef]
- Man, Y.B.; Chow, K.L.; Kang, Y.; Wong, M.H. Mutagenicity and genotoxicity of Hong Kong soils contaminated by polycyclic aromatic hydrocarbons and dioxins/furans. Mutat. Res. Genet. Toxicol. Environ. Mutagen. 2013, 752, 47–56. [Google Scholar] [CrossRef]
- Phillips, T.M.; Liu, S.; Seech, A.G. Bioremediation in Field Box Plots of a Soil Contaminated with Wood-Preservatives: A Comparison of Treatment Conditions using Toxicity Testing as a Monitoring Technique. Water Air Soil Pollut. 2000, 121, 173–187. [Google Scholar] [CrossRef]
- Steliga, T.; Jakubowicz, P.; Kapusta, P. Changes in toxicity during in situ bioremediation of weathered drill wastes contaminated with petroleum hydrocarbons. Bioresour. Technol. 2012, 125, 1–10. [Google Scholar] [CrossRef] [PubMed]
- Verma, N.; Singh, M. Biosensors for heavy metals. Biometals 2005, 18, 121–129. [Google Scholar] [CrossRef] [PubMed]
- White, D.C.; Flemming, C.A.; Leung, K.T.; Macnaughton, S.J. In situ microbial ecology for quantitative appraisal, monitoring, and risk assessment of pollution remediation in soils, the subsurface, the rhizosphere and in biofilms. J. Microbiol. Methods 1998, 32, 93–105. [Google Scholar] [CrossRef]
- Widada, J.; Nojiri, H.; Omori, T. Recent developments in molecular techniques for identification and monitoring of xenobiotic-degrading bacteria and their catabolic genes in bioremediation. Appl. Microbiol. Biotechnol. 2002, 60, 45–59. [Google Scholar]
- Barhoumi, B.; Elbarhoumi, A.; Clérandeau, C.; Al-Rawabdeh, A.M.; Atyaoui, A.; Touil, S.; Driss, M.R.; Cachot, J. Using an Integrated Approach to Assess the Sediment Quality of a Mediterranean Lagoon, the Bizerte Lagoon (Tunisia). Ecotoxicology 2016, 25, 1082–1104. [Google Scholar] [CrossRef]
- De Souza Pohren, R.; Rocha, J.A.V.; Horn, K.A.; Vargas, V.M.F. Bioremediation of soils contaminated by PAHs: Mutagenicity as a tool to validate environmental quality. Chemosphere 2019, 214, 659–668. [Google Scholar] [CrossRef]
- Dorn, P.B.; Salanitro, J.P. Temporal ecological assessment of oil contaminated soils before and after bioremediation. Chemosphere 2000, 40, 419–426. [Google Scholar] [CrossRef]
- Foucault, Y.; Durand, M.J.; Tack, K.; Schreck, E.; Geret, F.; Leveque, T.; Pradere, P.; Goix, S.; Dumat, C. Use of ecotoxicity test and ecoscores to improve the management of polluted soils: Case of a secondary lead smelter plant. J. Hazard. Mater. 2013, 246, 291–299. [Google Scholar] [CrossRef] [PubMed]
- Jouanneau, S.; Durand, M.J.; Lahmar, A.; Thouand, G. Main Technological Advancements in Bacterial Bioluminescent Biosensors Over the Last Two Decades. In Bioluminescence: Fundamentals and Applications in Biotechnology; Thouand, G., Marks, R., Eds.; Advances in Biochemical Engineering/Biotechnology; Springer: Berlin/Heidelberg, Germany, 2015; Volume 3, pp. 101–116. [Google Scholar]
- Kim, K.R.; Owens, G. Potential for enhanced phytoremediation of landfills using biosolids—A review. J. Environ. Manag. 2010, 91, 791–797. [Google Scholar] [CrossRef]
- Lei, Y.; Chen, W.; Mulchandani, A. Microbial biosensors. Anal. Chim. Acta 2006, 568, 200–210. [Google Scholar] [CrossRef]
- Leitgib, L.; Kálmán, J.; Gruiz, K. Comparison of bioassays by testing whole soil and their water extract from contaminated sites. Chemosphere 2007, 66, 428–434. [Google Scholar] [CrossRef] [PubMed]
- Płaza, G.; Nałęcz-Jawecki, G.; Ulfig, K.; Brigmon, R.L. The application of bioassays as indicators of petroleum-contaminated soil remediation. Chemosphere 2005, 59, 289–296. [Google Scholar] [CrossRef] [PubMed]
- Saterbak, A.; Toy, R.J.; Wong, D.C.; McMain, B.J.; Williams, M.P.; Dorn, P.B.; Brzuzy, L.P.; Chai, E.Y.; Salanitro, J.P. Ecotoxicological and analytical assessment of hydrocarbon-contaminated soils and application to ecological risk assessment. Environ. Toxicol. Chem. 1999, 18, 1591–1607. [Google Scholar] [CrossRef]
- Chi, X.Q.; Zhang, J.J.; Zhao, S.; Zhou, N.Y. Bioaugmentation with a consortium of bacterial nitrophenol-degraders for remediation of soil contaminated with three nitrophenol isomers. Environ. Pollut. 2013, 172, 33–41. [Google Scholar] [CrossRef] [PubMed]
- Edlund, A.; Jansson, J.K. Changes in active bacterial communities before and after dredging of highly polluted Baltic Sea sediments. Appl. Environ. Microbiol. 2006, 72, 6800–6807. [Google Scholar] [CrossRef]
- Hall, K.; Miller, C.D.; Sorensen, D.L.; Anderson, A.J.; Sims, R.C. Development of a catabolically significant genetic probe for polycyclic aromatic hydrocarbon-degrading Mycobacteria in soil. Biodegradation 2005, 16, 475–484. [Google Scholar] [CrossRef]
- Hendrickx, B.; Dejonghe, W.; Boënne, W.; Brennerova, M.; Cernik, M.; Lederer, T.; Bucheli-Witschel, M.; Bastiaens, L.; Verstraete, W.; Top, E.M.; et al. Dynamics of an oligotrophic bacterial aquifer community during contact with a groundwater plume contaminated with benzene, toluene, ethylbenzene, and xylenes: An in situ mesocosm study. Appl. Environ. Microbiol. 2005, 71, 3815–3825. [Google Scholar] [CrossRef]
- Junca, H.; Pieper, D.H. Functional gene diversity analysis in BTEX contaminated soils by means of PCR-SSCP DNA fingerprinting: Comparative diversity assessment against bacterial isolates and PCR-DNA clone libraries. Environ. Microbiol. 2004, 6, 95–110. [Google Scholar] [CrossRef]
- Paissé, S.; Coulon, F.; Goñi-Urriza, M.; Peperzak, L.; McGenity, T.J.; Duran, R. Structure of bacterial communities along a hydrocarbon contamination gradient in a coastal sediment. FEMS Microbiol. Ecol. 2008, 66, 295–305. [Google Scholar] [CrossRef]
- Rutgers, M.; Breure, A.M. Risk assessment, microbial communities, and pollution-induced community tolerance. Hum. Ecol. Risk Assess 1999, 5, 661–670. [Google Scholar] [CrossRef]
- Sabaté, J.; Vinas, M.; Solanas, A.M. Laboratory-scale bioremediation experiments on hydrocarbon-contaminated soils. Int. Biodeterior. Biodegrad. 2004, 54, 19–25. [Google Scholar] [CrossRef]
- Barhoumi, B.; LeMenach, K.; Devier, M.H.; Ben Ameur, W.; Etcheber, H.; Budzinski, H.; Cachot, J.; Driss, M.R. Polycyclic aromatic hydrocarbons (PAHs) in surface sediments from the Bizerte Lagoon, Tunisia: Levels, sources, and toxicological significance. Environ. Monit. Assess. 2014, 186, 2653–2669. [Google Scholar] [CrossRef] [PubMed]
- Barhoumi, B.; LeMenach, K.; Devier, M.H.; El Megdiche, Y.; Hammami, B.; Ben Ameur, W.; Ben Hassine, S.; Cachot, J.; Budzinski, H.; Driss, M.R. Distribution and ecological risk of polychlorinated biphenyls (PCBs) and organochlorine pesticides (OCPs) in surface sediments from the Bizerte lagoon, Tunisia. Environ. Sci. Pollut. Res. 2014, 21, 6290–6302. [Google Scholar] [CrossRef]
- Mansouri, A.; Abbes, C.; Ben Mouhoub, R.; Ben Hassine, S.; Landoulsi, A. Enhancement of mixture pollutant biodegradation efficiency using a bacterial consortium under static magnetic field. PLoS ONE 2019, 14, e0208431. [Google Scholar] [CrossRef] [PubMed]
- Jouanneau, S.; Durand, M.J.; Courcoux, P.; Blusseau, T.; Thouand, G. Improvement of the identification of four heavy metals in environmental samples by using predictive decision tree models coupled with a set of five bioluminescent bacteria. Env. Sci. Technol. 2011, 45, 2925–2931. [Google Scholar] [CrossRef]
- Atlas, R.M. Handbook of Microbiological Media, 2nd ed.; CRC Press: Boca Raton, FL, USA, 1997. [Google Scholar]
- Ben Garali, A.; Ouakad, M.; Gueddari, M. Contamination of superficial sediments by heavy metals and iron in the Bizerte lagoon, northern Tunisia. Arab. J. Geosci. 2010, 3, 295–306. [Google Scholar] [CrossRef]
- Trabelsi, S.; Driss, M.R. Polycyclic aromatic hydrocarbons in superficial coastal sediments from Bizerte Lagoon, Tunisia. Mar. Pollut. Bull. 2005, 50, 344–348. [Google Scholar] [CrossRef]
- OECD. Sediment-Water Chironomid Life-Cycle Toxicity Test Using Spiked Water or Spiked Sediment. In OECD Guidelines for the Testing of Chemicals Proposal for a New Guideline 2009; OECD: Paris, France, 2009. [Google Scholar]
- Charrier, T.; Chapeau, C.; Bendria, L.; Picart, P.; Daniel, P.; Thouand, G. A multi-channel bioluminescent bacterial biosensor for the on-line detection of metals and toxicity. Part II: Technical development and proof of concept of the biosensor. Anal. Bioanal. Chem. 2011, 400, 1061–1070. [Google Scholar] [CrossRef]
- Jouanneau, S.; Durand, M.J.; Thouand, G. Online detection of metals in environmental samples: Comparing two concepts of bioluminescent bacterial biosensors. Env. Sci. Technol. 2012, 46, 11979–11987. [Google Scholar] [CrossRef]
- Gueuné, H.; Thouand, G.; Durand, M.J. A new bioassay for the inspection and identification of TBT-containing antifouling paint. Mar. Pollut. Bull. 2009, 58, 1734–1738. [Google Scholar] [CrossRef]
- Durand, M.J.; Thouand, G.; Dancheva-Ivanova, T.; Vachon, P.; DuBow, M. Specific detection of organotin compounds with a recombinant luminescent bacteria. Chemosphere 2003, 52, 103–111. [Google Scholar] [CrossRef]
- Cregut, M.; Jouanneau, S.; Brillet, F.; Durand, M.J.; Sweetlove, C.; Chenèble, J.C.; L’Haridon, J.; Thouand, G. High throughput and miniaturised systems for biodegradability assessments. Environ. Sci. Pollut. Res. 2014, 21, 9545–9552. [Google Scholar] [CrossRef] [PubMed]
- François, B.; Maul, A.; Durand, M.J.; Thouand, G. From laboratory to environmental conditions: A new approach for chemical’s biodegradability assessment. Environ. Sci. Pollut. Res. 2016, 23, 18684–18693. [Google Scholar] [CrossRef] [PubMed]
- Bennasar, A.; Guasp, C.; Lalucat, J. Molecular methods for the detection and identification of Pseudomonas stutzeri in pure culture and environmental samples. Microbial. Ecol. 1998, 35, 22–33. [Google Scholar] [CrossRef]
- Ryan, M.P.; Adley, C.C. Specific PCR to identify the heavy-metal-resistant bacterium Cupriavidus metallidurans. J. Ind. Microbiol. Biotechnol. 2011, 38, 1613–1615. [Google Scholar] [CrossRef]
- Rodríguez-Lázaro, D.; Lewis, D.A.; Ocampo-Sosa, A.A.; Fogarty, U.; Makrai, L.; Navas, J.; Scortti, M.; Hernández, M.; Vázquez-Boland, J.A. Internally controlled real-time PCR method for quantitative species-specific detection and vapA genotyping of Rhodococcus equi. Appl. Environ. Microbiol. 2006, 72, 4256–4263. [Google Scholar] [CrossRef]
- ISO 11348-3:2007; Water Quality—Determination of the Inhibitory Effect of Water Samples on the Light Emission of Vibrio fischeri (Luminescent bacteria Test)—Part 3: Method Using Freeze-Dried Bacteria. ISO: Geneva, Switzerland, 2007.
- Ames, B.N.; McCann, J.; Yamasaki, E. Methods for detecting carcinogens and mutagens with the Salmonella/mammalian-microsome mutagenicity test. Mut. Res. Environ. Mutagenesis Relat. Subj. 1975, 31, 347–363. [Google Scholar] [CrossRef]
- Ceccanti, B.; Masciandaro, G.; Garcia, C.; Macci, C.; Doni, S. Soil bioremediation: Combination of earthworms and compost for the ecological remediation of a hydrocarbon polluted soil. Water Air Soil Pollut. 2006, 177, 383–397. [Google Scholar] [CrossRef]
- Kao, C.M.; Wang, C.C. Control of BTEX migration by intrinsic bioremediation at a gasoline spill site. Water Res. 2000, 34, 3413–3423. [Google Scholar] [CrossRef]
- Lin, T.C.; Pan, P.T.; Cheng, S.S. Ex situ bioremediation of oil-contaminated soil. J. Hazard. Mater. 2010, 176, 27–34. [Google Scholar] [CrossRef]
- Vinas, M.; Sabaté, J.; Espuny, M.J.; Solanas, A.M. Bacterial community dynamics and polycyclic aromatic hydrocarbon degradation during bioremediation of heavily creosote-contaminated soil. Appl. Environ. Microbiol. 2005, 71, 7008–7018. [Google Scholar] [CrossRef] [PubMed]
- Nocentini, M.; Pinelli, D.; Fava, F. Bioremediation of a soil contaminated by hydrocarbon mixtures: The residual concentration problem. Chemosphere 2000, 41, 1115–1123. [Google Scholar] [CrossRef]
- Betancur-Corredor, B.; Pino, N.J.; Cardona, S.; Peñuela, G.A. Evaluation of biostimulation and Tween 80 addition for the bioremediation of long-term DDT-contaminated soil. J. Environ. Sci. 2015, 28, 101–109. [Google Scholar] [CrossRef] [PubMed]
- Brohon, B.; Gourdon, R. Influence of soil microbial activity level on the determination of contaminated soil toxicity using Lumistox and MetPlate bioassays. Soil Biol. Biochem. 2000, 32, 853–857. [Google Scholar] [CrossRef]
- Pagnout, C.; Rast, C.; Veber, A.M.; Poupin, P.; Férard, J.F. Ecotoxicological assessment of PAHs and their dead-end metabolites after degradation by Mycobacterium sp. strain SNP11. Ecotoxicol. Environ. Saf. 2006, 65, 151–158. [Google Scholar] [CrossRef]



| Strains | Target Gene | Primers | Tm (°C) | References |
|---|---|---|---|---|
| Pseudomonas stutzeri | Fps158 | 5′-GTGGGGGACAACGTTTC-3′ 5′-CGATTATGACTGACTCCAC-3′ | 48 | [57] |
| Cupriavidus metallidurans | Cm | 5′-AGTTTCCTGGCCATGATGAG-3′ 5′-TCCGTTTCCTGTACCACCTC-3′ | 59 | [58] |
| Rhodococcus equi | ChoE | 5′-CGACAAGCGCTCGATGTG-3′ 3′-TGCCGAAGCCCATGAAGT-5′ | 56 | [59] |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mansouri, A.; Cregut, M.; Jouanneau, S.; Abbes, C.; Landoulsi, A.; Thouand, G.; Durand, M.-J. Evaluation of Biomonitoring Strategies to Assess Performance of a Bioremediation Bioprocess. Sustainability 2022, 14, 10932. https://doi.org/10.3390/su141710932
Mansouri A, Cregut M, Jouanneau S, Abbes C, Landoulsi A, Thouand G, Durand M-J. Evaluation of Biomonitoring Strategies to Assess Performance of a Bioremediation Bioprocess. Sustainability. 2022; 14(17):10932. https://doi.org/10.3390/su141710932
Chicago/Turabian StyleMansouri, Ahlem, Mickael Cregut, Sulivan Jouanneau, Chiraz Abbes, Ahmed Landoulsi, Gerald Thouand, and Marie-Jose Durand. 2022. "Evaluation of Biomonitoring Strategies to Assess Performance of a Bioremediation Bioprocess" Sustainability 14, no. 17: 10932. https://doi.org/10.3390/su141710932









