The Role of Cellulose in Microbial Diversity Changes in the Soil Contaminated with Cadmium
Abstract
:1. Introduction
2. Materials and Methods
2.1. Soil
2.2. Experimental Scheme
2.3. Soil and Plant Analysis
2.4. Indicators Valorizing the Usability of Plants for the Remediation of Cadmium-Contaminated Soil
2.5. Microbiological Analyses
2.6. Statistical Analyses
3. Results
3.1. Effect of Cadmium and Cellulose on Plants
3.2. Effect of Cadmium and Cellulose on Soil Microorganisms
4. Discussion
4.1. Plant Response to Soil Contamination with Cadmium and Fertilization with Cellulose
4.2. Response of Microorganisms to Soil Contamination with Cadmium and Fertilization with Cellulose
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Rajaganpathy, V.; Xavier, F.; Sreekumar, D.; Mandal, P.K. Heavy metal contamination in soil, water and fodder and their presence in livestock and products: A review. J. Environ. Sci. Technol. 2011, 4, 234–249. [Google Scholar] [CrossRef] [Green Version]
- Dhaliwal, S.S.; Taneja, P.K.; Singh, J.; Bhatti, S.S.; Singh, R. Cadmium accumulation potential of brassica species grown in metal spiked loamy sand soil. Soil Sediment. Contam. Int. J. 2020, 29, 638–649. [Google Scholar] [CrossRef]
- Setia, R.; Dhaliwal, S.S.; Kumar, V.; Singh, R.; Kukal, S.S.; Pateriya, B. Impact assessment of metal contamination in surface water of Sutlej River (India) on human health risks. Environ. Pollut. 2020, 265, 114907. [Google Scholar] [CrossRef]
- Setia, R.; Dhaliwal, S.S.; Singh, R.; Kumar, V.; Taneja, S.; Kukal, S.S.; Pateriya, B. Phytoavailability and human risk assessment of heavy metals in soils and food crops around Sutlej River, India. Chemosphere 2020, 263, 128321. [Google Scholar] [CrossRef]
- McLaughlin, M.J.; Smolders, E.; Zhao, F.J.; Grant, C.; Montalvo, D. Managing cadmium in agricultural systems. Adv. Agron. 2021, 166, 1–129. [Google Scholar] [CrossRef]
- Raza, A.; Habib, M.; Kakavand, S.N.; Zahid, Z.; Zahra, N.; Sharif, R.; Hasanuzzaman, M. Phytoremediation of cadmium: Physiological, biochemical, and molecular mechanisms. Biology 2020, 9, 177. [Google Scholar] [CrossRef]
- Bhatti, S.S.V.; Kumar, V.; Sambyal, J.; Singh, N.A.K. Comparative analysis of tissue compartmentalized heavy metal uptake by common forage crop: A field experiment. Catena 2018, 160, 185–193. [Google Scholar] [CrossRef]
- Kubier, A.; Wilkin, R.T.; Pichler, T. Cadmium in soils and groundwater: A review. Appl. Geochem. 2019, 108, 104388. [Google Scholar] [CrossRef]
- Moreno-Caselles, J.; Moral, R.; Perez-Espinosa, A.; Perez-Murcia, M.D. Cadmium accumulation and distribution in cucumber plant. J. Plant Nutr. 2000, 23, 243–250. [Google Scholar] [CrossRef]
- Watanabe, T.; Murata, Y.; Osaki, M. Amaranthus tricolor has the potential for phytoremediation of cadmium contaminated soils. Commum. Soil Sci. Plant Anal. 2009, 40, 3158–3169. [Google Scholar] [CrossRef]
- Ejnik, J.; Robinson, J.; Zhu, J.; Försterling, H.; FrankShaw, C.; Petering, D.H. Folding pathway of apo-metallothionein induced by Zn2+, Cd2+ and Co2+. J. Inorg. Biochem. 2002, 88, 144–152. [Google Scholar] [CrossRef]
- Pedro, F.; Feria-Cáceres, P.F.; Penagos-Velez, L.; Claudia, X.; Moreno-Herrera, C.X. Tolerance and cadmium (Cd) immobiliza-tion by native bacteria isolated in cocoa soils with increased metal content. Microbiol. Res. 2022, 13, 556–573. [Google Scholar] [CrossRef]
- Azizollahi, Z.; Ghaderian, S.M.; Ghotbi-Ravandi, A.A. Cadmium accumulation and its effects on physiological and biochemical characters of summer savory (Satureja hortensis L.). Int. J. Phytoremediat. 2019, 21, 1241–1253. [Google Scholar] [CrossRef]
- DalCorso, G.; Farinati, S.; Furini, A. Regulatory networks of cadmium stress in plants. Plant Signal. Behav. 2010, 5, 663–667. [Google Scholar] [CrossRef]
- Mendoza-Cozatl, D.G.; Butko, E.; Springer, F.; Torpey, J.W.; Komives, E.A.; Kehr, J.; Schroeder, J.I. Identification of high levels of phytochelatins, glutathione and cadmium in the phloem sap of Brassica napus. A role for thiol-peptides in the long-distance transport of cadmium and the effect of cadmium on iron translocation. Plant J. 2008, 54, 249–259. [Google Scholar] [CrossRef] [Green Version]
- Lux, A.; Martinka, M.; Vaculík, M.; White, P.J. Root responses to cadmium in the rhizosphere: A review. J. Exp. Bot. 2011, 62, 21–37. [Google Scholar] [CrossRef] [Green Version]
- Agents Classified by the IARC Monographs, Volumes 1–132. Available online: https://monographs.iarc.who.int/agents-classified-by-the-iarc/ (accessed on 12 August 2022).
- Conesa, H.M.; Wieser, M.; Gasser, M.; Hockmann, K.; Evangelou, M.W.H.; Studer, B. Effects of three amendments on extractability and fractionation of Pb, Cu, Ni and Sb in two shooting range soils. J. Hazard. Mater. 2010, 181, 845–850. [Google Scholar] [CrossRef]
- Bolan, N.B.; Kunhikrishnanc, A.; Thangarajana, R.; Kumpiened, J.; Parke, J.; Makinof, T.; Kirkhamg, M.B.; Scheckelh, K. Remediation of heavy metal(loid)s contaminated soils—To mobilize or to immobilize? J. Hazard. Mater. 2014, 266, 141–166. [Google Scholar] [CrossRef]
- Stritsis, B.; Steingrobe, B.; Claassen, N. Cadmium fractions in an acid sandy soil and Cd in soil solution as affected by plant growth. J. Plant Nutr. Soil Sci. 2014, 177, 431–437. [Google Scholar] [CrossRef]
- Oves, M.; Saghir Khan, M.; Zaidi, A.; Ahmad, E. Soil contamination, nutritive value, and human health risk assessment of heavy metals: An overview. In Toxicity of Heavy Metals to Legumes and Bioremediation; Zaidi, A., Wani, P.A., Khan, M.S., Eds.; Springer: Vienna, Austria, 2012; pp. 1–27. [Google Scholar] [CrossRef]
- Kabata-Pendias, A.; Pendias, H. Trace Elements in Soils and Plants; CRC Press: Boca Raton, FL, USA, 2010. [Google Scholar] [CrossRef]
- Li, M.; Mohamed, I.; Raleve, D.; Chen, W.; Huang, Q. Field evaluation of intensive compost application on Cd fractionation and phytoavailability in a mining-contaminated soil. Environ. Geochem. Health 2016, 38, 1193–1201. [Google Scholar] [CrossRef]
- Massaccesi, L.; Benucci, G.M.N.; Gigliotti, G.; Cocco, S.; Corti, G.; Agnelli, A. Rhizosphere effect of three plant species of environment under periglacial conditions (Majella Massif, central Italy). Soil Biol. Biochem. 2015, 89, 184–195. [Google Scholar] [CrossRef]
- Hütsch, B.W.; Augustin, J.; Merbach, W. Plant rhizodeposition—An important source for carbon turnover in soils. J. Plant. Nutr. Soil Sci. 2002, 165, 397–407. [Google Scholar] [CrossRef]
- Tahervand, S.; Jalali, M. Sorption, desorption, and speciation of Cd, Ni, and Fe by four calcareous soils as affected by pH. Environ. Monit. Assess. 2016, 188, 322. [Google Scholar] [CrossRef] [PubMed]
- Karimzadeh, L.; Nair, S.; Merkel, B.J. Effect of microbial siderophore DFOB on Pb, Zn, and Cd sorption onto zeolite. Aquat. Geochem. 2013, 19, 25–37. [Google Scholar] [CrossRef]
- Amabilis-Sosa, L.E.; Siebe, C.; Moeller-Chávez, G.; del Carmen Durán-Domínguez-de-Bazúa, M. Accumulation and distribution of lead and chromium in laboratory-scale constructed wetlands inoculated with metal-tolerant bacteria. Int. J. Phytoremed. 2015, 17, 1090–1096. [Google Scholar] [CrossRef]
- Lebeau, T.; Jesequel, K.; Braud, A. Bioaugmentation-assisted phytoextraction applied to metal-contaminated soils: State of the art and future prospects. In Microbes and Microbial Technology: Agricultural and Environmental Applications; Ahmad, I., Ahmad, F., Pichtel, J., Eds.; Microbes and Microbial Technology; Springer: New York, NY, USA, 2011. [Google Scholar] [CrossRef]
- Lange, B.; van der Ent, A.; Baker, A.J.M.; Echevarria, G.; Mahy, G.; Malaisse, F.; Meets, P.; Pourret, O.; Verbruggen, N.; Faucon, M.-P. Copper and cobalt accumulation in plants: A critical assessment of the current state of knowledge. New Phytol. 2017, 213, 537–551. [Google Scholar] [CrossRef]
- Sarwar, N.; Imran, M.; Shaheen, M.R.; Ishaque, W.; Kamran, M.A.; Matloob, A.; Rehim, A.; Hussain, S. Phytoremediation strategies for soils contaminated with heavy metals: Modifications and future perspectives. Chemosphere 2017, 171, 710–721. [Google Scholar] [CrossRef]
- Boros-Lajszner, E.; Wyszkowska, J.; Kucharski, J. Use of zeolite to neutralise nickel in a soil environment. Environ. Monit. Assess. 2018, 190, 54. [Google Scholar] [CrossRef] [Green Version]
- Burns, R.G.; DeForest, J.L.; Marxsen, J.; Sinsabaugh, R.L.; Stromberger, M.E.; Wallenstein, M.D.; Weintraub, M.N.; Zoppini, A. Soil enzymes in a changing environment: Current knowledge and future directions. Soil Biol. Biochem. 2013, 58, 216–234. [Google Scholar] [CrossRef]
- Gómez-Sagasti, M.T.; Alkorta, I.; Becerril, J.M.; Epelde, L.; Anza, M.; Garbisu, C. Microbial monitoring of the recovery of soil quality during heavy metal phytoremediation. Water Air Soil Pollut. 2012, 223, 3249–3262. [Google Scholar] [CrossRef]
- Haddad, S.A.; Lemanowicz, J.; Abd El-Azeim, M.M. Cellulose decomposition in clay and sandy soils contaminated with heavy metals. Int. J. Environ. Sci. Technol. 2019, 16, 3275–3290. [Google Scholar] [CrossRef]
- Chmolowska, D.; Hamda, N.; Laskowski, R. Cellulose decomposed faster in fallow soil than in meadow soil due to a shorter lag time. J. Soils Sediments 2017, 17, 299–305. [Google Scholar] [CrossRef] [Green Version]
- Xie, Y.; Fan, J.; Zhu, W.; Amombo, E.; Lou, Y.; Chen, L.; Fu, J. Effect of heavy metals pollution on soil microbial diversity and bermudagrass genetic variation. Front Plant Sci. 2016, 7, 755. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tica, D.; Udovic, M.; Lestan, D. Immobilization of potentially toxic metals using different soil amendments. Chemosphere 2011, 85, 577–583. [Google Scholar] [CrossRef] [PubMed]
- Chen, X.; Liu, L.; Luo, Z.; Shen, J.; Ni, Q.; Yao, J. Facile preparation of a cellulose-based bioadsorbent modified by hPEI in heterogeneous system for high-efficiency removal of multiple types of dyes. React. Funct. Polym. 2018, 125, 77–83. [Google Scholar] [CrossRef]
- Vasile, G.-G.; Tenea, A.-G.; Dinu, C.; Iordache, A.M.M.; Gheorghe, S.; Mureseanu, M.; Pascu, L.F. Bi-oavailability, accumulation and distribution of toxic metals (As, Cd, Ni and Pb) and their impact on Sinapis alba plant nutrient metabolism. Int. J. Environ. Res. Public Health 2021, 18, 12947. [Google Scholar] [CrossRef]
- Jan, R.; Asaf, S.; Numan, M.; Lubna; Kim, K.-M. Plant Secondary metabolite biosynthesis and transcriptional regulation in response to biotic and abiotic stress conditions. Agronomy 2021, 11, 968. [Google Scholar] [CrossRef]
- De Leij, F.A.A.M.; Whips, J.; Lynch, J.M. The use of colony development for the characterization of bacterial communities in soil and on roots. Microb. Ecol. 1993, 7, 81–97. [Google Scholar] [CrossRef]
- Sarathchandra, S.U.; Burch, G.; Cox, N.R. Growth patterns of bacterial communities in the rhizoplane and rhizosphere of white clover (Trifolium repens L.) and perennial ryegrass (Lolium perenne L.) in long-term pasture. Appl. Soil Ecol. 1997, 6, 293–299. [Google Scholar] [CrossRef]
- PN-R-04032; In Soil and Mineral materials—Sampling and Determination of Particle Size Distribution. Polish Committee for Standardization: Warsaw, Poland, 1998.
- ISO 11464; Soil Quality—Pre-Treatment of Samples for Physico-Chemical Analysis. International Organization for Standardization: Geneva, Switzerland, 2006.
- ISO 10390; Soil Quality—Determination of PH. International Organization for Standardization: Geneva, Switzerland, 2005.
- PN-EN 14084: 2004 (N); Nickel, Cadmium and Cobalt Content In Above-Ground Parts and in Roots Determined by Flame Atomic Absorption Spectrometry and by Graphite Furnace Atomic Absorption Spectroscopy (FAAS and GFAAS) following Microwave Mineralization. International Organization for Standardization: Geneva, Switzerland, 2016. Available online: https://pzn.pkn.pl/kt/info/published/9000128800 (accessed on 20 April 2021).
- Orwin, K.H.; Wardle, D.A. New indices for quantifying the resistance and resilience of soil biota to exogenous disturbance. Soil Biol. Biochem. 2004, 36, 1907–1912. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Boros-Lajszner, E.; Kucharski, J. Calorific value of Festuca rubra biomass in the phytostabilization of soil contaminated with nickel, cobalt and cadmium which disrupt the microbiological and biochemical properties of soil. Energies 2022, 15, 3445. [Google Scholar] [CrossRef]
- Boros-Lajszner, E.; Wyszkowska, J.; Borowik, A.; Kucharski, J. The response of the soil microbiome to contamination with cadmium, cobalt an nickel in soil sown with Brassica napus. Minerals 2021, 11, 498. [Google Scholar] [CrossRef]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Bacteria and soil enzymes supporting the valorization of forested soils. Materials 2022, 15, 3287. [Google Scholar] [CrossRef] [PubMed]
- Dell Inc. Dell Statistica (Data Analysis Software System), version 13.1; Dell Inc.: Tulsa, OK, USA, 2016.
- Parks, D.H.; Tyson, G.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef] [Green Version]
- Krzywiński, M.; Schein, J.; Birol, I.; Connors, J.; Gascoyne, R.; Horsman, D.; Jones, S.J.; Marra, M.A. Circos: An information aesthetic for comparative genomics. Genome Res. 2009, 19, 1639–1645. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- R Studio Team. R Studio: Integrated Development for R; R Studio Inc.: Boston, MA, USA, 2019; Available online: http://www.rstudio.com/ (accessed on 10 May 2022).
- R Core Team. A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019; Available online: https://www.R-project.org/ (accessed on 10 May 2022).
- Warnes, G.R.; Bolker, B.; Bonebakker, L.; Gentleman, R.; Huber, W.; Liaw, A.; Lumley, T.; Maechler, M.; Magnusson, M.; Moeller, S.; et al. Various R Programming Tools for Plotting Data. R Package Version 3.1.3. 2022. Available online: https://CRAN.R-project.org/package=gplots (accessed on 14 July 2022).
- Heberle, H.; Meirelles, G.V.; da Silva, F.R.; Telles, G.P.; Minghim, R. InteractiVenn: A web-based tool for the analysis of sets through Venn diagrams. BMC Bioinform. 2015, 16, 169. [Google Scholar] [CrossRef] [Green Version]
- Shannon, V.L.; Vanguelova, E.I.; Morison, J.I.L.; Shaw, L.J.; Clark, J.M. The contribution of deadwood to soil carbon dynamics in contrasting temperate forest ecosystems. Eur. J. For. Res. 2021, 141, 241–252. [Google Scholar] [CrossRef]
- Bibi, F.; Ali, Z. Measurement of diversity indices of avian communities at Taunsa Barrage Wildlife Sanctuary, Pakistan. J. Anim. Plant Sci. 2013, 23, 469–474. [Google Scholar]
- Nazar, R.; Iqbal, N.; Masood, A.; Khan, M.I.R.; Syeed, S.; Khan, N.A. Cadmium toxicity in plants and role of mineral nutrients in its alleviation. Am. J. Plant Sci. 2012, 3, 1476–1489. [Google Scholar] [CrossRef] [Green Version]
- Sobariu, D.L.; Fertu, D.I.T.; Diaconu, M.; Pavel, I.V.; Hlihor, R.-M.; Dragoi, E.N.; Curteanu, S.; Lenz, M.; Corvini, P.F.-X.; Gavrilescu, M. Rhizobacteria and plant symbiosis in heavy metals uptake and its implications for soil bioremediation. New Biotechnol. 2017, 39, 125–134. [Google Scholar] [CrossRef]
- Xu, P.; Wang, Z. Physiological mechanism of hypertolerance of cadmium in Kentucky bluegrass and tall fescue: Chemical forms and tissue distribution. Environ. Exp. Bot. 2013, 96, 35–42. [Google Scholar] [CrossRef]
- Zhang, X.; Xia, H.; Li, Z.; Zhung, P.; Gao, B. Potential of four grasses in remediation of Cd and Zn contaminated soils. Bioresour. Technol. 2010, 10, 2063–2066. [Google Scholar] [CrossRef] [PubMed]
- Rascio, N.; Navari-Izzo, F. Heavy metal hyperaccumulating plants: How and why do they do it? And what makes them so interesting? Plant Sci. 2011, 180, 169–181. [Google Scholar] [CrossRef] [PubMed]
- Zheng, R.; Li, H.; Jiang, R.; Römheld, V.; Zhang, F.; Zhao, F.-J. The role of root hairs in cadmium acquisition by barley. Environ. Pollut. 2011, 159, 408–415. [Google Scholar] [CrossRef]
- Deram, A.; Denayer, F.O.; Petit, D.; Van Haluvyn, C. Seasonal variation of cadmium and zinc in Arrhenatherum elatius, a perennial grass species from highly contaminated soils. Environ. Pollut. 2006, 140, 62–70. [Google Scholar] [CrossRef]
- Aibibu, N.; Liu, Y.; Zeng, G.; Wang, X.; Chen, B.; Song, H.; Xu, L. Cadmium accumulation in Vetiveria zizanioides and its effects on growth, physiological and biochemical characters. Bioresour. Technol. 2010, 101, 6297–6303. [Google Scholar] [CrossRef]
- Zhang, S.; Lin, H.; Deng, L.; Gong, G.; Jia, Y.; Xu, X.; Li, T.; Li, Y.; Chen, H. Cadmium tolerance and accumulation characteristic Siegesbeckia orientalis L. Ecol. Eng. 2013, 5, 133–139. [Google Scholar] [CrossRef]
- Guo, Q.; Meng, L.; Mao, P.C.; Tian, X.X. An assessment of Agropyron cristatum tolerance to cadmium contaminated soil. Biol. Plant. 2014, 58, 174–178. [Google Scholar] [CrossRef]
- Zhang, X.; Gao, B.; Xia, H. Effect of cadmium on growth, photosynthesis, mineral nutrition and metal accumulation of bana grass and vetiver grass. Ecotoxicol. Environ. Saf. 2014, 106, 102–108. [Google Scholar] [CrossRef]
- Quezada-Hinojosa, R.; Föllmi, K.B.; Gillet, F.; Matera, V. Cadmium accumulation in six common plant species associated with soils containing high geogenic cadmium concentrations at Le Gurnigel, Swiss Jura Mountains. Catena 2015, 124, 85–96. [Google Scholar] [CrossRef]
- Stanislawska-Glubiak, E.; Korzeniowska, J.; Kocon, A. Effect of the reclamation of heavy metal-contaminated soil on growth of energy willow. Pol. J. Environ. Stud. 2012, 21, 187–192. [Google Scholar]
- Masarovicova, E.; Kralova, K.; Kummerova, M. Principles of classification of medical plants as hyperaccumulators or excluders. Acta Physiol. Plant. 2010, 32, 823–829. [Google Scholar] [CrossRef]
- O’Connell, D.W.; Birkinshaw, C.; O’Dwyer, T.F. Heavy metal adsorbents prepared from the modification of cellulose: A review. Bioresour. Technol. 2008, 99, 6709–6724. [Google Scholar] [CrossRef] [PubMed]
- Sud, D.; Mahajan, G.; Kaur, M.P. Agricultural waste material as potential adsorbent for sequestering heavy metal ions from aqueous solutions—A review. Bioresour. Technol. 2008, 99, 6017–6027. [Google Scholar] [CrossRef]
- Hashim, M.A.; Mukhopadhyay, S.; Sahu, J.N.; Sengupta, B. Remediation technologies for heavy metal contaminated groundwater. J. Environ. Manag. 2011, 92, 2355–2388. [Google Scholar] [CrossRef]
- Zaborowska, M.; Kucharski, J.; Wyszkowska, J. Using basalt flour and brown algae to improve biological properties of soil contaminated with cadmium. Soil Water Res. 2015, 10, 181–188. [Google Scholar] [CrossRef] [Green Version]
- Wei, Y.; Zhao, Y.; Zhao, X.; Gao, X.; Zheng, Y.; Zuo, H.; Wei, Z. Roles of different humin and heavy-metal resistant bacteria from composting on heavy metal removal Yuquan. Bioresour. Technol. 2020, 296, 122375. [Google Scholar] [CrossRef]
- Boros-Lajszner, E.; Wyszkowska, J.; Borowik, A.; Kucharski, J. Energetic value of Elymus elongatus L. and Zea mays L. grown on soil polluted with Ni2+, Co2+, Cd2+, and Sensitivity of rhizospheric bacteria to heavy metals. Energies 2021, 14, 4903. [Google Scholar] [CrossRef]
- Abd El-Azeim, M.M.; Mohamed, W.S.; Hammam, A.A. Soil physiochemical properties in relation to heavy metals status of agricultural soils in El-Minia Governorate, Egypt. J. Soil Sci. Agric. Eng. Mansoura Univ. 2016, 7, 423–431. [Google Scholar] [CrossRef]
- Salman, S.A.; Elnazer, A.A.; El Nazer, H.A. Integrated mass balance of some heavy metals fuxes in Yaakob village, south Sohag, Egypt. Int. J. Environ. Sci. Technol. 2017, 14, 1011–1018. [Google Scholar] [CrossRef]
- Zeng, P.; Guo, Z.; Cao, X.; Xiao, X.; Liu, Y.; Shi, L. Phytostabilization potential of ornamental plants grown in soil contaminated with cadmium. Int. J. Phytoremed. 2018, 20, 311–320. [Google Scholar] [CrossRef] [PubMed]
- Hou, D.; Wang, K.; Liu, T.; Wang, H.; Lin, Z.; Qian, J.; Lu, L.; Tian, S. Unique rhizosphere micro-characteristics facilitate phytoextraction of multiple metals in soil by the hyperaccumulating plant Sedum alfredii. Environ. Sci. Technol. 2017, 51, 5675–5684. [Google Scholar] [CrossRef] [PubMed]
- Fu, Z.D.; Zhou, L.; Chen, P.; Du, Q.; Pang, T.; Song, C.; Wang, X.C.; Liu, W.G.; Yang, W.Y.; Yong, T.W. Effects of maize-soybean relay intercropping on crop nutrient uptake and soil bacterial community. J. Integr. Agric. 2019, 18, 2006–2018. [Google Scholar] [CrossRef]
- Wang, M.; Li, S.; Chen, S.; Meng, N.; Li, X.; Zheng, H.; Zhao, C.; Wang, D. Manipulation of the rhizosphere bacterial community by biofertilizers is associated with mitigation of cadmium phytotoxicity. Sci. Total Environ. 2019, 649, 413–421. [Google Scholar] [CrossRef]
- Min, X.; Wang, Y.; Chai, L.; Yang, Z.; Liao, Q. High-resolution analyses reveal structural diversity patterns of microbial communities in Chromite Ore Processing Residue (COPR) contaminated soils. Chemosphere 2017, 183, 266–276. [Google Scholar] [CrossRef] [PubMed]
- Li, X.; Sun, M.; Zhang, H.; Xu, N.; Sun, G. Use of mulberry–soybean intercropping in salt–alkali soil impacts the diversity of the soil bacterial community. Microb. Biotechnol. 2016, 9, 293–304. [Google Scholar] [CrossRef] [Green Version]
- Acosta-Martinez, V.; Bell, C.W.; Morris, B.E.L.; Zak, J.; Allen, V.G. Long-term soil microbial community and enzyme activity responses to an integrated cropping-livestock system in a semi-arid region. Agric. Ecosyst. Environ. 2010, 137, 231–240. [Google Scholar] [CrossRef]
- Wyszkowska, J.; Borowik, A.; Olszewski, J.; Kucharski, J. Soil Bacterial Community and soil enzyme activity depending on the cultivation of Triticum aestivum, Brassica napus, and Pisum sativum ssp. arvense. Diversity 2019, 11, 246. [Google Scholar] [CrossRef] [Green Version]
- Maron, P.A.; Sarr, A.; Kaisermann, A.; Lévêque, J.; Mathieu, O.; Guigue, J.; Karimi, B.; Bernard, L.; Dequiedt, S.; Terrat, S.; et al. High microbial diversity promotes soil ecosystem functioning. Appl. Environ. Microbiol. 2018, 84, 1–13. [Google Scholar] [CrossRef]
- Pascault, N.; Ranjard, L.; Kaisermann, A.; Bachar, D.; Christen, R.; Terrat, S.; Mathieu, O.; Lévêque, J.; Mougel, C.; Henault, C.; et al. Stimulation of different functional groups of bacteria by various plant residues as a driver of soil priming effect. Ecosystems 2013, 16, 810–822. [Google Scholar] [CrossRef]
- Liu, X.; Huang, Z.; Havrilla, C.A.; Liu, Y.; Wu, G. Plant litter crust role in nutrients cycling potentials by bacterial communities in a sandy land ecosystem. Land Degrad. Dev. 2021, 32, 3194–3203. [Google Scholar] [CrossRef]
- Eichorst, S.A.; Kuske, C.R.; Schmidt, T.M. Influence of plant polymers on the distribution and cultivation of bacteria in the phylum Acidobacteria. Appl. Environ. Microbiol. 2011, 77, 586–596. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Duan, C.; Liu, Y.; Zhang, H.; Chen, G.; Song, J. Cadmium pollution impact on the bacterial community of haplic cambisols in Northeast China and inference of resistant genera. J. Soil Sci. Plant Nutr. 2020, 20, 1156–1170. [Google Scholar] [CrossRef]
- Liang, J.; Tang, S.Q.; Gong, J.L.; Zeng, G.M.; Tang, W.W.; Song, B.; Zhang, P.; Yang, Z.X.; Luo, Y. Responses of enzymatic activity and microbial communities to biochar/compost amendment in sulfamethoxazole polluted wetland soil. J. Hazard. Mater. 2020, 385, 121533. [Google Scholar] [CrossRef] [PubMed]
- Borowik, A.; Wyszkowska, J.; Kucharski, M.; Kucharski, J. Implications of soil pollution with diesel oil and BP petroleum with active technology for soil health. Int. J. Environ. Res. Public Health 2019, 16, 2474. [Google Scholar] [CrossRef] [Green Version]
- Young, C.C.; Kämpfer, P.; Chen, W.M.; Yen, W.S.; Arun, A.B.; Lai, W.A.; Shen, F.T.; Rekha, P.D.; Lin, K.Y.; Chou, J.H. Luteimonas composti sp. nov., a moderately thermophilic bacterium isolated from food waste. Int. J. Syst. Evol. Microbiol. 2007, 57, 741–744. [Google Scholar] [CrossRef]
- Siddiqi, M.Z.; Yeon, J.M.; Choi, H.; Lee, J.H.; Kim, S.Y.; Wee, J.H.; Im, W.T. Luteimonas granuli sp. nov., isolated from granules of the wastewater treatment plant. Curr. Microbiol. 2020, 77, 2002–2007. [Google Scholar] [CrossRef]
- Zhou, J.; Chen, J.; Ma, J.; Xu, N.; Xin, F.; Zhang, W.; Zhang, H.; Dong, W.; Jiang, M. Luteimonas wenzhouensis sp. nov., a chitinolytic bacterium isolated from a landfill soil. Curr. Microbiol. 2021, 78, 383–388. [Google Scholar] [CrossRef]
- Wang, L.; Lilburn, M.; Yu, Z. Intestinal Microbiota of Broiler Chickens As Affected by Litter Management Regimens. Front. Microbiol. 2016, 7, 593. [Google Scholar] [CrossRef]
- Tayyab, M.; Islam, W.; Arafat, Y.; Pang, Z.; Zhang, C.; Lin, Y.; Waqas, M.; Lin, S.; Lin, W.; Zhang, H. Effect of sugarcane straw and goat manure on soil nutrient transformation and bacterial communities. Sustainability 2018, 10, 2361. [Google Scholar] [CrossRef] [Green Version]
- Galitskaya, P.; Biktasheva, L.; Blagodatsky, S.; Selivanovskaya, S. Response of bacterial and fungal communities to high petroleum pollution in different soils. Sci. Rep. 2021, 11, 164. [Google Scholar] [CrossRef] [PubMed]
- Novakovskiy, A.B.; Kanev, V.A.; Markarova, M.Y. Long-term dynamics of plant communities after biological remediation of oil-contaminated soils in Far North. Sci. Rep. 2021, 11, 4888. [Google Scholar] [CrossRef] [PubMed]
- Liu, N.; Lv, J.-L.; Cai, Y.-L.; Yao, Y.-Y.; Zhang, K.; Ma, C.; Li, J.-X.; Ren, X.-Y.; Hu, J.-J.; Zhao, J.-H. Study on gaseous chlorobenzene treatment by a bio-trickling filter: Degradation mechanism and microbial community. Processes 2022, 10, 1483. [Google Scholar] [CrossRef]
- Ruan, M.; Zhang, Y.; Chai, T. Rhizosphere soil microbial properties on tetraena mongolica in the arid and semi-arid regions, China. Int. J. Environ. Res. Public Health 2020, 17, 5142. [Google Scholar] [CrossRef]
- Wang, X.; Tian, L.; Li, Y.; Zhong, C.; Tian, C. Effects of exogenous cellulose-degrading bacteria on humus formation and bacterial community stability during composting. Bioresour. Technol. 2022, 359, 127458. [Google Scholar] [CrossRef] [PubMed]
- Liu, X.; Zhang, S.; Jiang, Q.; Bai, Y.; Shen, G.; Li, S.; Ding, W. Using community analysis to explore bacterial indicators for disease suppression of tobacco bacterial wilt. Sci. Rep. 2016, 6, 36773. [Google Scholar] [CrossRef] [Green Version]
- Borowik, A.; Wyszkowska, J.; Kucharski, J. Microbiological study in petrol-spiked soil. Molecules 2021, 26, 2664. [Google Scholar] [CrossRef] [PubMed]
- Gao, Y.; Lu, Y.; Lin, W.; Tian, J.; Cai, K. Biochar suppresses bacterial wilt of tomato by improving soil chemical properties and shifting soil microbial community. Microorganisms 2019, 7, 676. [Google Scholar] [CrossRef] [Green Version]
- Estendorfer, J.; Stempfhuber, B.; Vestergaard, G.; Schulz, S.; Rillig, M.C.; Joshi, J.; Schröder, P.; Schloter, M. Definition of core bacterial taxa in different root compartments of Dactylis glomerata, grown in soil under different levels of land use intensity. Diversity 2020, 12, 392. [Google Scholar] [CrossRef]
- Altimira, F.; Yáñez, C.; Bravo, G.; González, M.; Rojas, L.; Seeger, M. Characterization of copper-resistant bacteria and bacterial communities from copper-polluted agricultural soils of central Chile. BMC Microbiol. 2012, 12, 193. [Google Scholar] [CrossRef] [Green Version]
- Nilgiriwala, K.S.; Alahari, A.; Rao, A.S.; Apte, S.K. Cloning and overexpression of alkaline phosphatase PhoK from Sphingomonas sp. strain BSAR-1 for bioprecipitation of uranium from alkaline solutions. Appl. Environ. Microbiol. 2008, 74, 5516–5523. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Haas, D.; Défago, G. Biological control of soil-borne pathogens by fluorescent pseudomonads. Nat. Rev. Microbiol. 2005, 3, 307–319. [Google Scholar] [CrossRef] [PubMed]
- Lalucat, J.; Bennasar, A.; Bosch, R.; Garcia-Valdes, E.; Palleroni, N.J. Biology of Pseudomonas stutzeri. Microbiol. Mol. Biol. Rev. 2006, 70, 510–547. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gomez-Balderas, C.D.C.; Cochet, N.; Bert, V.; Tarnaud, E.; Sarde, C. 16S rDNA analysis of bacterial communities associated with the hyperaccumulator Arabidopsis halleri grown on a Zn and Cd polluted soil. Eur. J. Soil Biol. 2014, 60, 16–23. [Google Scholar] [CrossRef]
- Qiong, W.; Fengshan, P.; Xiaomeng, X.; Rafiq, M.T.; Xiao’e, Y.; Bao, C.; Ying, F. Cadmium level and soil type played a selective role in the endophytic bacterial community of hyperaccumulator Sedum alfredii Hance. Chemosphere 2021, 263, 127986. [Google Scholar] [CrossRef]

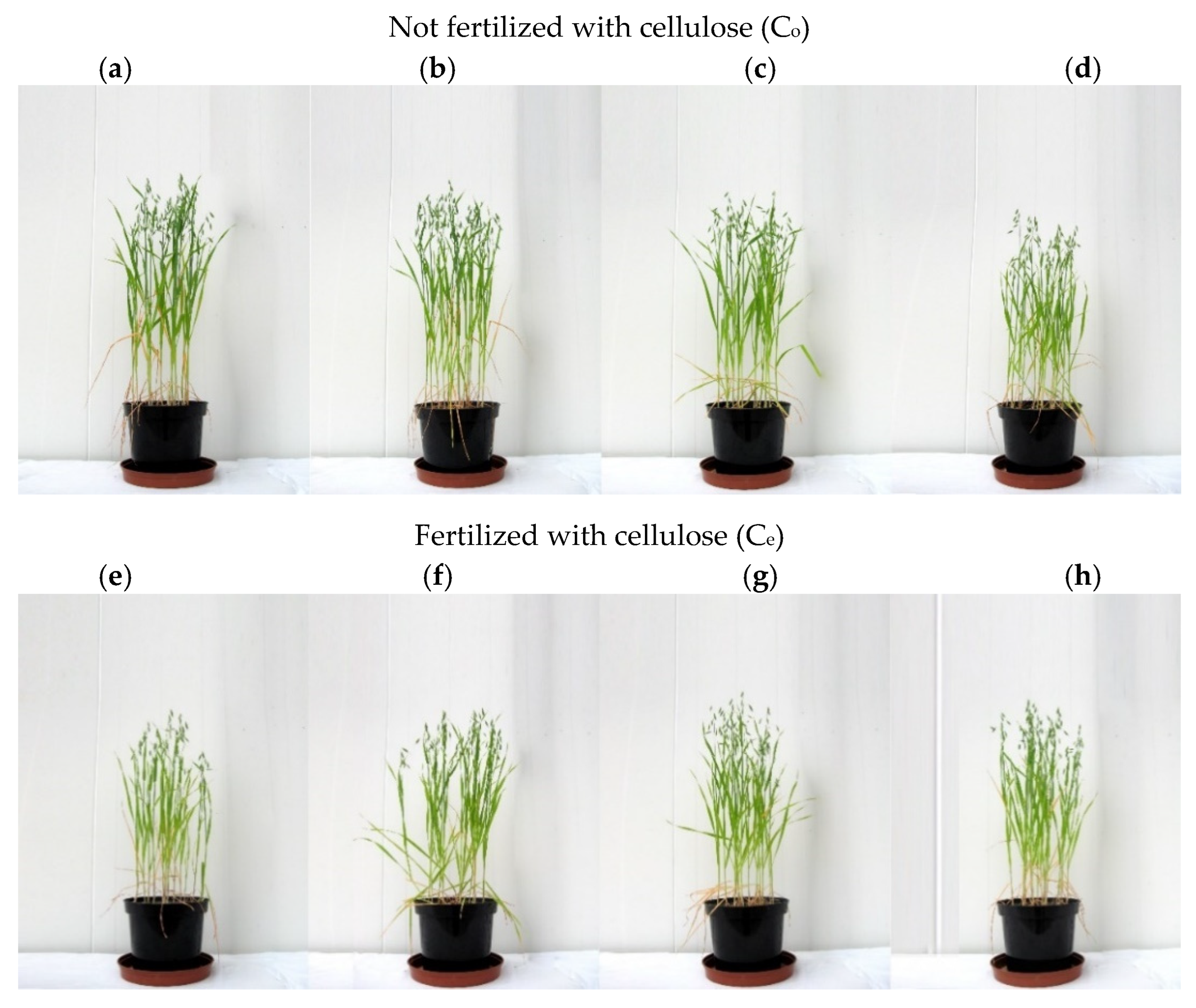
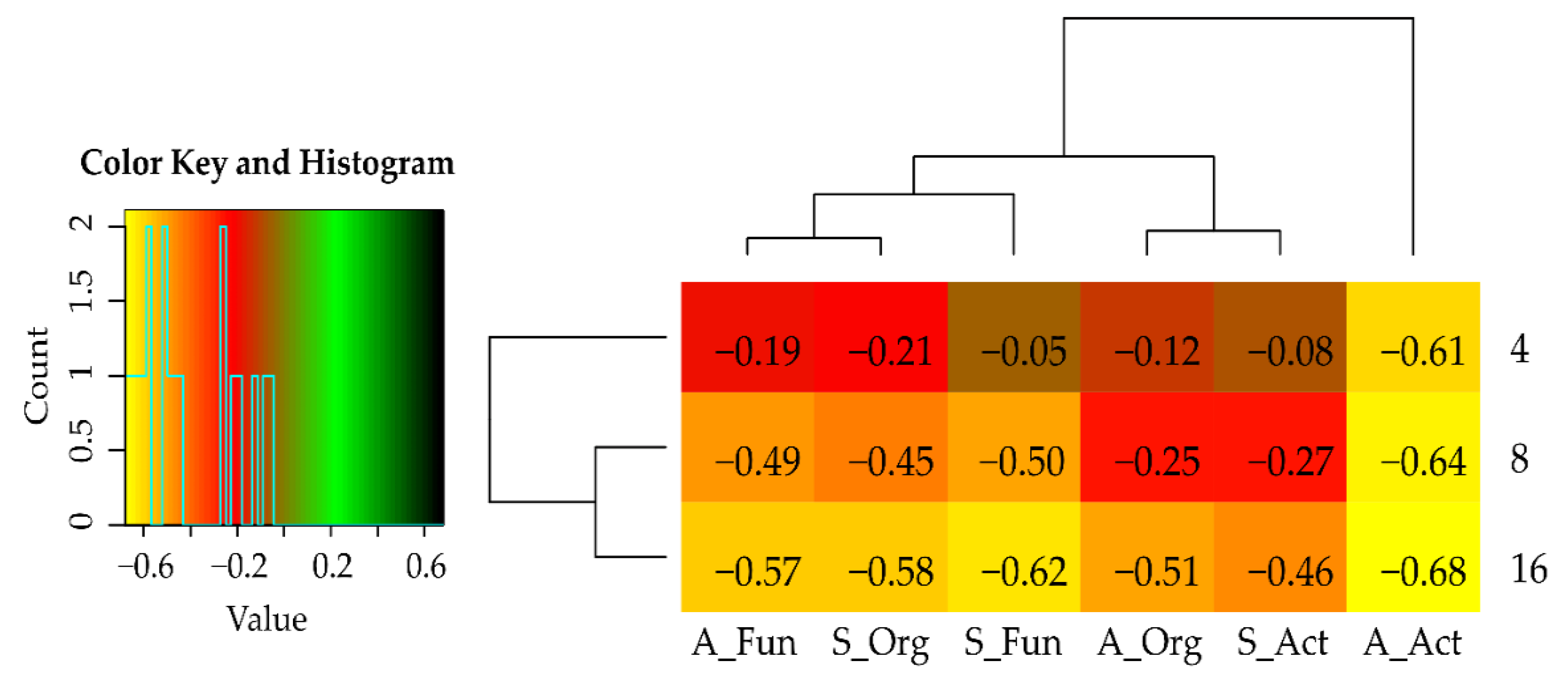
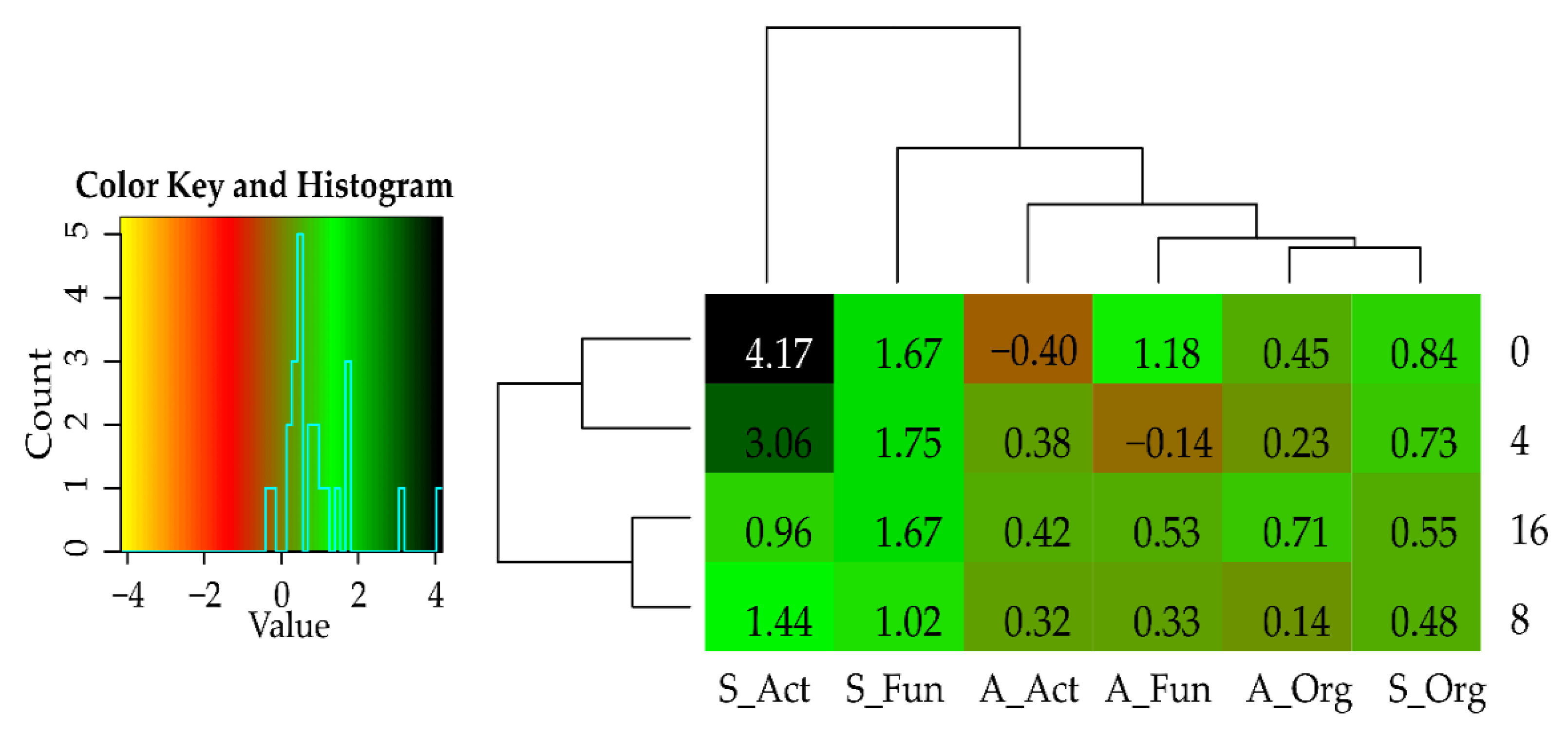
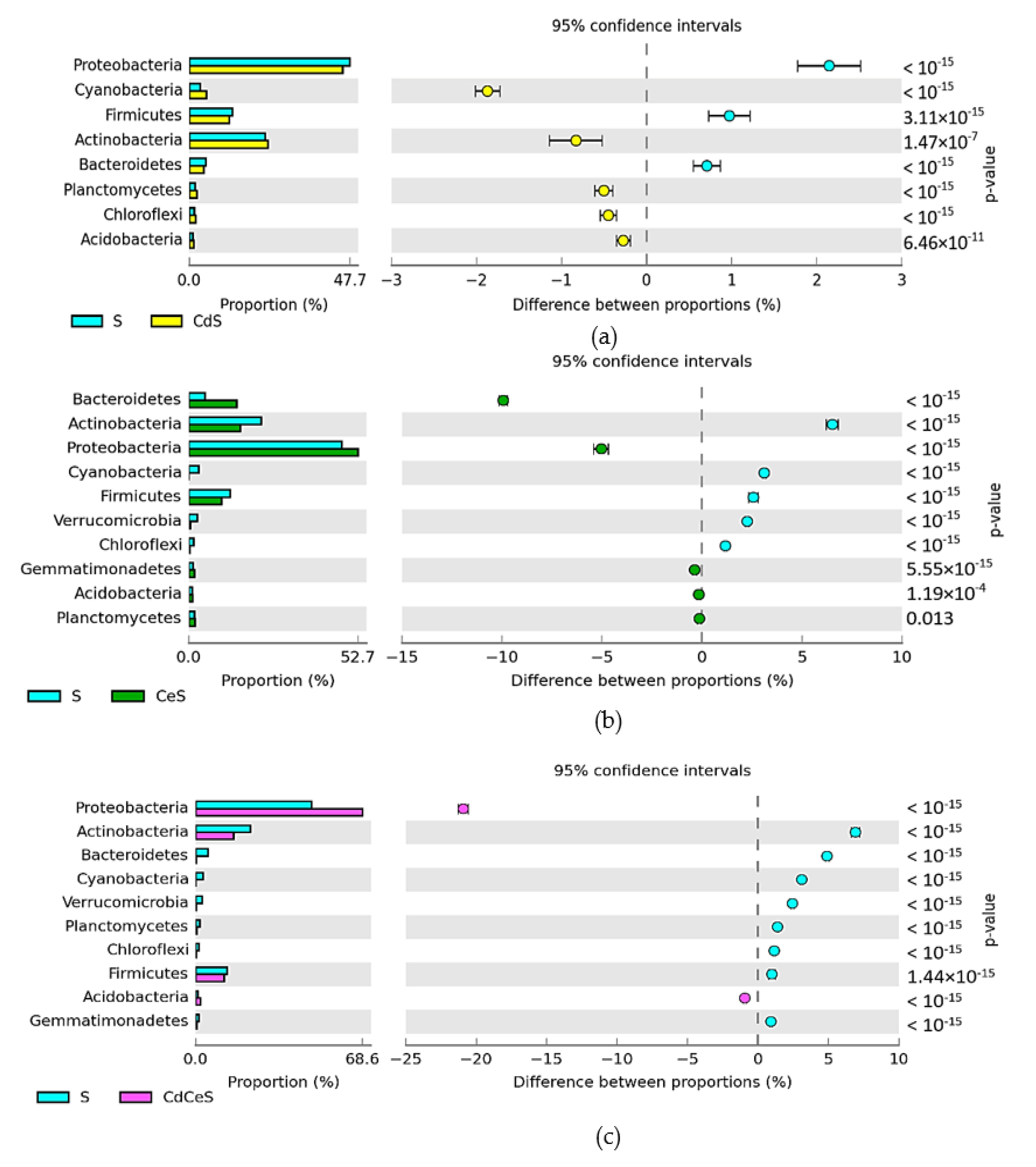
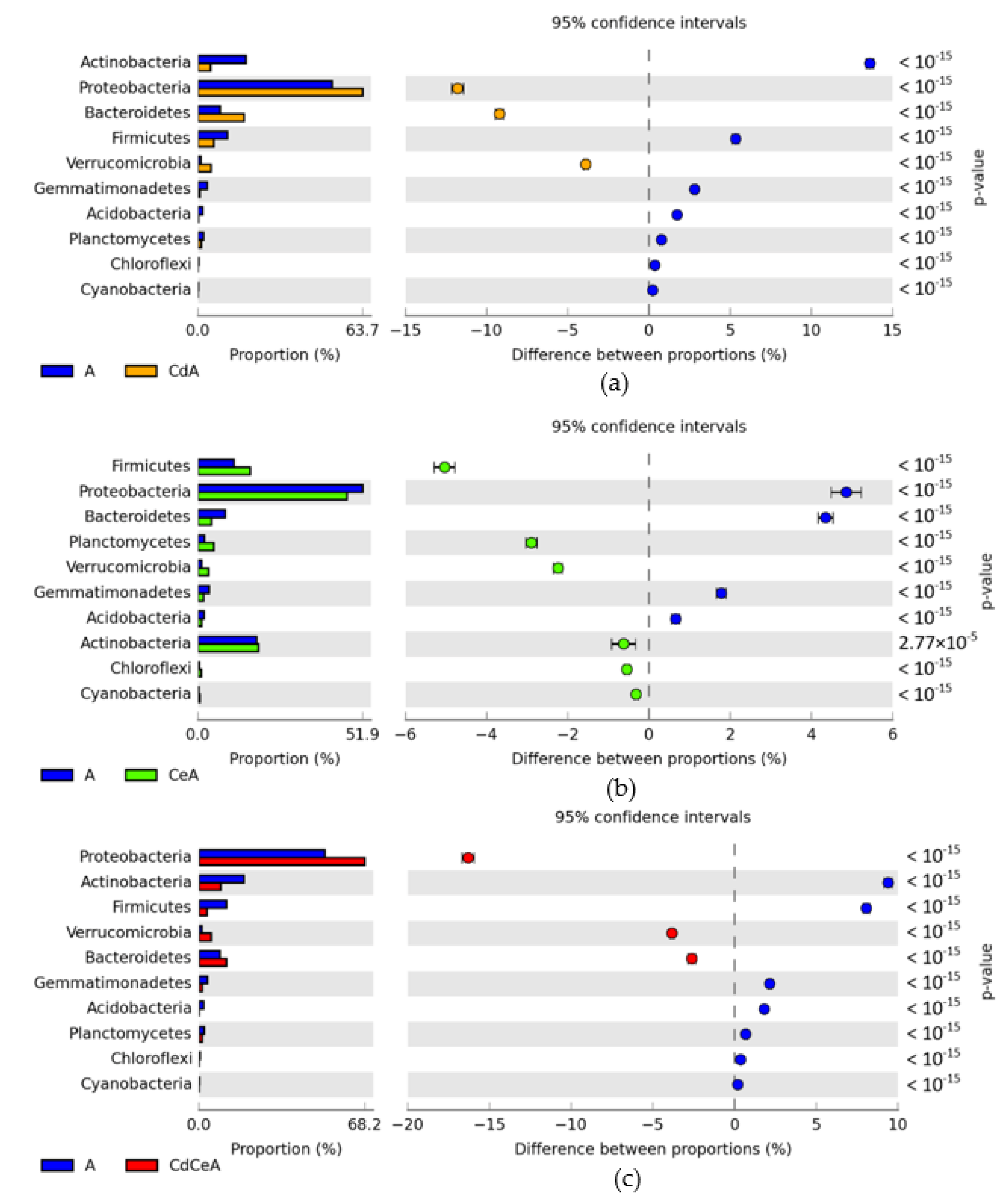

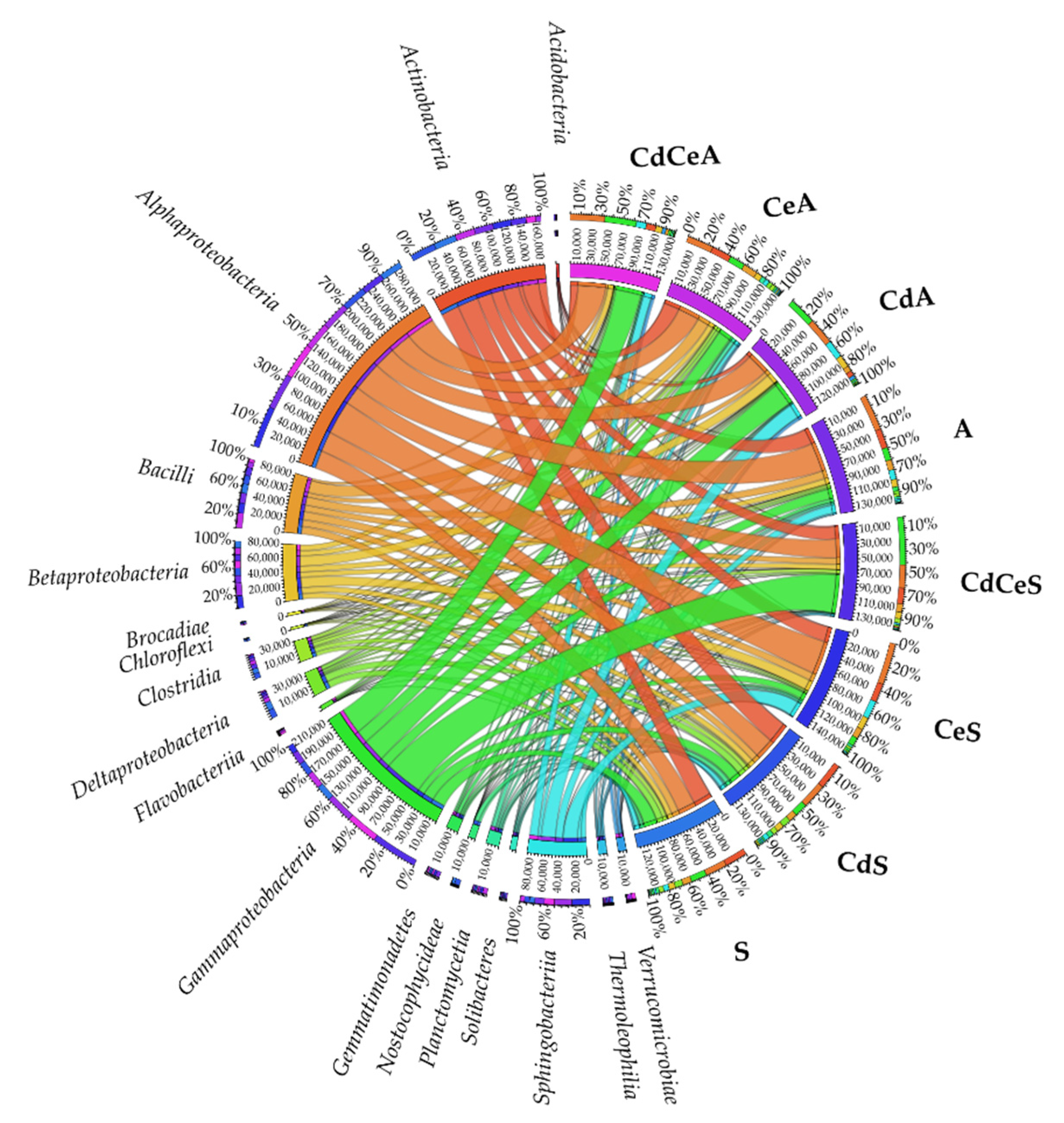
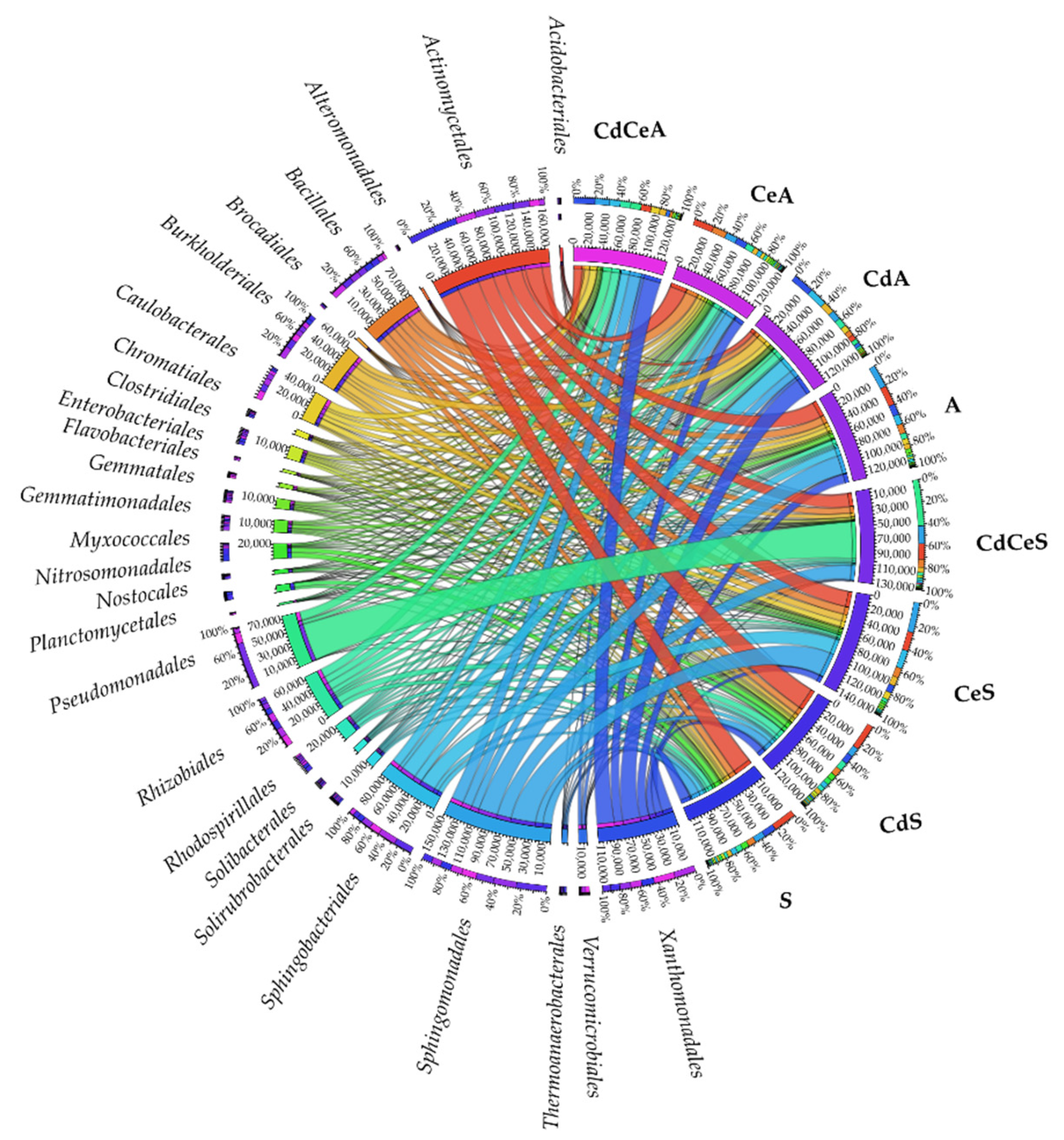
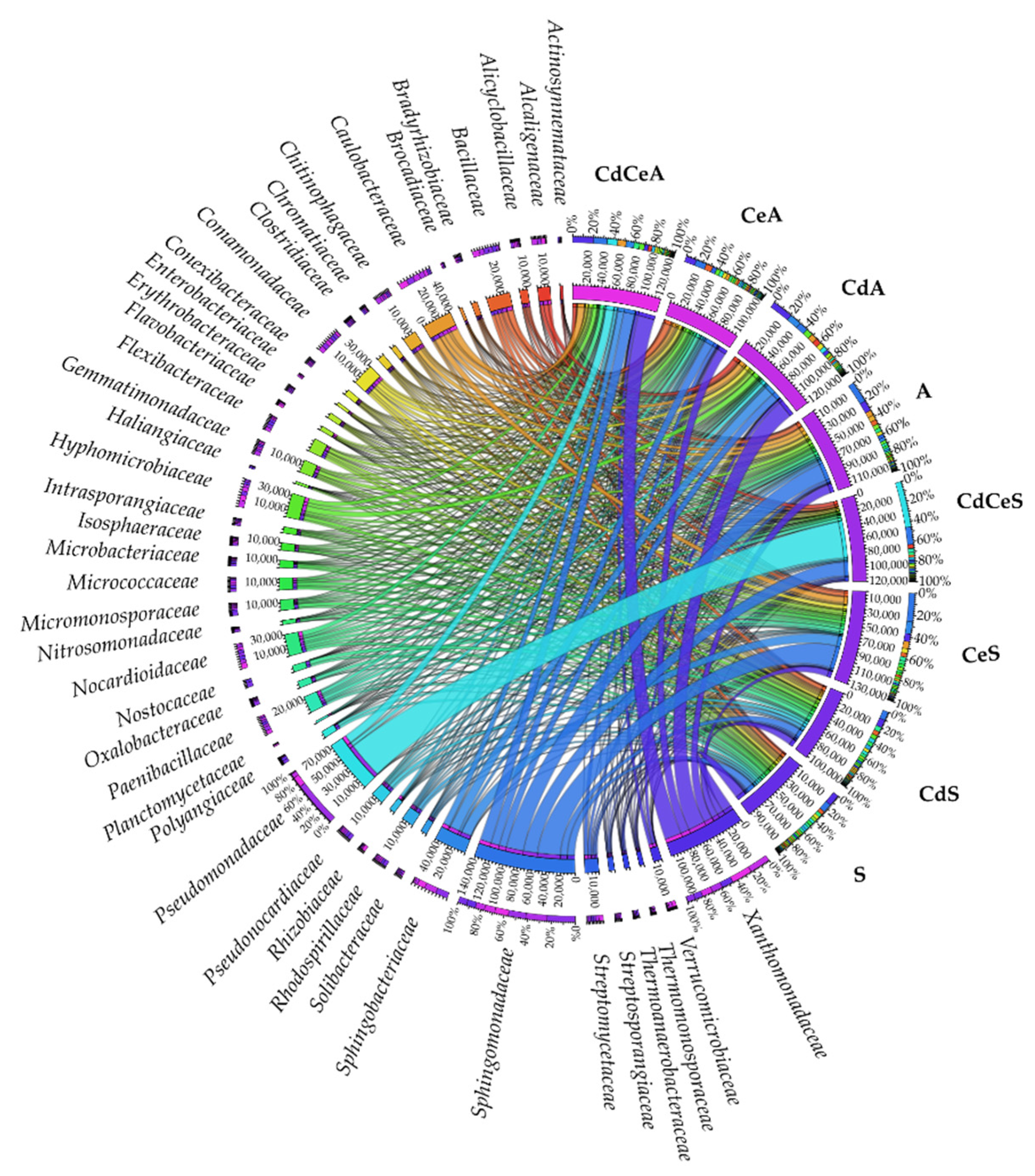
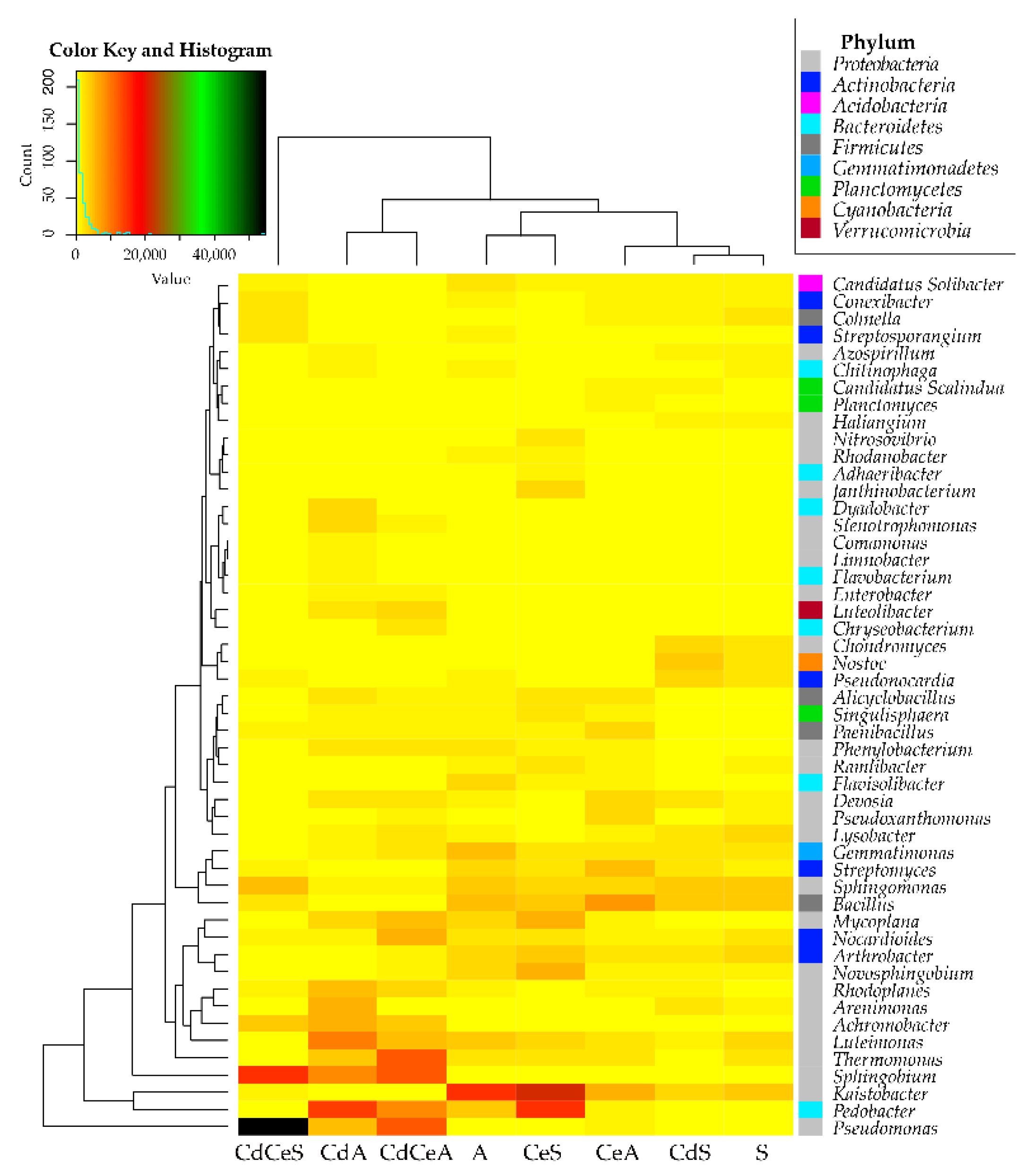
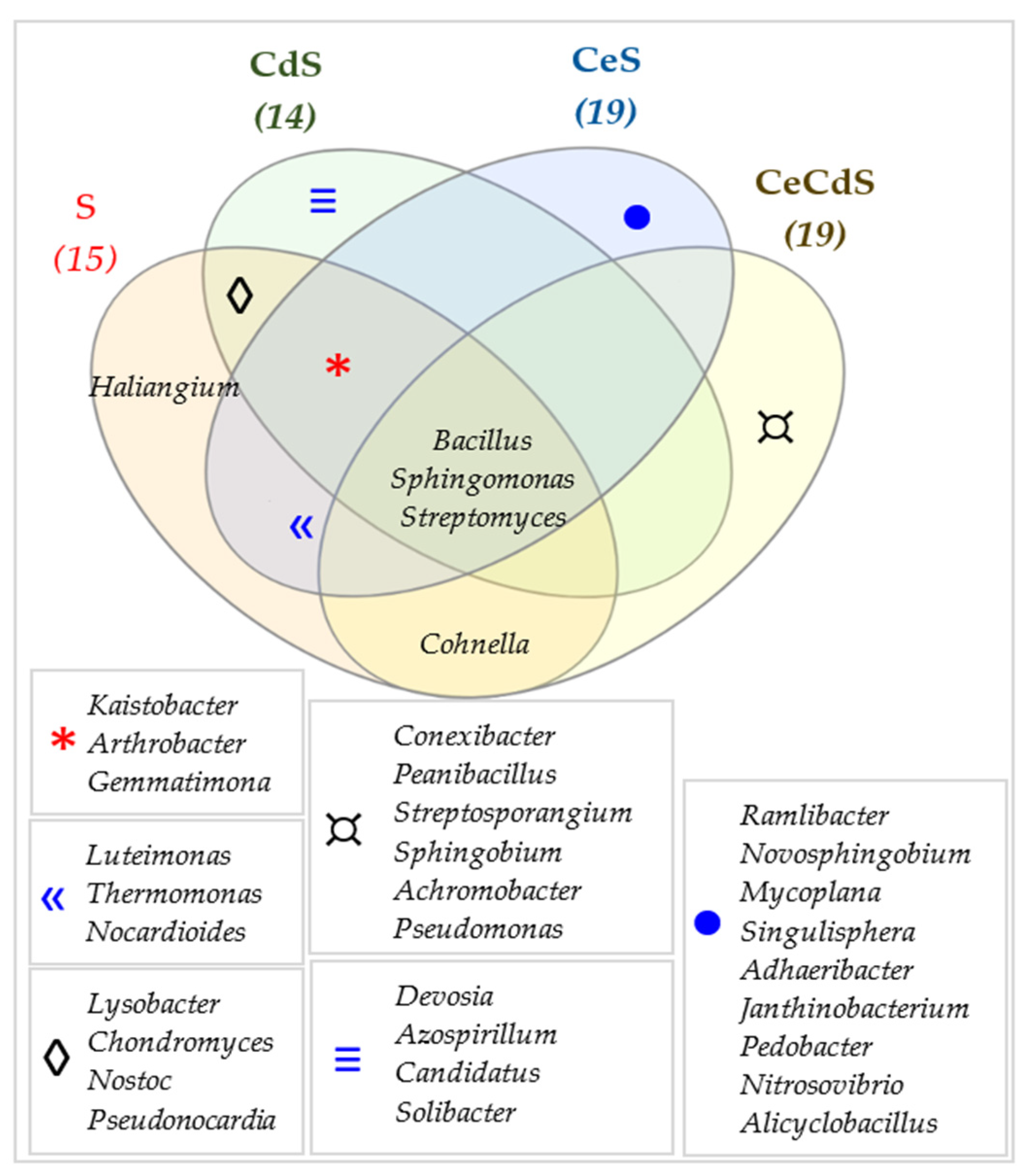
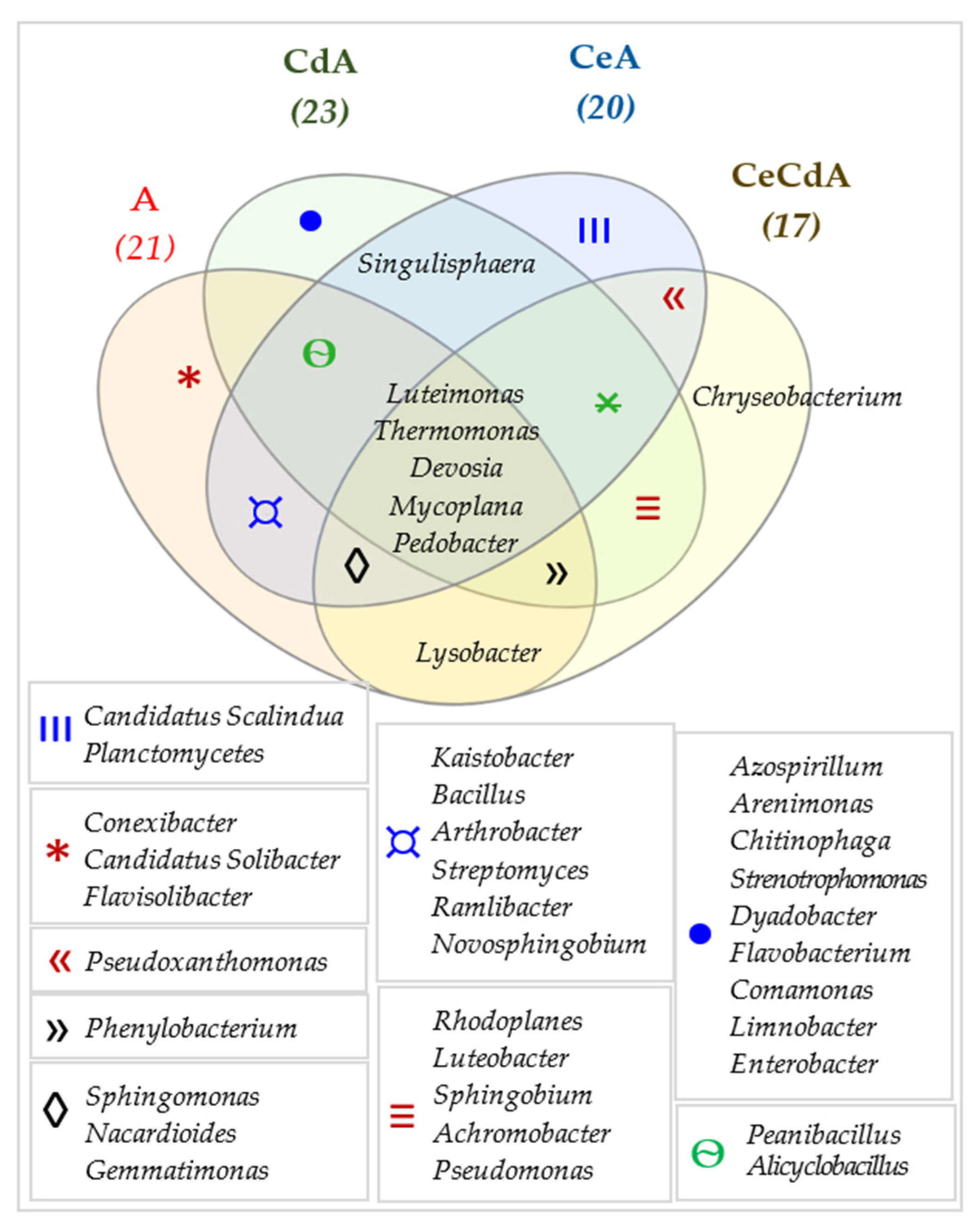
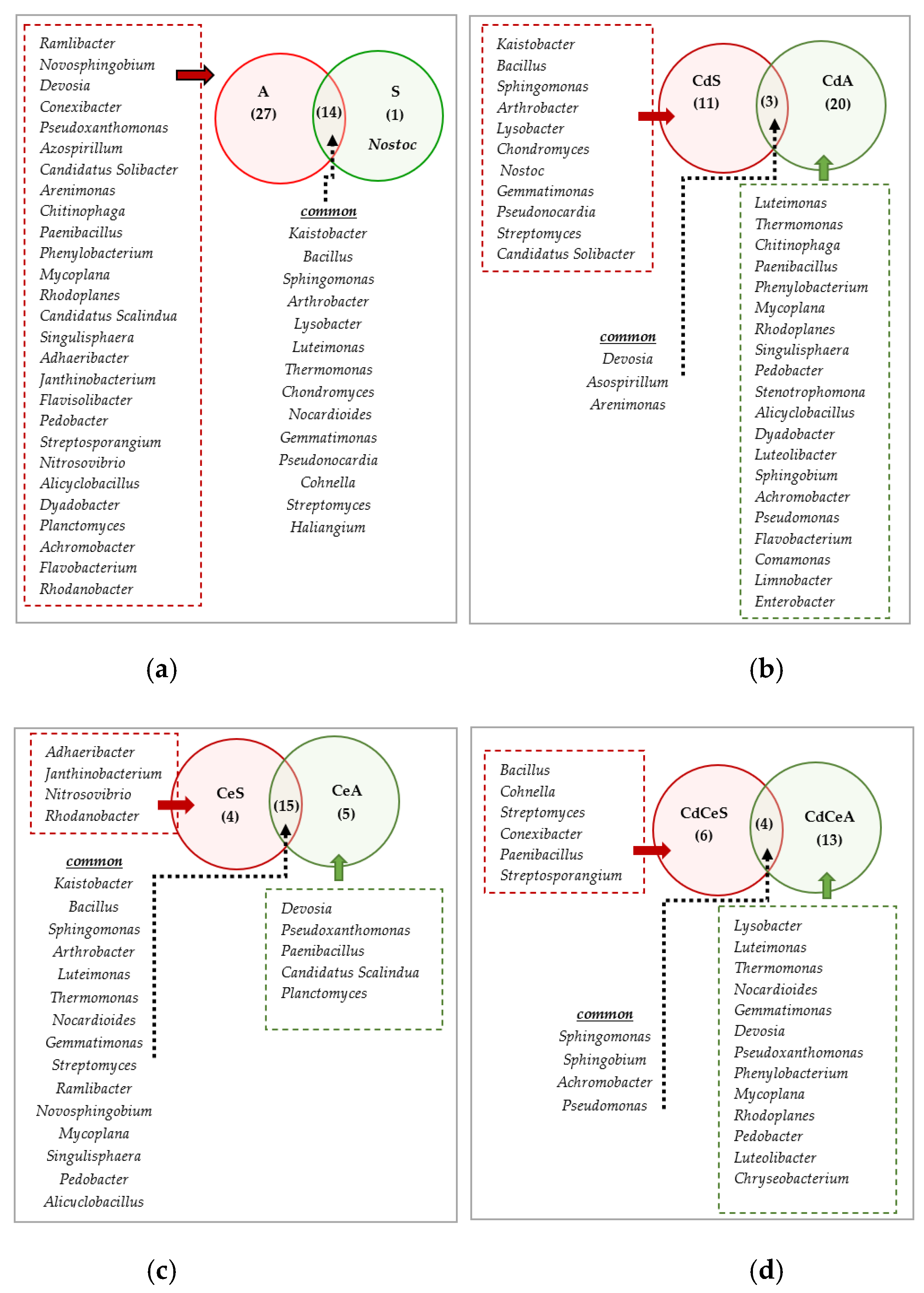
| Dose Cd2+ (mg kg−1 soil) | Sinapis alba L. | Avena sativa L. | ||
|---|---|---|---|---|
| Aerial Parts | Roots | Aerial Parts | Roots | |
| not fertilized with cellulose (Co) | ||||
| 4 | 0.735 b | 0.644 a | 0.870 a | 0.920 a |
| 8 | 0.697 c | 0.523 bc | 0.771 c | 0.655 c |
| 16 | 0.642 c | 0.469 d | 0.718 c | 0.655 c |
| 0.691 B | 0.545 C | 0.786 A | 0.743 B | |
| fertilized with cellulose (Ce) | ||||
| 4 | 0.959 a | 0.564 b | 0.829 b | 0.792 b |
| 8 | 0.955 a | 0.506 c | 0.815 b | 0.755 b |
| 16 | 0.792 b | 0.312 e | 0.529 d | 0.578 d |
| 0.902 A | 0.461 D | 0.724 B | 0.708 C | |
| Dose Cd2+ (mg kg−1 soil) | Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | ||
|---|---|---|---|---|---|---|
| D | RA | RR | RA | RR | ||
| not fertilized with cellulose (Co) | ||||||
| 0 | 17.141 e | 0.519 g | 13.035 d | 86.965 c | 10.069 e | 89.931 c |
| 16 | 196.392 a | 43.137 c | 29.080 a | 70.920 f | 6.741 f | 93.259 ab |
| fertilized with cellulose (Ce) | ||||||
| 0 | 4.822 f | 5.396 f | 24.670 b | 75.330 e | 3.203 g | 96.797 a |
| 16 | 90.824 b | 31.069 d | 20.703 c | 79.297 d | 9.120 e | 90.880 bc |
| Dose Cd2+ (mg kg−1 soil) | Organotrophic bacteria cfu × 108 | Actinobacteria cfu × 108 | Fungi cfu × 106 | |||
|---|---|---|---|---|---|---|
| Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | |
| not fertilized with cellulose (Co) | ||||||
| 0 | 12.382 c | 14.441 c | 2.430 d | 10.260 a | 5.373 b | 9.120 b |
| 4 | 9.757 d | 12.774 d | 2.226 d | 3.960 e | 5.115 b | 7.353 c |
| 8 | 6.822 f | 10.766 e | 1.776 de | 3.745 ef | 2.710 c | 4.634 ef |
| 16 | 5.263 g | 7.029 f | 1.316 e | 3.302 f | 2.021 c | 3.963 f |
| 8.556 D | 11.253 C | 1.937 C | 5.317 B | 3.805 D | 6.268 C | |
| r | −0.954 * | −0.999 * | −0.990 * | −0.741 | −0.926 * | −0.927 * |
| fertilized with cellulose (Ce) | ||||||
| 0 | 22.759 a | 20.973 a | 12.571 a | 6.193 b | 14.347 a | 19.847 a |
| 4 | 16.848 b | 15.750 b | 9.043 b | 5.470 c | 14.087 a | 6.319 d |
| 8 | 10.062 d | 12.262 d | 4.339 c | 4.933 d | 5.484 b | 6.155 d |
| 16 | 8.153 e | 11.997 de | 2.577 d | 4.703 d | 5.388 b | 6.047 d |
| 14.456 B | 15.246 A | 7.132 A | 5.325 B | 9.827 A | 9.592 B | |
| r | −0.924 * | −0.861 * | −0.942 * | −0.921 * | −0.854 * | −0.694 |
| Dose Cd2+ (mg kg−1 soil) | Organotrophic Bacteria | Actinobacteria | Fungi | |||
|---|---|---|---|---|---|---|
| Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | |
| not fertilized with cellulose (Co) | ||||||
| 4 | 0.650 a | 0.793 a | 0.845 a | 0.239 d | 0.908 a | 0.675 a |
| 8 | 0.380 b | 0.594 b | 0.576 b | 0.223 d | 0.337 b | 0.341 b |
| 16 | 0.270 cd | 0.322 d | 0.371 c | 0.192 e | 0.232 c | 0.278 c |
| 0.433 B | 0.570 A | 0.597 B | 0.218 D | 0.492 A | 0.431 B | |
| fertilized with cellulose (Ce) | ||||||
| 4 | 0.588 a | 0.601 b | 0.562 b | 0.791 a | 0.964 a | 0.189 d |
| 8 | 0.284 c | 0.413 c | 0.209 d | 0.662 b | 0.236 c | 0.184 d |
| 16 | 0.218 d | 0.401 c | 0.114 e | 0.612 bc | 0.231 c | 0.180 d |
| 0.382 C | 0.472 B | 0.295 C | 0.688 A | 0.477 A | 0.184 C | |
| Dose Cd2+ (mg kg−1 soil) | Organotrophic Bacteria | Actinobacteria | Fungi | |||
|---|---|---|---|---|---|---|
| Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | |
| not fertilized with cellulose (Co) | ||||||
| 0 | 44.394 bc | 36.931 b | 21.644 a | 21.627 ab | 30.128 d | 35.389 d |
| 4 | 48.026 b | 37.690 b | 21.926 a | 19.508 c | 30.059 de | 37.399 cd |
| 8 | 38.606 c | 38.453 ab | 21.908 a | 22.942 a | 33.388 cd | 34.278 d |
| 16 | 67.508 a | 42.346 a | 20.603 b | 20.852 b | 28.876 e | 35.379 d |
| 49.634 A | 38.855 C | 21.520 A | 21.232 A | 30.613 D | 35.611 C | |
| fertilized with cellulose (Ce) | ||||||
| 0 | 33.972 c | 36.927 bc | 21.069 ab | 21.537 ab | 50.389 a | 50.680 a |
| 4 | 44.827 bc | 32.589 c | 21.574 a | 20.329 b | 47.700 b | 46.712 b |
| 8 | 49.403 b | 33.680 c | 20.416 b | 20.205 b | 45.342 b | 39.479 c |
| 16 | 46.905 b | 38.941 ab | 19.047 c | 20.510 b | 29.621 e | 44.952 b |
| 43.777 B | 35.534 D | 20.527 B | 20.645 B | 43.263 B | 45.456 A | |
| Dose Cd2+ (mg kg−1 soil) | Organotrophic Bacteria | Actinobacteria | Fungi | |||
|---|---|---|---|---|---|---|
| Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | Sinapis alba L. | Avena sativa L. | |
| not fertilized with cellulose (Co) | ||||||
| 0 | 0.858 b | 0.856 b | 0.873 a | 0.871 ab | 0.689 b | 0.697 b |
| 4 | 0.833 c | 0.824 c | 0.887 a | 0.872 ab | 0.716 b | 0.640 c |
| 8 | 0.842 bc | 0.846 bc | 0.811 d | 0.853 b | 0.739 ab | 0.781 ab |
| 16 | 0.645 e | 0.840 bc | 0.851 b | 0.893 a | 0.772 a | 0.824 a |
| 0.795 B | 0.842 A | 0.856 A | 0.872 A | 0.729 A | 0.736 A | |
| fertilized with cellulose (Ce) | ||||||
| 0 | 0.895 a | 0.762 d | 0.843 b | 0.842 b | 0.502 d | 0.521 d |
| 4 | 0.851 b | 0.914 a | 0.885 a | 0.896 a | 0.569 c | 0.567 cd |
| 8 | 0.780 d | 0.915 a | 0.885 a | 0.850 b | 0.695 b | 0.712 b |
| 16 | 0.846 bc | 0.841 b | 0.826 c | 0.827 c | 0.764 a | 0.624 c |
| 0.843 A | 0.858 A | 0.860 A | 0.854 A | 0.633 B | 0.606 B | |
| Taxa | S | CdS | CeS | CdCeS | A | CdA | CeA | CdCeA |
|---|---|---|---|---|---|---|---|---|
| Phylum | 1.777 e | 1.845 e | 1.493 e | 1.095 c | 1.635 e | 1.248 e | 1.785 e | 1.220 d |
| Class | 2.605 d | 2.614 d | 2.118 d | 1.922 b | 2.333 d | 2.125 d | 2.562 d | 2.035 c |
| Order | 3.138 c | 3.146 c | 2.573 c | 2.280 b | 2.865 cd | 2.755 cd | 3.198 c | 2.728 bc |
| Family | 4.019 b | 3.999 b | 3.346 b | 2.799 ab | 3.631 b | 3.254 b | 4.001 b | 3.187 b |
| Genus | 4.608 a | 4.559 a | 3.988 a | 3.137 a | 4.324 a | 4.036 a | 4.614 a | 3.902 a |
| Taxa | S | CdS | CeS | CdCeS | A | CdA | CeA | CdCeA |
|---|---|---|---|---|---|---|---|---|
| Phylum | 0.738 c | 0.762 d | 0.675 d | 0.515 d | 0.698 d | 0.565 d | 0.742 c | 0.528 d |
| Class | 0.891 b | 0.892 c | 0.824 c | 0.773 c | 0.850 c | 0.831 c | 0.883 b | 0.800 c |
| Order | 0.925 ab | 0.923 bc | 0.881 b | 0.805 b | 0.907 b | 0.904 b | 0.930 ab | 0.908 b |
| Family | 0.963 a | 0.958 a | 0.923 ab | 0.824 a | 0.943 ab | 0.929 ab | 0.963 a | 0.925 ab |
| Genus | 0.959 a | 0.945 a | 0.950 a | 0.832 a | 0.963 a | 0.965 a | 0.967 a | 0.958 a |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wyszkowska, J.; Boros-Lajszner, E.; Borowik, A.; Kucharski, J. The Role of Cellulose in Microbial Diversity Changes in the Soil Contaminated with Cadmium. Sustainability 2022, 14, 14242. https://doi.org/10.3390/su142114242
Wyszkowska J, Boros-Lajszner E, Borowik A, Kucharski J. The Role of Cellulose in Microbial Diversity Changes in the Soil Contaminated with Cadmium. Sustainability. 2022; 14(21):14242. https://doi.org/10.3390/su142114242
Chicago/Turabian StyleWyszkowska, Jadwiga, Edyta Boros-Lajszner, Agata Borowik, and Jan Kucharski. 2022. "The Role of Cellulose in Microbial Diversity Changes in the Soil Contaminated with Cadmium" Sustainability 14, no. 21: 14242. https://doi.org/10.3390/su142114242
APA StyleWyszkowska, J., Boros-Lajszner, E., Borowik, A., & Kucharski, J. (2022). The Role of Cellulose in Microbial Diversity Changes in the Soil Contaminated with Cadmium. Sustainability, 14(21), 14242. https://doi.org/10.3390/su142114242









