Maize Seedling Leave Counting Based on Semi-Supervised Learning and UAV RGB Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Image Acquisition and Preprocessing
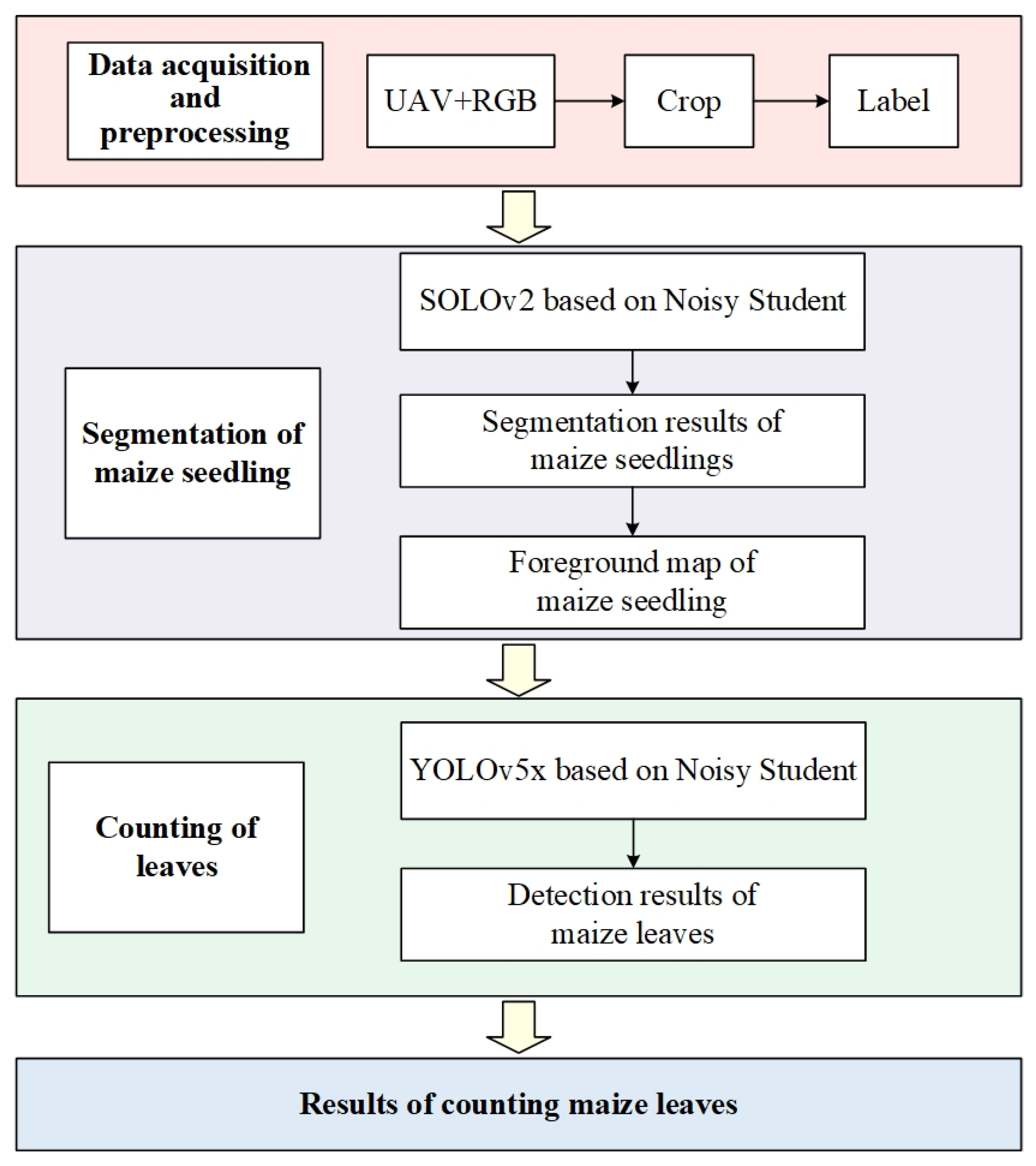
2.2. Overall Design of Leaf Counting
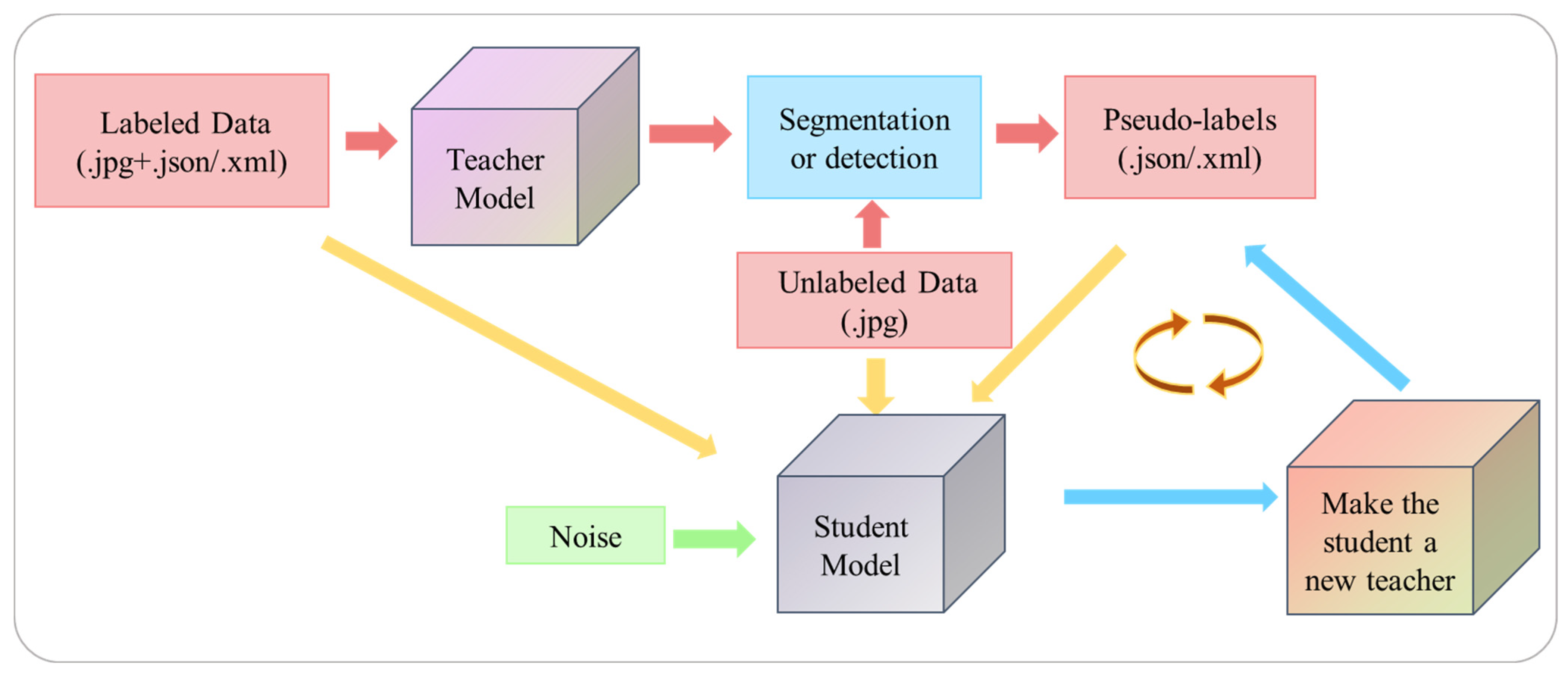
2.3. Semi-Supervised Learning Framework: Noisy Student
- (1)
- Using a small number of labeled images to initialize the teacher models (SOLOv2 and YOLOv5x) training;
- (2)
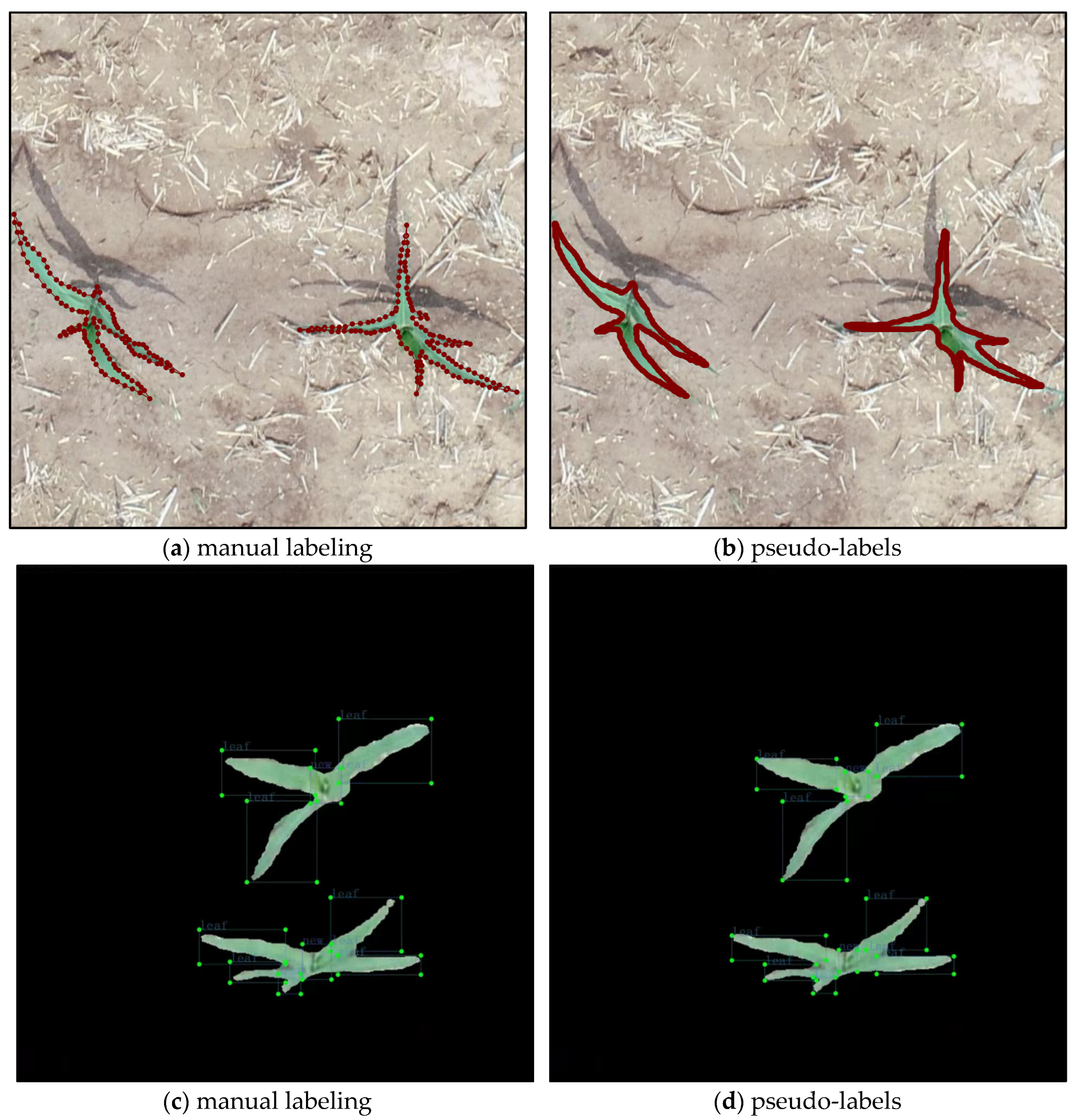
- Predicting the unlabeled data using the trained teacher models, filtering out masks and bounding boxes with confidence less than 0.35 from the unlabeled data, and creating pseudo-labels based on the model’s predicted mask or bounding box. The file format for the maize seedling segmentation dataset was .json, and the file format for the leaf detection dataset was .xml;
- (3)
- Training the student model (SOLOv2 and YOLOv5x) by merging labeled and pseudo-labeled data. The test images were used to obtain predicted values from the student model. To improve the model’s generalization, input noise and model noise (Noise) were added to the training process. Input noise refers to data augmentation, and model noise refers to the stochastic gradient descent optimizer (SGD);
- (4)
- The student model became the new teacher model, and pseudo-labels are predicted again to train teacher models;
- (5)
- Repeat steps (2)–(4) for a total of three cycles.
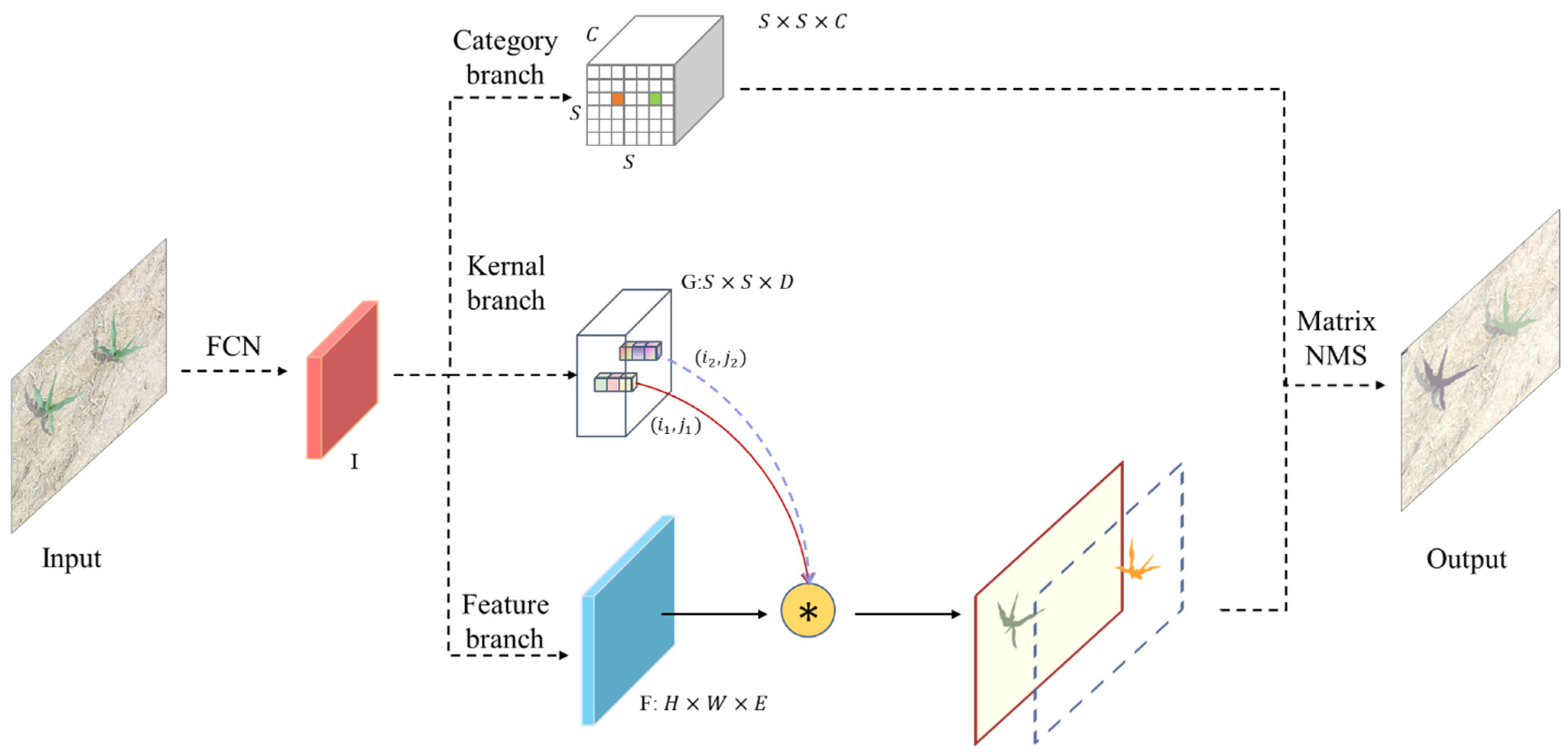
2.4. SOLOv2 of Segment Maize Seedlings
2.5. YOLOv5x of Count Maize Leaves
2.6. Parameter Setting for Training
2.7. Evaluation Metrics
3. Results
3.1. Maize Seedling Segmentation Results
3.2. Leaf Counting Results
4. Discussion
5. Conclusions
- (1)
- The maize seedling segmentation performance of SOLOv2 Resnet101 was better than that of SOLOv2 Resnet50, achieving a mAP of 94.2% and a single-image inference time of 0.15 s. However, when the labeled data proportion was 30%, the student model SOLOv2 Resnet101 achieved the best segmentation performance with a mAP of 93.6% and a single-image inference time of 0.16 s.
- (2)
- When the labeled data proportion was 40%, the leaf detection performance of the student model YOLOv5x was comparable to that of the fully supervised model. The precision for detecting fully unfolded leaves and newly appearing leaves was 89.1% and 57.5%, respectively, with recall rates of 87.2% and 56.6% and average precision rates of 89.6% and 57.4%. The counting accuracy for newly appearing leaves and fully unfolded leaves was 72.9% and 69.4%, respectively.
Author Contributions
Funding
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Miao, T.; Zhu, C.; Xu, T.; Yang, T.; Li, N.; Zhou, Y.; Deng, H. Automatic stem-leaf segmentation of maize shoots using three-dimensional point cloud. Comput. Electron. Agric. 2021, 187, 106310. [Google Scholar] [CrossRef]
- Shu, M.Y.; Dong, Q.Z.; Fei, S.P.; Yang, X.H.; Zhu, J.Y.; Meng, L.; Li, B.G.; Ma, Y.T. Improved estimation of canopy water status in maize using UAV-based digital and hyperspectral images. Comput. Electron. Agric. 2022, 197, 106982. [Google Scholar]
- Wang, Y.-H.; Su, W.-H. Convolutional Neural Networks in Computer Vision for Grain Crop Phenotyping: A Review. Agronomy 2022, 12, 2659. [Google Scholar] [CrossRef]
- Amarasingam, N.; Ashan Salgadoe, A.S.; Powell, K.; Gonzalez, L.F.; Natarajan, S. A review of UAV platforms, sensors, and applications for monitoring of sugarcane crops. Remote Sens. Appl. Soc. Environ. 2022, 26, 100712. [Google Scholar] [CrossRef]
- Ji, Y.; Chen, Z.; Cheng, Q.; Liu, R.; Li, M.; Yan, X.; Li, G.; Wang, D.; Fu, L.; Ma, Y.; et al. Estimation of plant height and yield based on UAV imagery in faba bean (Vicia faba L.). Plant Methods 2022, 18, 26. [Google Scholar] [CrossRef]
- Oehme, L.H.; Reineke, A.-J.; Weiß, T.M.; Würschum, T.; He, X.; Müller, J. Remote Sensing of Maize Plant Height at Different Growth Stages Using UAV-Based Digital Surface Models (DSM). Agronomy 2022, 12, 958. [Google Scholar] [CrossRef]
- Gao, M.; Yang, F.; Wei, H.; Liu, X. Individual Maize Location and Height Estimation in Field from UAV-Borne LiDAR and RGB Images. Remote Sens. 2022, 14, 2292. [Google Scholar] [CrossRef]
- Gan, Y.; Wang, Q.; Iio, A. Tree Crown Detection and Delineation in a Temperate Deciduous Forest from UAV RGB Imagery Using Deep Learning Approaches: Effects of Spatial Resolution and Species Characteristics. Remote Sens. 2023, 15, 778. [Google Scholar] [CrossRef]
- Terryn, L.; Calders, K.; Bartholomeus, H.; Bartolo, R.E.; Brede, B.; D’Hont, B.; Disney, M.; Herold, M.; Lau, A.; Shenkin, A.; et al. Quantifying tropical forest structure through terrestrial and UAV laser scanning fusion in Australian rainforests. Remote Sens. Environ. 2022, 271, 112912. [Google Scholar] [CrossRef]
- Panagiotidis, D.; Abdollahnejad, A.; Slavík, M. 3D point cloud fusion from UAV and TLS to assess temperate managed forest structures. Int. J. Appl. Earth Obs. Geoinf. 2022, 112, 102917. [Google Scholar] [CrossRef]
- Du, L.; Yang, H.; Song, X.; Wei, N.; Yu, C.; Wang, W.; Zhao, Y. Estimating leaf area index of maize using UAV-based digital imagery and machine learning methods. Sci. Rep. 2022, 12, 15937. [Google Scholar] [CrossRef] [PubMed]
- Wu, S.; Deng, L.; Guo, L.; Wu, Y. Wheat leaf area index prediction using data fusion based on high-resolution unmanned aerial vehicle imagery. Plant Methods 2022, 18, 68. [Google Scholar] [CrossRef]
- Shu, M.; Shen, M.; Zuo, J.; Yin, P.; Wang, M.; Xie, Z.; Tang, J.; Wang, R.; Li, B.; Yang, X.; et al. The Application of UAV-Based Hyperspectral Imaging to Estimate Crop Traits in Maize Inbred Lines. Plant Phenomics 2021, 2021, 9890745. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Che, Y.; Shao, K.; Zhu, J.; Wang, R.; Sui, Y.; Guo, Y.; Li, B.; Meng, L.; Ma, Y. Estimation of sugar content in sugar beet root based on UAV multi-sensor data. Comput. Electron. Agric. 2022, 203, 107433. [Google Scholar] [CrossRef]
- Wu, Q.; Zhang, Y.; Zhao, Z.; Xie, M.; Hou, D. Estimation of Relative Chlorophyll Content in Spring Wheat Based on Multi-Temporal UAV Remote Sensing. Agronomy 2023, 13, 211. [Google Scholar] [CrossRef]
- Fu, H.; Chen, J.; Lu, J.; Yue, Y.; Xu, M.; Jiao, X.; Cui, G.; She, W. A Comparison of Different Remote Sensors for Ramie Leaf Area Index Estimation. Agronomy 2023, 13, 899. [Google Scholar] [CrossRef]
- Hu, P.; Zhang, R.; Yang, J.; Chen, L. Development Status and Key Technologies of Plant Protection UAVs in China: A Review. Drones 2022, 6, 354. [Google Scholar] [CrossRef]
- Liu, Y.; Feng, H.; Yue, J.; Li, Z.; Yang, G.; Song, X.; Yang, X.; Zhao, Y. Remote-sensing estimation of potato above-ground biomass based on spectral and spatial features extracted from high-definition digital camera images. Comput. Electron. Agric. 2022, 198, 107089. [Google Scholar] [CrossRef]
- Syazwani, R.W.N.; Asraf, H.M.; Amin, M.M.S.; Dalila, K.N. Automated image identification, detection and fruit counting of top-view pineapple crown using machine learning. Alex. Eng. J. 2022, 61, 1265–1276. [Google Scholar] [CrossRef]
- Liu, S.; Yin, D.; Feng, H.; Li, Z.; Xu, X.; Shi, L.; Jin, X. Estimating maize seedling number with UAV RGB images and advanced image processing methods. Precis. Agric. 2022, 23, 1604–1632. [Google Scholar] [CrossRef]
- Kim, Y.H.; Park, K.R. MTS-CNN: Multi-task semantic segmentation-convolutional neural network for detecting crops and weeds. Comput. Electron. Agric. 2022, 199, 107146. [Google Scholar] [CrossRef]
- Wang, P.; Zhang, Y.; Jiang, B.; Hou, J. An maize leaf segmentation algorithm based on image repairing technology. Comput. Electron. Agric. 2020, 172, 105349. [Google Scholar] [CrossRef]
- Xu, X.; Wang, L.; Shu, M.; Liang, X.; Ghafoor, A.Z.; Liu, Y.; Ma, Y.; Zhu, J. Detection and Counting of Maize Leaves Based on Two-Stage Deep Learning with UAV-Based RGB Image. Remote Sens. 2022, 14, 5388. [Google Scholar] [CrossRef]
- Song, C.-Y.; Zhang, F.; Li, J.-S.; Xie, J.-Y.; Yang, C.; Zhou, H.; Zhang, J.-X. Detection of maize tassels for UAV remote sensing image with an improved YOLOX model. J. Integr. Agric. 2022, 22, 1671–1683. [Google Scholar] [CrossRef]
- Lac, L.; Da Costa, J.-P.; Donias, M.; Keresztes, B.; Bardet, A. Crop stem detection and tracking for precision hoeing using deep learning. Comput. Electron. Agric. 2022, 192, 106606. [Google Scholar] [CrossRef]
- Barreto, A.; Lottes, P.; Yamati, F.R.I.; Baumgarten, S.; Wolf, N.A.; Stachniss, C.; Mahlein, A.-K.; Paulus, S. Automatic UAV-based counting of seedlings in sugar-beet field and extension to maize and strawberry. Comput. Electron. Agric. 2021, 191, 106493. [Google Scholar] [CrossRef]
- Yu, X.; Yin, D.; Nie, C.; Ming, B.; Xu, H.; Liu, Y.; Bai, Y.; Shao, M.; Cheng, M.; Liu, Y.; et al. Maize tassel area dynamic monitoring based on near-ground and UAV RGB images by U-Net model. Comput. Electron. Agric. 2022, 203, 107477. [Google Scholar] [CrossRef]
- Gan, H.; Ou, M.; Li, C.; Wang, X.; Guo, J.; Mao, A.; Ceballos, M.C.; Parsons, T.D.; Liu, K.; Xue, Y. Automated detection and analysis of piglet suckling behaviour using high-accuracy amodal instance segmentation. Comput. Electron. Agric. 2022, 199, 107162. [Google Scholar] [CrossRef]
- Mendoza, A.; Trullo, R.; Wielhorski, Y. Descriptive modeling of textiles using FE simulations and deep learning. Compos. Sci. Technol. 2021, 213, 108897. [Google Scholar] [CrossRef]
- Wagner, F.H.; Dalagnol, R.; Tarabalka, Y.; Segantine, T.Y.F.; Thomé, R.; Hirye, M.C.M. U-Net-Id, an Instance Segmentation Model for Building Extraction from Satellite Images—Case Study in the Joanópolis City, Brazil. Remote Sens. 2020, 12, 1544. [Google Scholar] [CrossRef]
- Gao, X.; Zan, X.; Yang, S.; Zhang, R.; Chen, S.; Zhang, X.; Liu, Z.; Ma, Y.; Zhao, Y.; Li, S. Maize seedling information extraction from UAV images based on semi-automatic sample generation and Mask R-CNN model. Eur. J. Agron. 2023, 147, 126845. [Google Scholar] [CrossRef]
- Sun, X.; Fang, W.; Gao, C.; Fu, L.; Majeed, Y.; Liu, X.; Gao, F.; Yang, R.; Li, R. Remote estimation of grafted apple tree trunk diameter in modern orchard with RGB and point cloud based on SOLOv2. Comput. Electron. Agric. 2022, 199, 107209. [Google Scholar] [CrossRef]
- Soetedjo, A.; Hendriarianti, E. Plant Leaf Detection and Counting in a Greenhouse during Day and Nighttime Using a Raspberry Pi NoIR Camera. Sensors 2021, 21, 6659. [Google Scholar] [CrossRef] [PubMed]
- Tu, Y.-L.; Lin, W.-Y.; Lin, Y.-C. Automatic Leaf Counting Using Improved YOLOv3. In Proceedings of the International Symposium on Computer, Consumer and Control (IS3C) 2020, Taichung, Taiwan, 13–16 November 2020; pp. 197–200. [Google Scholar] [CrossRef]
- Lu, S.; Song, Z.; Chen, W.; Qian, T.; Zhang, Y.; Chen, M.; Li, G. Counting Dense Leaves under Natural Environments via an Improved Deep-Learning-Based Object Detection Algorithm. Agriculture 2021, 11, 1003. [Google Scholar] [CrossRef]
- Li, X.; Fan, W.Q.; Wang, Y.; Zhang, L.K.; Liu, Z.X.; Xia, C.L. Detecting Plant Leaves Based on Vision Transformer Enhanced YOLOv5. In Proceedings of the 2022 3rd International Conference on Pattern Recognition and Machine Learning (PRML), Chengdu, China, 22–24 July 2022; pp. 32–37. [Google Scholar]
- Zhang, C.; Ding, H.; Shi, Q.; Wang, Y. Grape Cluster Real-Time Detection in Complex Natural Scenes Based on YOLOv5s Deep Learning Network. Agriculture 2022, 12, 1242. [Google Scholar] [CrossRef]
- Wang, L.; Zhao, Y.; Xiong, Z.; Wang, S.; Li, Y.; Lan, Y. Fast and precise detection of litchi fruits for yield estimation based on the improved YOLOv5 model. Front. Plant Sci. 2022, 13, 965425. [Google Scholar] [CrossRef]
- Xu, L.; Chen, C.P.; Han, R. Graph-based sparse bayesian broad learning system for semi-supervised learning. Inf. Sci. 2022, 597, 193–210. [Google Scholar] [CrossRef]
- Najafian, K.; Ghanbari, A.; Stavness, I.; Jin, L.; Shirdel, G.H.; Maleki, F. A Semi-self-supervised Learning Approach for Wheat Head Detection using Extremely Small Number of Labeled Samples. In Proceedings of the IEEE/CVF International Conference on Computer Vision, Montreal, BC, Canada, 11–17 October 2021; pp. 1342–1351. [Google Scholar] [CrossRef]
- Khan, S.; Tufail, M.; Khan, M.T.; Khan, Z.A.; Iqbal, J.; Alam, M. A novel semi-supervised framework for UAV based crop/weed classification. PLoS ONE 2021, 16, e0251008. [Google Scholar] [CrossRef]
- Nong, C.; Fan, X.; Wang, J. Semi-supervised Learning for Weed and Crop Segmentation Using UAV Imagery. Front. Plant Sci. 2022, 13, 927368. [Google Scholar] [CrossRef]
- Xie, Q.Z.; Luong, M.T.; Hovy, E.; Le, Q.V. Self-training with noisy student improves imagenet classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 13–19 June 2020; pp. 10687–10698. [Google Scholar]
- Blok, P.M.; Van Evert, F.K.; Tielen, A.P.M.; Van Henten, E.J.; Kootstra, G. The effect of data augmentation and network simplification on the image-based detection of broccoli heads with Mask R-CNN. J. Field Robot. 2021, 38, 85–104. [Google Scholar] [CrossRef]
- Pang, Y.; Shi, Y.; Gao, S.; Jiang, F.; Veeranampalayam-Sivakumar, A.-N.; Thompson, L.; Luck, J.; Liu, C. Improved crop row detection with deep neural network for early-season maize stand count in UAV imagery. Comput. Electron. Agric. 2020, 178, 105766. [Google Scholar] [CrossRef]
- Pańka, D.; Jeske, M.; Łukanowski, A.; Baturo-Cieśniewska, A.; Prus, P.; Maitah, M.; Maitah, K.; Malec, K.; Rymarz, D.; Muhire, J.D.D.; et al. Can Cold Plasma Be Used for Boosting Plant Growth and Plant Protection in Sustainable Plant Production? Agronomy 2022, 12, 841. [Google Scholar] [CrossRef]
- Liang, Z.; van der Werf, W.; Xu, Z.; Cheng, J.; Wang, C.; Cong, W.-F.; Zhang, C.; Zhang, F.; Groot, J.C. Identifying exemplary sustainable cropping systems using a positive deviance approach: Wheat-maize double cropping in the North China Plain. Agric. Syst. 2022, 201, 103471. [Google Scholar] [CrossRef]
- Whang, S.E.; Roh, Y.; Song, H.; Lee, J.-G. Data collection and quality challenges in deep learning: A data-centric AI perspective. VLDB J. 2023, 32, 791–813. [Google Scholar] [CrossRef]
- Chung, P.-C.; Yang, W.-J.; Wu, T.-H.; Huang, C.-R.; Hsu, Y.-Y. Emerging Research Directions of Deep Learning for Pathology Image Analysis. In Proceedings of the 2022 IEEE Biomedical Circuits and Systems Conference (BioCAS), Taipei, Taiwan, 13–15 October 2022; pp. 100–104. [Google Scholar] [CrossRef]
- Gul, F.; Ahmed, I.; Ashfaq, M.; Jan, D.; Fahad, S.; Li, X.; Wang, D.; Fahad, M.; Fayyaz, M.; Shah, S.A. Use of crop growth model to simulate the impact of climate change on yield of various wheat cultivars under different agro-environmental conditions in Khyber Pakhtunkhwa, Pakistan. Arab. J. Geosci. 2020, 13, 112. [Google Scholar] [CrossRef]
- Han, C.; Zhang, B.; Chen, H.; Wei, Z.; Liu, Y. Spatially distributed crop model based on remote sensing. Agric. Water Manag. 2019, 218, 165–173. [Google Scholar] [CrossRef]
- Xie, D.L.; Yang, R.H.; Qiao, Y.C.; Zhang, J.B. Intelligent Identification of Landslide Based on Deep Semi-Supervised Learning. In Proceedings of the 2022 5th International Conference on Pattern Recognition and Artificial Intelligence (PRAI), Chengdu, China, 19–21 August 2022; pp. 264–269. [Google Scholar]
- Wang, J.; Jia, Y.; Wang, D.; Xiao, W.; Wang, Z. Weighted IForest and siamese GRU on small sample anomaly detection in healthcare. Comput. Methods Programs Biomed. 2022, 218, 106706. [Google Scholar] [CrossRef]

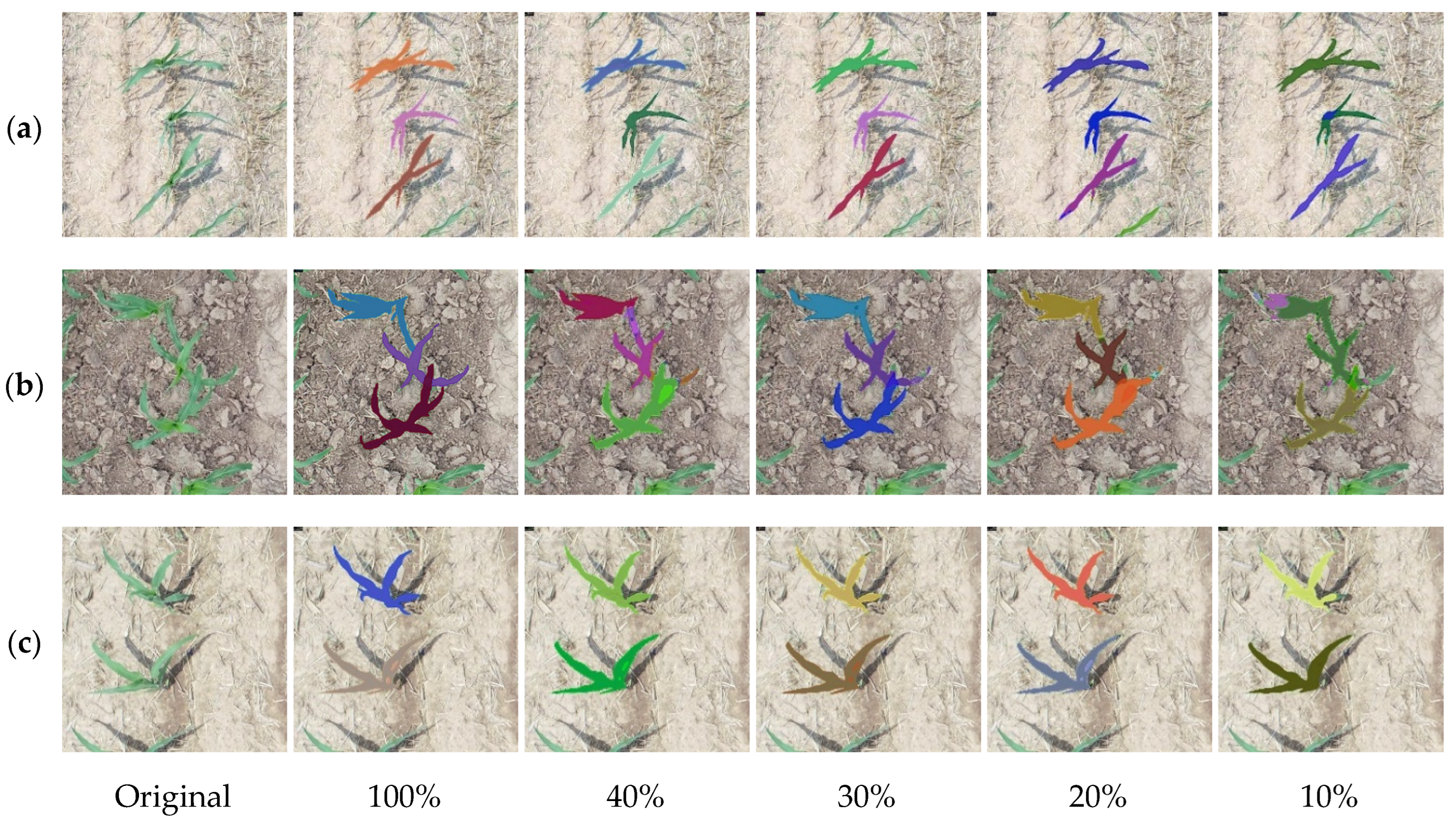
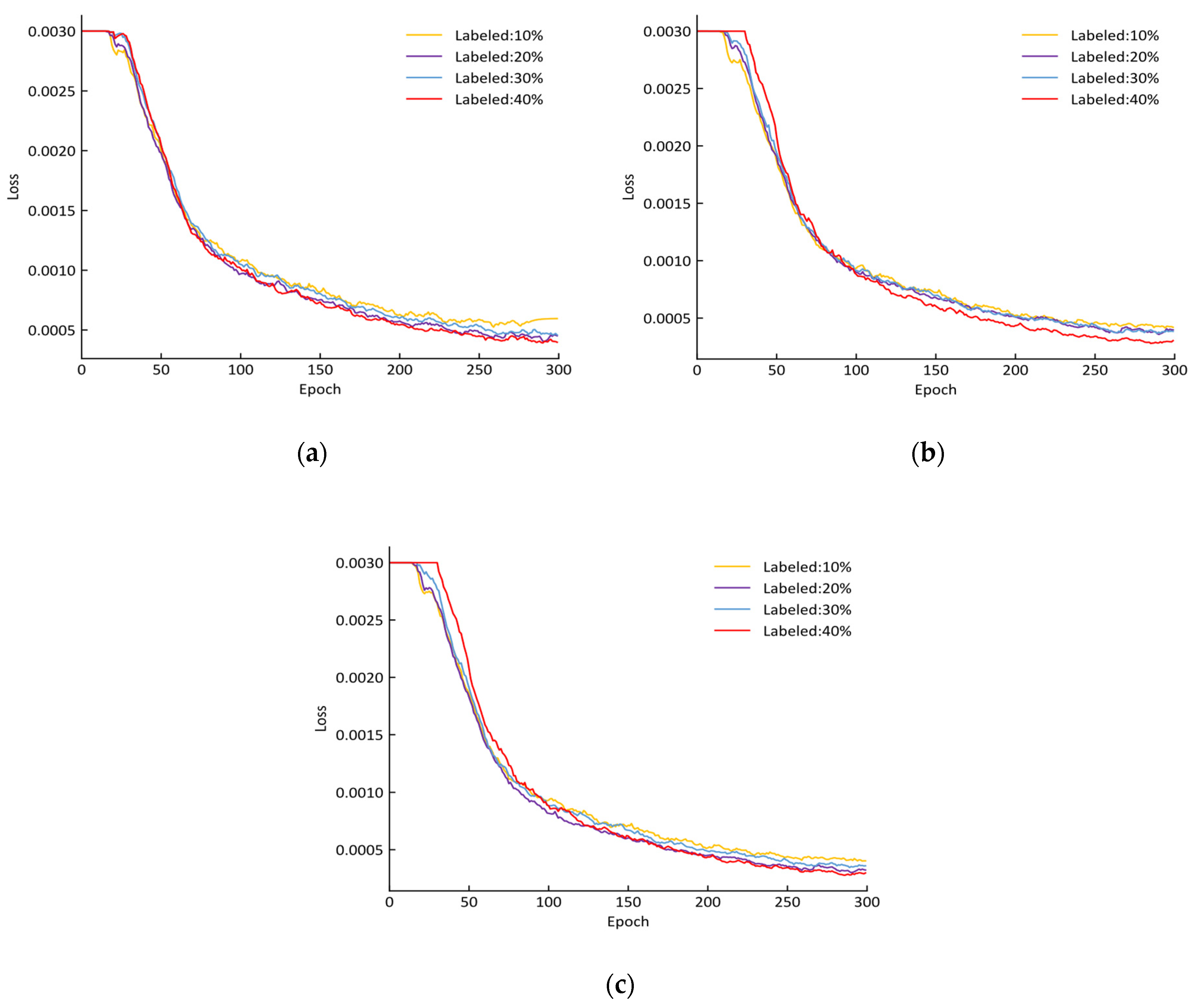
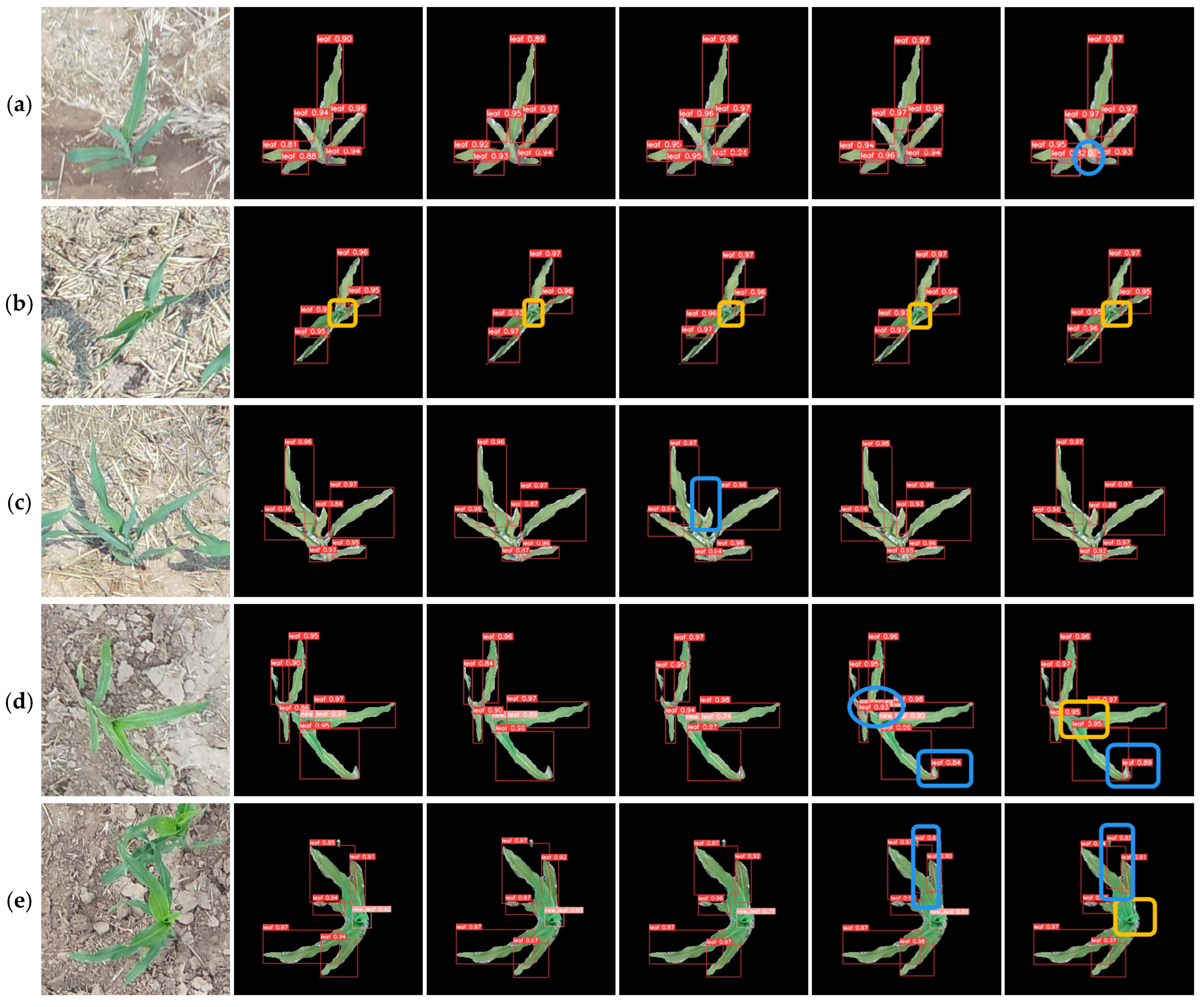
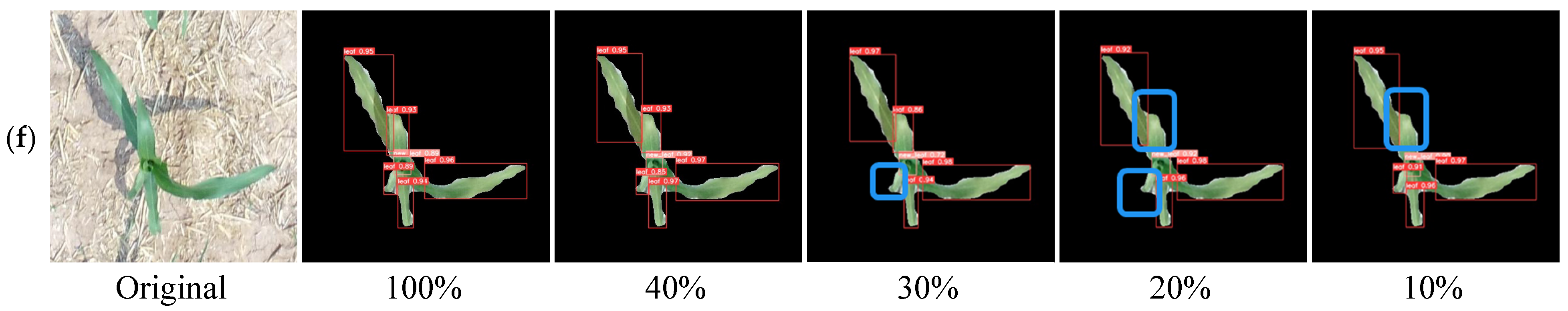
| Labeled Ratio (%) | Number of Labeled Data | Number of Unlabeled Data | After Data Enhancement | ||
|---|---|---|---|---|---|
| Teacher Model | Student Model | ||||
| Segmentation of maize seedlings | 40 | 362 | 542 | 1267 | 3338 |
| 30 | 272 | 632 | 952 | 3338 | |
| 20 | 181 | 723 | 634 | 3338 | |
| 10 | 90 | 814 | 315 | 3338 | |
| Detection of leaves | 40 | 362 | 542 | 1521 | 3810 |
| 30 | 272 | 632 | 1143 | 3810 | |
| 20 | 181 | 723 | 761 | 3810 | |
| 10 | 90 | 814 | 378 | 3810 | |
| Model | Batch-Size | Learning Rate | Weight Decay | Momentum | Epoch |
|---|---|---|---|---|---|
| SOLOv2 | 16 | 0.02 | 0.0001 | 0.9 | 100 |
| YOLOv5x | 16 | 0.01 | 0.0005 | 0.937 | 300 |
| Model | Backbone | Mask AP | Time (s/img) | ||
|---|---|---|---|---|---|
| mAP0.50 (%) | mAP0.75 (%) | mAP0.5:0.95 (%) | |||
| SOLOv2 | Resnet 50 | 91.6 | 81.6 | 63.1 | 0.15 |
| Resnet 101 | 94.2 | 85.1 | 65.6 | 0.15 | |
| Labeled Proportion | Round | mAP0.5(%) | mAP0.75(%) | mAP0.5:0.95(%) | Time(s/img) |
|---|---|---|---|---|---|
| 100% | - | 94.2 | 85.1 | 65.6 | 0.15 |
| 40% | Round-1 | 91.3 | 84.1 | 62.6 | 0.15 |
| Round-2 | 91.9 | 76.9 | 59.2 | 0.15 | |
| Round-3 | 88.4 | 65.8 | 53.4 | 0.15 | |
| 30% | Round-1 | 93.6 | 76.0 | 60.4 | 0.16 |
| Round-2 | 92.1 | 70.4 | 56.7 | 0.17 | |
| Round-3 | 91.0 | 65.2 | 55.3 | 0.16 | |
| 20% | Round-1 | 89.6 | 65.7 | 55.0 | 0.16 |
| Round-2 | 86.5 | 58.5 | 49.5 | 0.23 | |
| Round-3 | 86.3 | 53.4 | 48.2 | 0.16 | |
| 10% | Round-1 | 88.2 | 60.4 | 51.9 | 0.17 |
| Round-2 | 85.9 | 48.0 | 47.0 | 0.16 | |
| Round-3 | 79.0 | 35.9 | 40.1 | 0.17 |
| Labeled Proportion | Round | Precision (%) | Recall (%) | AP (%) | mAP (%) | |||
|---|---|---|---|---|---|---|---|---|
| Fully Unfolded leaf | Newly Appearing Leaf | Fully Unfolded Leaf | Newly Appearing Leaf | Fully Unfolded Leaf | Newly Appearing Leaf | |||
| 100% | — | 92.0 | 68.8 | 84.4 | 50.0 | 89.6 | 54.0 | 71.8 |
| 40% | Round-1 | 87.8 | 57.0 | 84.5 | 46.7 | 88.3 | 47.7 | 68.0 |
| Round-2 | 90.4 | 58.0 | 86.0 | 47.5 | 90.1 | 48.7 | 69.4 | |
| Round-3 | 89.1 | 57.5 | 87.2 | 56.6 | 89.6 | 57.4 | 73.5 | |
| 30% | Round-1 | 88.1 | 53.6 | 83.5 | 48.4 | 87.3 | 47.8 | 67.5 |
| Round-2 | 88.1 | 56.3 | 83.9 | 47.5 | 87.7 | 49.4 | 68.5 | |
| Round-3 | 87.8 | 54.4 | 84.6 | 45.1 | 88.6 | 44.8 | 66.7 | |
| 20% | Round-1 | 88.8 | 50.9 | 83.7 | 41.0 | 88.2 | 43.1 | 65.7 |
| Round-2 | 88.4 | 50.0 | 85.4 | 43.4 | 88.4 | 45.2 | 66.8 | |
| Round-3 | 86.8 | 50.0 | 85.4 | 43.4 | 87.4 | 42.1 | 64.7 | |
| 10% | Round-1 | 90.3 | 62.2 | 78.7 | 47.3 | 85.5 | 51.4 | 68.8 |
| Round-2 | 87.9 | 51.1 | 82.2 | 47.5 | 86.7 | 45.3 | 66.0 | |
| Round-3 | 87.9 | 51.1 | 81.4 | 46.7 | 85.7 | 42.5 | 64.1 | |
| Labeled Proportion | Differential Value | Accuracy Rate | |||||
|---|---|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | 2 | 3 | ||
| 100% | 2 | 16 | 124 | 28 | - | - | 72.9% |
| 40% | 1 | 23 | 118 | 27 | 1 | - | 69.4% |
| 30% | 3 | 18 | 116 | 28 | 5 | - | 68.2% |
| 20% | 3 | 29 | 106 | 28 | 2 | 1 | 62.4% |
| 10% | 3 | 22 | 102 | 39 | 4 | - | 60.0% |
| Labeled Proportion | Differential Value | Accuracy Rate | |||
|---|---|---|---|---|---|
| −2 | −1 | 0 | 1 | ||
| 100% | - | 11 | 128 | 31 | 75.3% |
| 40% | - | 13 | 124 | 33 | 72.9% |
| 30% | - | 8 | 120 | 42 | 70.6% |
| 20% | 1 | 15 | 123 | 31 | 72.4% |
| 10% | - | 16 | 119 | 35 | 70.0% |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xu, X.; Wang, L.; Liang, X.; Zhou, L.; Chen, Y.; Feng, P.; Yu, H.; Ma, Y. Maize Seedling Leave Counting Based on Semi-Supervised Learning and UAV RGB Images. Sustainability 2023, 15, 9583. https://doi.org/10.3390/su15129583
Xu X, Wang L, Liang X, Zhou L, Chen Y, Feng P, Yu H, Ma Y. Maize Seedling Leave Counting Based on Semi-Supervised Learning and UAV RGB Images. Sustainability. 2023; 15(12):9583. https://doi.org/10.3390/su15129583
Chicago/Turabian StyleXu, Xingmei, Lu Wang, Xuewen Liang, Lei Zhou, Youjia Chen, Puyu Feng, Helong Yu, and Yuntao Ma. 2023. "Maize Seedling Leave Counting Based on Semi-Supervised Learning and UAV RGB Images" Sustainability 15, no. 12: 9583. https://doi.org/10.3390/su15129583
APA StyleXu, X., Wang, L., Liang, X., Zhou, L., Chen, Y., Feng, P., Yu, H., & Ma, Y. (2023). Maize Seedling Leave Counting Based on Semi-Supervised Learning and UAV RGB Images. Sustainability, 15(12), 9583. https://doi.org/10.3390/su15129583








