Metabolomics for Plant Health Biosecurity Diagnostics and Response
Abstract
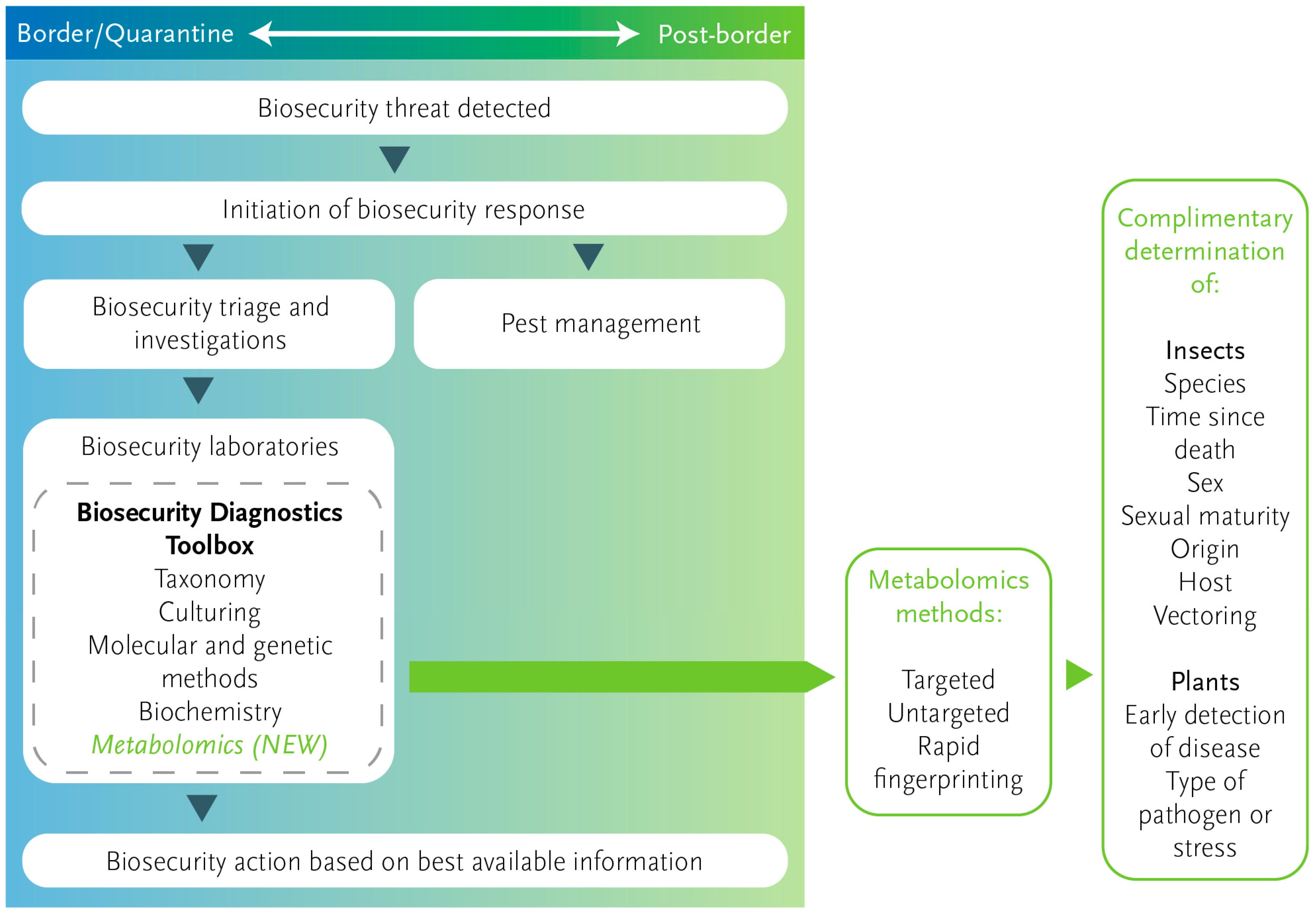
:1. Introduction
The Evolving Role of Diagnostics for Biosecurity
2. Metabolomics
2.1. An Overview of Metabolomics
2.2. Metabolomics Methodologies
3. Exploring Metabolomics for Plant Health Biosecurity
3.1. Metabolomics Applied to Insect Traits Relevant to Biosecurity
3.1.1. Entometabolomics
3.1.2. Species Identification
3.1.3. Sex Differentiation
3.1.4. Age and Sexual Maturity
3.1.5. Time since Death and Storage
3.1.6. Geographical Origin
3.1.7. Plant Host
3.1.8. Insecticide Resistance
3.1.9. Insects as Vectors of Plant Pathogens
3.2. Metabolomics Applied to the Biosecurity Detection of Plant Pathogens
3.3. Other Potential Biosecurity Applications
3.4. Translational Metabolomics for Biosecurity: Direct Analysis Mass Spectrometry
3.5. The Interface between Diagnostics and Underpinning Metabolomics Science
4. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Abbreviations and Glossary
References
- Waage, J.K.; Mumford, J.D. Agricultural biosecurity. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2008, 363, 863–876. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Armstrong, K.; Ball, S. DNA barcodes for biosecurity: Invasive species identification. Philos. Trans. R. Soc. B Biol. Sci. 2005, 360, 1813–1823. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Olmos, A.; Boonham, N.; Candresse, T.; Gentit, P.; Giovani, B.; Kutnjak, D.; Liefting, L.; Maree, H.J.; Minafra, A.; Moreira, A.; et al. High-throughput sequencing technologies for plant pest diagnosis: Challenges and opportunities. EPPO Bull. 2018, 48, 219–224. [Google Scholar] [CrossRef] [Green Version]
- Boykin, L.M.; Sseruwagi, P.; Alicai, T.; Ateka, E.; Mohammed, I.U.; Stanton, J.-A.L.; Kayuki, C.; Mark, D.; Fute, T.; Erasto, J.; et al. Tree Lab: Portable Genomics for Early Detection of Plant Viruses and Pests in Sub-Saharan Africa. Genes 2019, 10, 632. [Google Scholar] [CrossRef] [Green Version]
- Cleary, M.; Oskay, F.; Doğmuş, H.; Lehtijärvi, A.; Woodward, S.; Vettraino, A.M. Cryptic Risks to Forest Biosecurity Associated with the Global Movement of Commercial Seed. Forests 2019, 10, 459. [Google Scholar] [CrossRef] [Green Version]
- Congrains, C.; Zucchi, R.A.; de Brito, R.A. Phylogenomic approach reveals strong signatures of introgression in the rapid diversification of neotropical true fruit flies (Anastrepha: Tephritidae). Mol. Phylogenet. Evol. 2021, 162, 107200. [Google Scholar] [CrossRef]
- Holder, P.W.; Van Hale, R.; Frew, R.; George, S.; Armstrong, K.F. Natal origin of the invasive biosecurity pest, brown marmorated stink bug (Halyomorpha halys: Penatomidae), determined by dual-element stable isotope-ratio mass spectrometry. Pest Manag. Sci. 2020, 76, 1456–1463. [Google Scholar] [CrossRef] [Green Version]
- Reese, K.L.; Rasley, A.; Avila, J.R.; Jones, A.D.; Frank, M. Metabolic Profiling of Volatile Organic Compounds (VOCs) Emitted by the Pathogens Francisella tularensis and Bacillus anthracis in Liquid Culture. Sci. Rep. 2020, 10, 9333. [Google Scholar] [CrossRef]
- Trifonova, O.P.; Maslov, D.; Balashova, E.; Lokhov, P.G. Mass spectrometry-based metabolomics diagnostics-myth or reality? Expert Rev. Proteom. 2021, 18, 7–12. [Google Scholar] [CrossRef]
- Blakebrough-Hall, C.; Dona, A.; D’Occhio, M.J.; McMeniman, J.; A González, L. Diagnosis of Bovine Respiratory Disease in feedlot cattle using blood 1H NMR metabolomics. Sci. Rep. 2020, 10, 115. [Google Scholar] [CrossRef] [Green Version]
- Castro-Moretti, F.R.; Gentzel, I.N.; Mackey, D.; Alonso, A.P. Metabolomics as an Emerging Tool for the Study of Plant–Pathogen Interactions. Metabolites 2020, 10, 52. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Villas-Bôas, S.G.; Rasmussen, S.; Lane, G.A. Metabolomics or metabolite profiles? Trends Biotechnol. 2005, 23, 385–386. [Google Scholar] [CrossRef] [PubMed]
- Fiehn, O. Metabolomics—The link between genotypes and phenotypes. Plant Mol. Biol. 2002, 48, 155–171. [Google Scholar] [CrossRef]
- Wishart, D.S.; Guo, A.; Oler, E.; Wang, F.; Anjum, A.; Peters, H.; Dizon, R.; Sayeeda, Z.; Tian, S.; Lee, B.L.; et al. HMDB 5.0: The Human Metabolome Database for 2022. Nucleic Acids Res. 2021, 50, D622–D631. [Google Scholar] [CrossRef]
- Orešič, M.; McGlinchey, A.; Wheelock, C.E.; Hyötyläinen, T. Metabolic Signatures of the Exposome—Quantifying the Impact of Exposure to Environmental Chemicals on Human Health. Metabolites 2020, 10, 454. [Google Scholar] [CrossRef]
- Soltis, N.E.; Kliebenstein, D.J. Natural Variation of Plant Metabolism: Genetic Mechanisms, Interpretive Caveats, and Evolutionary and Mechanistic Insights. Plant Physiol. 2015, 169, 1456–1468. [Google Scholar] [CrossRef] [Green Version]
- Chong, J.; Yamamoto, M.; Xia, J. MetaboAnalystR 2.0: From raw spectra to biological insights. Metabolites 2019, 9, 57. [Google Scholar] [CrossRef] [Green Version]
- Draper, S.L.; McCarney, E.R. Benchtop nuclear magnetic resonance spectroscopy in forensic chemistry. Org. Magn. Reson. 2021, 61, 106–129. [Google Scholar] [CrossRef] [PubMed]
- Barding, G.A., Jr.; Béni, S.; Fukao, T.; Bailey-Serres, J.; Larive, C.K. Comparison of GC-MS and NMR for Metabolite Profiling of Rice Subjected to Submergence Stress. J. Proteome Res. 2013, 12, 898–909. [Google Scholar] [CrossRef] [PubMed]
- Balog, J.; Szaniszlo, T.; Schaefer, K.-C.; Denes, J.; Lopata, A.; Godorhazy, L.; Szalay, D.; Balogh, L.; Sasi-Szabo, L.; Toth, M.; et al. Identification of Biological Tissues by Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 2010, 82, 7343–7350. [Google Scholar] [CrossRef]
- Balog, J.; Sasi-Szabó, L.; Kinross, J.; Lewis, M.; Muirhead, L.; Veselkov, K.; Mirnezami, R.; Dezső, B.; Damjanovich, L.; Darzi, A.; et al. Intraoperative tissue identification using rapid evaporative ionization mass spectrometry. Sci. Transl. Med. 2013, 5, 194ra193. [Google Scholar] [CrossRef] [PubMed]
- Beyramysoltan, S.; Giffen, J.; Rosati, J.; Musah, R.A. Direct Analysis in Real Time-Mass Spectrometry and Ko-honen Artificial Neural Networks for Species Identification of Larva, Pupa and Adult Life Stages of Carrion Insects. Anal. Chem. 2018, 90, 9206–9217. [Google Scholar] [CrossRef] [PubMed]
- Ross, A.; Brunius, C.; Chevallier, O.; Dervilly, G.; Elliott, C.; Guitton, Y.; Prenni, J.E.; Savolainen, O.; Hemeryck, L.; Vidkjær, N.H.; et al. Making complex measurements of meat composition fast: Application of rapid evaporative ionisation mass spectrometry to measuring meat quality and fraud. Meat Sci. 2020, 181, 108333. [Google Scholar] [CrossRef]
- Snart, C.J.; Hardy, I.C.; Barrett, D.A. Entometabolomics: Applications of modern analytical techniques to insect studies. Èntomol. Exp. Et Appl. 2015, 155, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Phillips, C.B.; Iline, I.I.; Richards, N.K.; Novoselov, M.; McNeill, M. Development and validation of a quick easily used biochemical assay for evaluating the viability of small immobile arthropods. J. Econ. Èntomol. 2013, 106, 2006–2019. [Google Scholar] [CrossRef] [Green Version]
- Holze, H.; Schrader, L.; Buellesbach, J. Advances in deciphering the genetic basis of insect cuticular hydrocarbon biosynthesis and variation. Heredity 2020, 126, 219–234. [Google Scholar] [CrossRef]
- Kather, R.; Martin, S.J. Cuticular hydrocarbon profiles as a taxonomic tool: Advantages, limitations and technical aspects. Physiol. Èntomol. 2012, 37, 25–32. [Google Scholar] [CrossRef]
- Wang, Q.; Goodger, J.; Woodrow, I.; Elgar, M.A. Location-specific cuticular hydrocarbon signals in a social insect. Proc. Biol. Sci. 2016, 283, 20160310. [Google Scholar] [CrossRef] [Green Version]
- Würf, J.; Pokorny, T.; Wittbrodt, J.; Millar, J.G.; Ruther, J. Cuticular Hydrocarbons as Contact Sex Pheromone in the Parasitoid Wasp Urolepis rufipes. Front. Ecol. Evol. 2020, 8, 180. [Google Scholar] [CrossRef]
- Bien, T.; Gadau, J.; Schnapp, A.; Yew, J.Y.; Sievert, C.; Dreisewerd, K. Detection of very long-chain hydrocarbons by laser mass spectrometry reveals novel species-, sex-, and age-dependent differences in the cuticular profiles of three Nasonia species. Anal. Bioanal. Chem. 2019, 411, 2981–2993. [Google Scholar] [CrossRef]
- Souza, N.M.; Schröder, M.; Hayes, R.; Bello, J.; Nahrung, H.F. Cuticular hydrocarbons of Gonipterus weevils: Are there species differences? Chemoecology 2021, 31, 159–167. [Google Scholar] [CrossRef]
- Aliferis, K.A.; Copley, T.; Jabaji, S. Gas chromatography–mass spectrometry metabolite profiling of worker honey bee (Apis mellifera L.) hemolymph for the study of Nosema ceranae infection. J. Insect Physiol. 2012, 58, 1349–1359. [Google Scholar] [CrossRef] [PubMed]
- Zhou, L.; Li, H.; Hao, F.; Li, N.; Liu, X.; Wang, G.; Wang, Y.; Tang, H. Developmental Changes for the Hemolymph Metabolome of Silkworm (Bombyx mori L.). J. Proteome Res. 2015, 14, 2331–2347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Borsuk, G.; Ptaszyńska, A.A.; Olszewski, K.; Domaciuk, M.; Krutmuang, P.; Paleolog, J. A New Method for Quick and Easy Hemolymph Collection from Apidae Adults. PLoS ONE 2017, 12, e0170487. [Google Scholar] [CrossRef] [PubMed]
- Bozic, J.; Di Cesare, J.; Wells, H.; Abramson, C.I. Ethanol levels in honeybee hemolymph resulting from alcohol in-gestion. Alcohol 2007, 41, 281–284. [Google Scholar] [CrossRef]
- Stevens, V.L.; Hoover, E.; Wang, Y.; Zanetti, K.A. Pre-Analytical Factors that Affect Metabolite Stability in Human Urine, Plasma, and Serum: A Review. Metabolites 2019, 9, 156. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- González-Domínguez, R.; González-Domínguez, Á.; Sayago, A.; Fernández-Recamales, Á. Recommendations and Best Practices for Standardizing the Pre-Analytical Processing of Blood and Urine Samples in Metabolomics. Metabolites 2020, 10, 229. [Google Scholar] [CrossRef]
- Hulme, P.E. One Biosecurity: A unified concept to integrate human, animal, plant, and environmental health. Emerg. Top. Life Sci. 2020, 4, 539–549. [Google Scholar] [CrossRef]
- Vaníčková, L.; Virgilio, M.; Tomčala, A.; Břízová, R.; Ekesi, S.; Hoskovec, M.; Kalinová, B.; Nascimento, R.D.; De Meyer, M. Resolution of three cryptic agricultural pests (Ceratitis fasciventris, C. anonae, C. rosa, Diptera: Tephritidae) using cuticular hydrocarbon profiling. Bull. Entomol. Res. 2014, 104, 631–638. [Google Scholar] [CrossRef]
- De Meyer, M.; Delatte, H.; Ekesi, S.; Jordaens, K.; Kalinova, B.; Manrakhan, A.; Mwatawala, M.; Steck, G.; Van Cann, J.; Vancikova, L.; et al. An integrative approach to unravel the Ceratitis FAR (Diptera, Tephritidae) cryptic species complex: A review. Zookeys 2015, 540, 405–427. [Google Scholar] [CrossRef] [Green Version]
- Copren, K.A.; Nelson, L.J.; Vargo, E.L.; Haverty, M.I. Phylogenetic analyses of mtDNA sequences corroborate taxonomic designations based on cuticular hydrocarbons in subterranean termites. Mol. Phylogenet. Evol. 2005, 35, 689–700. [Google Scholar] [CrossRef] [PubMed]
- Moore, H.E.; Hall, M.; Drijfhout, F.; Cody, R.; Whitmore, D. Cuticular hydrocarbons for identifying Sar-cophagidae (Diptera). Sci. Rep. 2021, 11, 7732. [Google Scholar] [CrossRef]
- Wagner, I.; Koch, N.; Sarsby, J.; White, N.; Price, T.; Jones, S.; Hurst, J.; Beynon, R.J. The application of rapid evaporative ionization mass spectrometry in the analysis of Drosophila species—A potential new tool in ento-mology. Open Biol. 2020, 10, 200196. [Google Scholar] [CrossRef]
- Chabi, J.; Van’t Hof, A.; N’Dri, L.K.; Datsomor, A.; Okyere, D.; Njoroge, H.; Pipini, D.; Hadi, M.P.; de Souza, D.K.; Suzuki, T.; et al. Rapid high throughput SYBR green assay for identifying the malaria vectors Anopheles ara-biensis, Anopheles coluzzii and Anopheles gambiae s.s. Giles. PLoS ONE 2019, 14, e0215669. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wagner, I. Exploration of Rapid Evaporative Ionisation Mass Spectrometry as a Novel Tool for Insect Identification and Characterisation. Ph.D. Thesis, University of Liverpool, Liverpool, UK, 2021. [Google Scholar]
- Cameron, S.J.S.; Alexander, J.L.; Bolt, F.; Burke, A.; Ashrafian, H.; Teare, J.P.; Marchesi, J.R.; Kinross, J.M.; Li, J.V.; Takáts, Z. Evaluation of Direct from Sample Metabolomics of Human Feces Using Rapid Evaporative Ionization Mass Spectrometry. Anal. Chem. 2019, 91, 13448–13457. [Google Scholar] [CrossRef]
- Otte, T.; Hilker, M.; Geiselhardt, S. Phenotypic Plasticity of Cuticular Hydrocarbon Profiles in Insects. J. Chem. Ecol. 2018, 44, 235–247. [Google Scholar] [CrossRef]
- Kuo, T.-H.; Yew, J.; Fedina, T.; Dreisewerd, K.; Dierick, H.; Pletcher, S.D. Aging modulates cuticular hydro-carbons and sexual attractiveness in Drosophila melanogaster. J. Exp. Biol. 2012, 215 Pt 5, 814–821. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kapranas, A.; Snart, C.J.P.; Williams, H.; Hardy, I.C.W.; Barrett, D.A. Metabolomics of aging assessed in individual parasitoid wasps. Sci. Rep. 2016, 6, 34848. [Google Scholar] [CrossRef] [Green Version]
- Dawidowska, J.; Krzyżanowska, M.; Markuszewski, M.; Kaliszan, M. The Application of Metabolomics in Forensic Science with Focus on Forensic Toxicology and Time-of-Death Estimation. Metabolites 2021, 11, 801. [Google Scholar] [CrossRef]
- Pesko, B.K.; Weidt, S.; McLaughlin, M.; Wescott, D.J.; Torrance, H.; Burgess, K.; Burchmore, R. Postmortomics: The Potential of Untargeted Metabolomics to Highlight Markers for Time Since Death. OMICS A J. Integr. Biol. 2020, 24, 649–659. [Google Scholar] [CrossRef]
- Bonadio, R.S.; Nunes, L.B.; Moretti, P.N.S.; Mazzeu, J.F.; Cagnin, S.; Pic-Taylor, A.; de Oliveira, S.F. Insights into how environment shapes post-mortem RNA transcription in mouse brain. Sci. Rep. 2021, 11, 13008. [Google Scholar] [CrossRef]
- Barr, N.; Ruiz-Arce, R.; Armstrong, K. Using Molecules to Identify the Source of Fruit Fly Invasions. Trapping and the Detection, Control, and Regulation of Tephritid Fruit Flies: Lures, Area-Wide Programs, and Trade Implications; Shelly, T., Epsky, N., Jang, E.B., Reyes-Flores, J., Vargas, R., Eds.; Springer: Dordrecht, The Netherlands, 2014; pp. 321–378. [Google Scholar] [CrossRef]
- Holder, P.W.; Armstrong, K.; Van Hale, R.; Millet, M.-A.; Frew, R.; Clough, T.; Baker, J. Isotopes and Trace Elements as Natal Origin Markers of Helicoverpa armigera—An Experimental Model for Biosecurity Pests. PLoS ONE 2014, 9, e92384. [Google Scholar] [CrossRef] [Green Version]
- Riach, A.C.; Perera, M.V.L.; Florance, H.V.; Robinson, L.A.; Penfield, S.D.; Hill, J.K. Metabolic fingerprints reveal how an insect metabolome is affected by different larval host plant species. Arthropod-Plant Interact. 2019, 13, 571–579. [Google Scholar] [CrossRef]
- Papantoniou, D.; Vergara, F.; Weinhold, A.; Quijano, T.; Khakimov, B.; Pattison, D.; Bak, S.; van Dam, N.; Mar-tínez-Medina, A. Cascading Effects of Root Microbial Symbiosis on the Development and Metabolome of the Insect Her-bivore Manduca sexta L. Metabolites 2021, 11, 731. [Google Scholar] [CrossRef]
- Candas, M.; Loseva, O.; Oppert, B.; Kosaraju, P.; Bulla, L.A., Jr. Insect Resistance to Bacillus thuringiensis: Alterations in the Indianmeal Moth Larval Gut Proteome. Mol. Cell. Proteom. 2003, 2, 19–28. [Google Scholar] [CrossRef] [Green Version]
- Abougamos, H.R.; Sadler, R.; White, B. Managing evolving insecticide resistance in stored grain pests within Avon Region, Western Australia. J. Stored Prod. Postharvest Res. 2017, 8, 16–30. [Google Scholar]
- Bass, C.; Denholm, I.; Williamson, M.S.; Nauen, R. The global status of insect resistance to neonicotinoid insecticides. Pestic. Biochem. Physiol. 2015, 121, 78–87. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, J.; Wang, H.; Liu, S.; Liu, L.; Tay, W.T.; Walsh, T.K.; Yang, Y.; Wu, Y. CRISPR/Cas9 mediated genome editing of Helicoverpa armigera with mutations of an ABC transporter gene HaABCA2 confers resistance to Bacillus thuringiensis Cry2A toxins. Insect Biochem. Mol. Biol. 2017, 87, 147–153. [Google Scholar] [CrossRef] [PubMed]
- Van Leeuwen, T.; Dermauw, W.; Mavridis, K.; Vontas, J. Significance and interpretation of molecular diagnostics for insecticide resistance management of agricultural pests. Curr. Opin. Insect Sci. 2020, 39, 69–76. [Google Scholar] [CrossRef] [PubMed]
- Li, Y.; Li, Y.; Wang, G.; Li, J.; Zhang, M.; Wu, J.; Liang, C.; Zhou, H.; Tang, J.; Zhu, G. Differential metabolome responses to deltamethrin between resistant and susceptible Anopheles sinensis. Ecotoxicol. Environ. Saf. 2022, 237, 113553. [Google Scholar] [CrossRef]
- Pusz-Bochenska, K.; Perez-Lopez, E.; Dumonceaux, T.; Olivier, C.; Wist, T.J. A Rapid, Simple, Laboratory and Field-Adaptable DNA Extraction and Diagnostic Method Suitable for Insect-Transmitted Plant Pathogen and Insect Identifi-cation. Plant Health Prog. 2020, 21, 63–68. [Google Scholar] [CrossRef] [Green Version]
- Killiny, N.; Jones, S.E. Metabolic alterations in the nymphal instars of Diaphorina citri induced by Candidatus Liberibacter asiaticus, the putative pathogen of huanglongbing. PLoS ONE 2018, 13, e0191871. [Google Scholar] [CrossRef] [PubMed]
- Liu, K.; He, J.; Guan, Z.; Zhong, M.; Pang, R.; Han, Q. Transcriptomic and Metabolomic Analyses of Diaphorina citri Kuwayama Infected and Non-infected With Candidatus Liberibacter Asiaticus. Front. Physiol. 2021, 11, 630037. [Google Scholar] [CrossRef] [PubMed]
- Killiny, N.; Nehela, Y.; Hijaz, F.; Vincent, C.I. A plant pathogenic bacterium exploits the tricarboxylic acid cycle metabolic pathway of its insect vector. Virulence 2017, 9, 99–109. [Google Scholar] [CrossRef] [Green Version]
- Smith, G.R.; Fletcher, J.; Marroni, V.; Kean, J.; Stringer, L.; Vereijssen, J. Plant pathogen eradication: Determi-nants of successful programs. Australas. Plant Pathol. 2017, 46, 277–284. [Google Scholar] [CrossRef]
- Adeniji, A.A.; Babalola, O.O.; Loots, D.T. Metabolomic applications for understanding complex tripartite plant-microbes interactions: Strategies and perspectives. Biotechnol. Rep. 2020, 25, e00425. [Google Scholar] [CrossRef]
- Vo, K.T.X.; Rahman, M.; Rahman, M.; Trinh, K.T.T.; Kim, S.T.; Jeon, J.-S. Proteomics and Metabolomics Studies on the Biotic Stress Responses of Rice: An Update. Rice 2021, 14, 30. [Google Scholar] [CrossRef]
- Gupta, S.; Schillaci, M.; Roessner, U. Metabolomics as an emerging tool to study plant–microbe interactions. Emerg. Top. Life Sci. 2022, 6, 175–183. [Google Scholar] [CrossRef]
- De Torres-Zabala, M.; Truman, W.; Bennett, M.H.; Lafforgue, G.; Mansfield, J.W.; Egea, P.R.; Bögre, L.; Grant, M. Pseudomonas syringae pv. tomato hijacks the Arabidopsis abscisic acid signalling pathway to cause disease. EMBO J. 2007, 26, 1434–1443. [Google Scholar] [CrossRef] [PubMed]
- Villate, A.; Nicolas, M.S.; Gallastegi, M.; Aulas, P.-A.; Olivares, M.; Usobiaga, A.; Etxebarria, N.; Aizpurua-Olaizola, O. Review: Metabolomics as a prediction tool for plants performance under environmental stress. Plant Sci. 2020, 303, 110789. [Google Scholar] [CrossRef] [PubMed]
- Tenenboim, H.; Brotman, Y. Omic Relief for the Biotically Stressed: Metabolomics of Plant Biotic Interactions. Trends Plant Sci. 2016, 21, 781–791. [Google Scholar] [CrossRef]
- Ren, Z.; Fang, M.; Muhae-Ud-Din, G.; Gao, H.; Yang, Y.; Liu, T.; Chen, W.; Gao, L. Metabolomics analysis of grains of wheat infected and noninfected with Tilletia controversa Kühn. Sci. Rep. 2021, 11, 18876. [Google Scholar] [CrossRef]
- Dai, T.; Chang, X.; Hu, Z.; Liang, L.; Sun, M.; Liu, P.; Liu, X. Untargeted Metabolomics Based on GC-MS and Chemometrics: A New Tool for the Early Diagnosis of Strawberry Anthracnose Caused by Colletotrichum theobromicola. Plant Dis. 2019, 103, 2541–2547. [Google Scholar] [CrossRef] [PubMed]
- Hu, Z.; Chang, X.; Dai, T.; Li, L.; Liu, P.; Wang, G.; Liu, P.; Huang, Z.; Liu, X. Metabolic Profiling to Identify the Latent Infection of Strawberry by Botrytis cinerea. Evol. Bioinform. 2019, 15. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, Y.; Liu, Q.; Du, L.; Hallerman, E.; Li, Y. Transcriptomic and Metabolomic Responses of Rice Plants to Cnaphalocrocis medinalis Caterpillar Infestation. Insects 2020, 11, 705. [Google Scholar] [CrossRef] [PubMed]
- Sanchez-Arcos, C.; Kai, M.; Svatoš, A.; Gershenzon, J.; Kunert, G. Untargeted Metabolomics Approach Reveals Differences in Host Plant Chemistry before and after Infestation with Different Pea Aphid Host Races. Front. Plant Sci. 2019, 10, 188. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Drès, M.; Mallet, J. Host races in plant–feeding insects and their importance in sympatric speciation. Philos. Trans. R. Soc. B Biol. Sci. 2002, 357, 471–492. [Google Scholar] [CrossRef] [Green Version]
- Gai, Y.P.; Han, X.; Li, Y.; Yuan, C.; Mo, Y.; Guo, F.; Liu, Q.; Ji, X.L. Metabolomic analysis reveals the po-tential metabolites and pathogenesis involved in mulberry yellow dwarf disease. Plant Cell Environ. 2014, 37, 1474–1490. [Google Scholar] [CrossRef]
- Blouin, A.G.; Greenwood, D.R.; Chavan, R.R.; Pearson, M.N.; Clover, G.R.; MacDiarmid, R.M.; Cohen, D. A generic method to identify plant viruses by high-resolution tandem mass spectrometry of their coat proteins. J. Virol. Methods 2010, 163, 49–56. [Google Scholar] [CrossRef]
- Schmidt, R.; Durling, M.; de Jager, V.; Menezes, R.; Nordkvist, E.; Svatoš, A.; Dubey, M.; Lauterbach, L.; Dickschat, J.; Karlsson, M.; et al. Deciphering the genome and secondary metabolome of the plant pathogen Fusarium culmorum. FEMS Microbiol. Ecol. 2018, 94, fiy078. [Google Scholar] [CrossRef] [Green Version]
- Hall, R.D.; D’Auria, J.; Ferreira, A.S.; Gibon, Y.; Kruszka, D.; Mishra, P.; van de Zedde, R. High-throughput plant phenotyping: A role for metabolomics? Trends Plant Sci. 2022, 27, 549–563. [Google Scholar] [CrossRef]
- Simó, C.; Ibáñez, C.; Valdés, A.; Cifuentes, A.; García-Cañas, V. Metabolomics of genetically modified crops. Inter-Natl. J. Mol. Sci. 2014, 15, 18941–18966. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Bedair, M.; Glenn, K.C. Evaluation of the use of untargeted metabolomics in the safety assessment of genetically modified crops. Metabolomics 2020, 16, 111. [Google Scholar] [CrossRef]
- Fraser, P.D.; Aharoni, A.; Hall, R.; Huang, S.; Giovannoni, J.; Sonnewald, U.; Fernie, A.R. Metabolomics should be deployed in the identification and characterization of gene-edited crops. Plant J. 2020, 102, 897–902. [Google Scholar] [CrossRef] [Green Version]
- Buddenhagen, C.E.; James, T.K.; Ngow, Z.; Hackell, D.L.; Rolston, M.P.; Chynoweth, R.J.; Gunnarsson, M.; Li, F.; Harrington, K.C.; Ghanizadeh, H. Resistance to post-emergent herbicides is becoming common for grass weeds on New Zealand wheat and barley farms. PLoS ONE 2021, 16, e0258685. [Google Scholar] [CrossRef]
- Pomyen, Y.; Wanichthanarak, K.; Poungsombat, P.; Fahrmann, J.; Grapov, D.; Khoomrung, S. Deep metabolome: Applications of deep learning in metabolomics. Comput. Struct. Biotechnol. J. 2020, 18, 2818–2825. [Google Scholar] [CrossRef] [PubMed]
- Pinu, F.R.; Goldansaz, S.A.; Jaine, J. Translational Metabolomics: Current Challenges and Future Opportunities. Metabolites 2019, 9, 108. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Davidson, N.B.; Koch, N.; Sarsby, J.; Jones, E.; Hurst, J.L.; Beynon, R.J. Rapid identification of species, sex and maturity by mass spectrometric analysis of animal faeces. BMC Biol. 2019, 17, 66. [Google Scholar] [CrossRef] [Green Version]
- Lesiak, A.D.; Cody, R.B.; Dane, A.J.; Musah, R.A. Plant Seed Species Identification from Chemical Fingerprints: A High-Throughput Application of Direct Analysis in Real Time Mass Spectrometry. Anal. Chem. 2015, 87, 8748–8757. [Google Scholar] [CrossRef] [PubMed]
- Subbaraj, A.K.; Barrett, B.; Wakelin, S.; Fraser, K. Using non-targeted direct analysis in real time-mass spec-trometry (DART-MS) to discriminate seeds based on endogenous or exogenous chemicals. Anal. Bioanal. Chem. 2015, 407, 8047–8058. [Google Scholar] [CrossRef]
- Musah, R.A.; Espinoza, E.; Cody, R.; Lesiak, A.; Christensen, E.; Moore, H.; Maleknia, S.; Drijfhout, F.P. A High Throughput Ambient Mass Spectrometric Approach to Species Identification and Classification from Chemical Finger-print Signatures. Sci. Rep. 2015, 5, 11520. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Beyramysoltan, S.; Ventura, M.; Rosati, J.; Giffen-Lemieux, J.; Musah, R.A. Identification of the Species Con-stituents of Maggot Populations Feeding on Decomposing Remains—Facilitation of the Determination of Post Mortem Interval and Time Since Tissue Infestation through Application of Machine Learning and Direct Analysis in Real Time-Mass Spec-trometry. Anal. Chem. 2020, 92, 5439–5446. [Google Scholar] [PubMed]
- Gekenidis, M.-T.; Studer, P.; Wüthrich, S.; Brunisholz, R.; Drissner, D. Beyond the matrix-assisted laser desorption ionization (MALDI) biotyping workflow: In search of microorganism-specific tryptic peptides enabling discrimination of sub-species. Appl. Environ. Microbiol. 2014, 80, 4234–4241. [Google Scholar] [CrossRef] [Green Version]
- Liu, Y.; Xue, A.; Ding, L.; Hao, Y.; Liu, H.; Cui, M.; Liu, L.; Nie, Z.; Luo, L. Direct identification and metabolomic analysis of Huanglongbing associated with Candidatus Liberibacter spp. in navel orange by MALDI-TOF-MS. Anal. Bioanal. Chem. 2020, 412, 3091–3101. [Google Scholar] [CrossRef] [PubMed]
- Gullion, J.D.; Gullion, T. Solid-State NMR Study of the Cicada Wing. J. Phys. Chem. B 2017, 121, 7646–7651. [Google Scholar] [CrossRef]


Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Ross, A.B.; Muller, H.; Subbaraj, A.; Homewood, I.; Mas, F.; Hardwick, S.; Stringer, L.; Vereijssen, J.; Visnovsky, S.; Najar-Rodriguez, A.; et al. Metabolomics for Plant Health Biosecurity Diagnostics and Response. Sustainability 2023, 15, 4654. https://doi.org/10.3390/su15054654
Ross AB, Muller H, Subbaraj A, Homewood I, Mas F, Hardwick S, Stringer L, Vereijssen J, Visnovsky S, Najar-Rodriguez A, et al. Metabolomics for Plant Health Biosecurity Diagnostics and Response. Sustainability. 2023; 15(5):4654. https://doi.org/10.3390/su15054654
Chicago/Turabian StyleRoss, Alastair B., Hadley Muller, Arvind Subbaraj, Ines Homewood, Flore Mas, Scott Hardwick, Lloyd Stringer, Jessica Vereijssen, Sandra Visnovsky, Adriana Najar-Rodriguez, and et al. 2023. "Metabolomics for Plant Health Biosecurity Diagnostics and Response" Sustainability 15, no. 5: 4654. https://doi.org/10.3390/su15054654




