Physiological Characterization of Tripidium arundinaceum and Sugarcane (Saccharum spp.) Germplasm for Salinity Stress Tolerance at the Formative Stage
Abstract
:1. Introduction
2. Materials and Methods
2.1. Plant Material and Treatments
2.2. Screening of T. arundinaceum Accessions at the Germination Stage
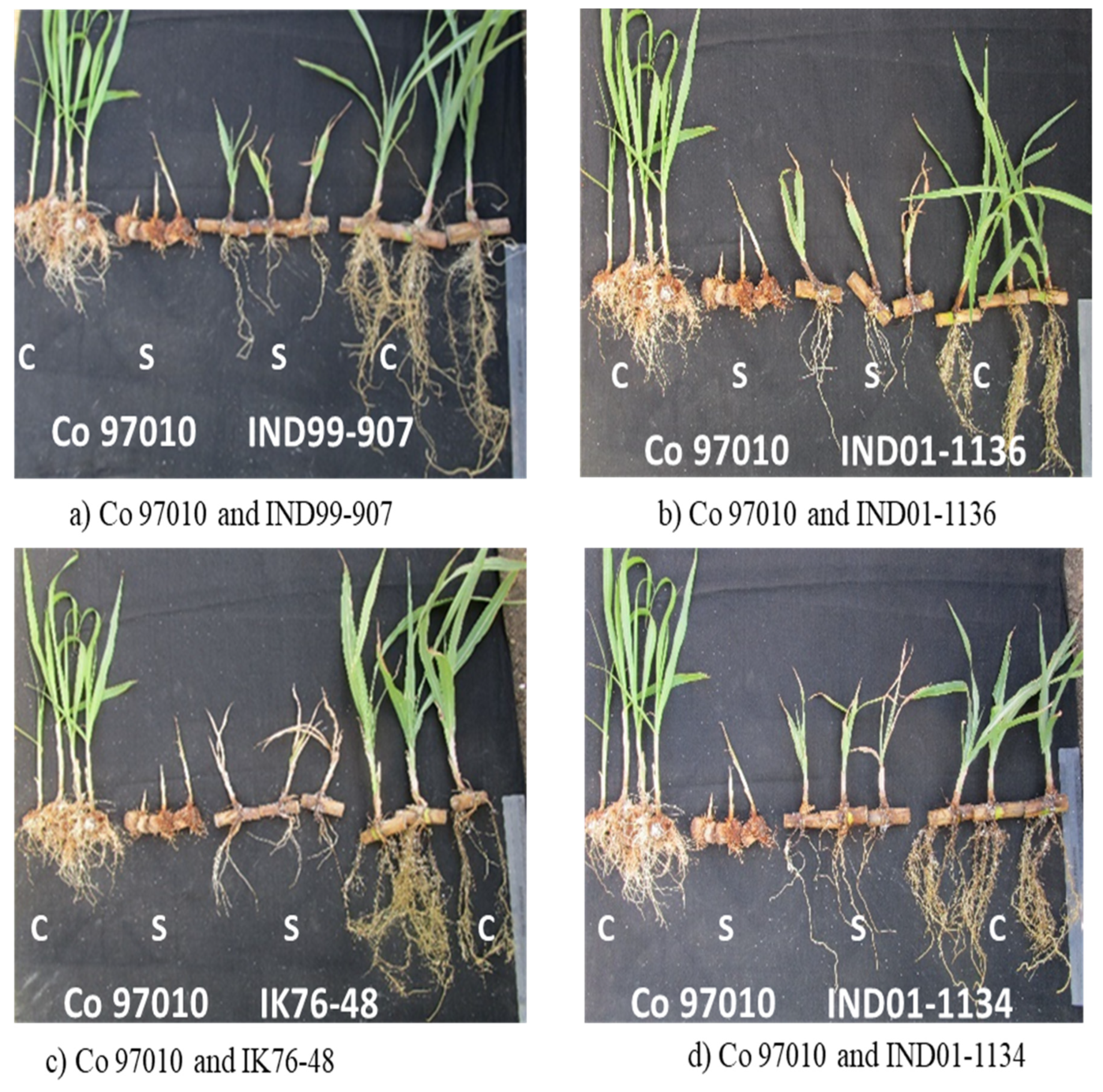
2.3. Assessing the Salinity Tolerance Level at the Formative Stage
2.4. Root Anatomy during Various Levels of Salinity Stress
2.5. Estimation of Sodium and Potassium in Root Samples
2.6. Relative Water Content
2.7. Biochemical Assay
3. Results
3.1. Effect of Salinity Stress on the Germination of T. arundinaceum
3.2. Leaf Elongation Rate (LER) of T. arundinaceum and Sugarcane Genotypes under Salinity Stress
3.3. The Effect of Various Levels of Salinity Stress on the Na/K Ratio in Roots
3.4. Root Anatomy under Salinity Stress
3.5. Chlorophyll a, Chlorphyll b, and Carotenoids Contents in Leaves
3.6. Relative Water Content (RWC) in Leaves under Salinity Stress
3.7. The Effect of Various Levels of Salinity Stress on the Accumulation of Proline
3.8. Accumulation of Total Protein under Various Levels of Salinity Stress
3.9. Superoxide Dismutase Activities under Various Levels of Salinity Stress
3.10. Peroxidase Activity under Various Levels of Salinity Stress in Roots
3.11. Lipid Peroxidation in Roots
3.12. Changes in Na/K Ratio under Salinity Stress Studied Using ICP-OES
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Acknowledgments
Conflicts of Interest
References
- Ladeiro, B. Saline Agriculture in the 21st Century: Using Salt Contaminated Resources to Cope Food Requirements. J. Bot. 2012, 2012, 310705. [Google Scholar] [CrossRef]
- Latef, A.A.H.A.; Omer, A.M.; Badawy, A.A.; Osman, M.S.; Ragaey, M.M. Strategy of Salt Tolerance and Interactive Impact of Azotobacter Chroococcum and/or Alcaligenes Faecalis Inoculation on Canola (Brassica napus L.) Plants Grown in Saline Soil. Plants 2021, 10, 110. [Google Scholar] [CrossRef] [PubMed]
- Omer, A.M.; Osman, M.S.; Badawy, A.A. Inoculation with Azospirillum Brasilense and/or Pseudomonas Geniculata Reinforces Flax (Linum usitatissimum) Growth by Improving Physiological Activities under Saline Soil Conditions. Bot. Stud. 2022, 63, 15. [Google Scholar] [CrossRef] [PubMed]
- Mahajan, S.; Tuteja, N. Cold, Salinity and Drought Stresses: An Overview. Arch. Biochem. Biophys. 2005, 444, 139–158. [Google Scholar] [CrossRef]
- Tuteja, N. Mechanisms of High Salinity Tolerance in Plants. Methods Enzymol. 2007, 428, 419–438. [Google Scholar] [CrossRef]
- Li, C.; Lei, J.; Zhao, Y.; Xu, X.; Li, S. Effect of Saline Water Irrigation on Soil Development and Plant Growth in the Taklimakan Desert Highway Shelterbelt. Soil Tillage Res. 2015, 146, 99–107. [Google Scholar] [CrossRef]
- IISR. ICAR-Indian Institute of Sugarcane Research Vision 2050; Indian Council of Agriculture Research: New Delhi, India, 2015. Available online: https://iisr.icar.gov.in/iisr/download/publications/iisrvision2050.pdf (accessed on 19 March 2023).
- Jain, R. India’s Green Push for Second-Generation Bioethanol. Available online: https://www.im4change.org/latest-news-updates/india-s-green-push-for-second-generation-bioethanol-rahul-jain.html (accessed on 19 March 2023).
- Sun, B.; Liu, G.L.; Phan, T.T.; Yang, L.T.; Li, Y.R.; Xing, Y.X. Effects of Cold Stress on Root Growth and Physiological Metabolisms in Seedlings of Different Sugarcane Varieties. Sugar Tech. 2017, 19, 165–175. [Google Scholar] [CrossRef]
- FAOSTAT. Crops and Livestock Products, Food and Agriculture Organization of the United Nations. Available online: https://fenix.fao.org/faostat/internal/en/#data/QCL (accessed on 19 March 2023).
- Wahid, A. Analysis of Toxic and Osmotic Effects of Sodium Chloride on Leaf Growth and Economic Yield of Sugarcane. Bot. Bull. Acad. Sin. 2004, 45, 133–141. [Google Scholar]
- Saxena, P.; Srivastava, R.P.; Sharma, M.L. Studies on Salinity Stress Tolerance in Sugarcane Varieties. Sugar Tech. 2010, 12, 59–63. [Google Scholar] [CrossRef]
- Wahid, A.; Rao, A.-U.-R.; Rasul, E. Field Crops Research Identification of Salt Tolerance Traits in Sugarcane Lines. Field Crop. Res. 1997, 54, 9–17. [Google Scholar] [CrossRef]
- Mangrauthia, S.K.; Bhogireddy, S.; Agarwal, S.; Prasanth, V.V.; Voleti, S.R.; Neelamraju, S.; Subrahmanyam, D. Genome-Wide Changes in MicroRNA Expression during Short and Prolonged Heat Stress and Recovery in Contrasting Rice Cultivars. J. Exp. Bot. 2017, 68, 2399–2412. [Google Scholar] [CrossRef] [PubMed]
- Shalata, A.; Tal, M. The Effect of Salt Stress on Lipid Peroxidation and Antioxidants in the Leaf of the Cultivated Tomato and Its Wild Salt-Tolerant Relative Lycopersicon Pennellii. Physiol. Plant. 1998, 104, 169–174. [Google Scholar] [CrossRef]
- Bor, M.; Özdemir, F.; Türkan, I. The Effect of Salt Stress on Lipid Peroxidation and Antioxidants in Leaves of Sugar Beet Beta Vulgaris L. and Wild Beet Beta Maritima L. Plant Sci. 2003, 164, 77–84. [Google Scholar] [CrossRef]
- Sekmen, A.H.; Türkan, I.; Takio, S. Differential Responses of Antioxidative Enzymes and Lipid Peroxidation to Salt Stress in Salt-Tolerant Plantago Maritima and Salt-Sensitive Plantago Media. Physiol. Plant. 2007, 131, 399–411. [Google Scholar] [CrossRef] [PubMed]
- Shabala, S.; Pottosin, I. Regulation of Potassium Transport in Plants under Hostile Conditions: Implications for Abiotic and Biotic Stress Tolerance. Physiol. Plant. 2014, 151, 257–279. [Google Scholar] [CrossRef] [PubMed]
- Gupta, B.; Huang, B. Mechanism of Salinity Tolerance in Plants: Physiological, Biochemical, and Molecular Characterization. Int. J. Genom. 2014, 2014, 701596. [Google Scholar] [CrossRef]
- Ji, H.; Pardo, J.M.; Batelli, G.; Van Oosten, M.J.; Bressan, R.A.; Li, X. The Salt Overly Sensitive (SOS) Pathway: Established and Emerging Roles. Mol. Plant 2013, 6, 275–286. [Google Scholar] [CrossRef]
- Welker, C.A.D.; McKain, M.R.; Vorontsova, M.S.; Peichoto, M.C.; Kellogg, E.A. Plastome Phylogenomics of Sugarcane and Relatives Confirms the Segregation of the Genus Tripidium (Poaceae: Andropogoneae). Taxon 2019, 68, 246–267. [Google Scholar] [CrossRef]
- NCBI-Taxonomy-Browser. Taxonomy Browser-Tripidium Arundinaceum. Available online: https://www.ncbi.nlm.nih.gov/Taxonomy/Browser/wwwtax.cgi?id=50346 (accessed on 19 March 2023).
- Augustine, S.M.; Cherian, A.V.; Syamaladevi, D.P.; Subramonian, N. Erianthus Arundinaceus HSP70 (EaHSP70) Acts as a Key Regulator in the Formation of Anisotropic Interdigitation in Sugarcane (Saccharum spp. Hybrid) in Response to Drought Stress. Plant Cell Physiol. 2015, 56, 2368–2380. [Google Scholar] [CrossRef]
- Augustine, S.M.; Ashwin Narayan, J.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Introduction of Pea DNA Helicase 45 into Sugarcane (Saccharum spp. Hybrid) Enhances Cell Membrane Thermostability and Upregulation of Stress-Responsive Genes Leads to Abiotic Stress Tolerance. Mol. Biotechnol. 2015, 57, 475–488. [Google Scholar] [CrossRef]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Tuteja, N.; Subramonian, N. Overexpression of EaDREB2 and Pyramiding of EaDREB2 with the Pea DNA Helicase Gene (PDH45) Enhance Drought and Salinity Tolerance in Sugarcane (Saccharum spp. Hybrid). Plant Cell Rep. 2015, 34, 247–263. [Google Scholar] [CrossRef] [PubMed]
- Augustine, S.M.; Narayan, J.A.; Syamaladevi, D.P.; Appunu, C.; Chakravarthi, M.; Ravichandran, V.; Subramonian, N. Erianthus Arundinaceus HSP70 (EaHSP70) Overexpression Increases Drought and Salinity Tolerance in Sugarcane (Saccharum spp. Hybrid). Plant Sci. 2015, 232, 23–34. [Google Scholar] [CrossRef] [PubMed]
- Wang, W.; Li, R.; Wang, H.; Qi, B.; Jiang, X.; Zhu, Q.; Cai, D.; Tang, X.; Zhao, Q. Sweetcane (Erianthus arundinaceus) as a Native Bioenergy Crop with Environmental Remediation Potential in Southern China: A Review. GCB Bioenergy 2019, 11, 1012–1025. [Google Scholar] [CrossRef]
- Mohanan, M.V.; Pushpanathan, A.; Padmanabhan, S.; Sasikumar, T.; Jayanarayanan, A.N.; Selvarajan, D.; Ramalingam, S.; Ram, B.; Chinnaswamy, A. Overexpression of Glyoxalase III Gene in Transgenic Sugarcane Confers Enhanced Performance under Salinity Stress. J. Plant Res. 2021, 134, 1083–1094. [Google Scholar] [CrossRef] [PubMed]
- Peter, S.C.; Murugan, N.; Mohanan, M.V.; Sasikumar, S.P.T.; Selvarajan, D.; Jayanarayanan, A.N.; Shivalingamurthy, S.G.; Chennappa, M.; Ramanathan, V.; Govindakurup, H.; et al. Isolation, Characterization and Expression Analysis of Stress Responsive Plant Nuclear Transcriptional Factor Subunit (NF-YB2) from Commercial Saccharum Hybrid and Wild Relative Erianthus arundinaceus. 3 Biotech 2020, 10, 304. [Google Scholar] [CrossRef] [PubMed]
- Pachakkil, B.; Terajima, Y.; Ohmido, N.; Ebina, M.; Irei, S.; Hayashi, H.; Takagi, H. Cytogenetic and Agronomic Characterization of Intergeneric Hybrids between Saccharum spp. Hybrid and Erianthus arundinaceus. Sci. Rep. 2019, 9, 1748. [Google Scholar] [CrossRef]
- Nair, N.V. Sugarcane Breeding Institute, Coimbatore: A Perspective. Sugar Tech. 2008, 10, 285–292. [Google Scholar] [CrossRef]
- Snecdecor, G.W.; Cochran, W.G. Statistical Methods, 8th ed.; Iowa State University Press: Ames, IA, USA, 1989; ISBN 978-0-813-81561-9. [Google Scholar]
- Lichtenthaler, H.K.; Wellburn, A.R. Determination of Total Carotenoids and Chlorophylls a and b of Leaf Extracts in Different Solvents. Biochem. Soc. Trans. 1983, 11, 591–592. [Google Scholar] [CrossRef]
- Bates, L.S.; Waldren, R.P.; Teare, I.D. Rapid Determination of Free Proline for Water-Stress Studies. Plant Soil 1973, 39, 205–207. [Google Scholar] [CrossRef]
- Bradford, M.M. A Rapid and Sensitive Method for the Quantitation of Microgram Quantities of Protein Utilizing the Principle of Protein-Dye Binding. Anal. Biochem. 1976, 72, 248–254. [Google Scholar] [CrossRef]
- Boaretto, L.F.; Carvalho, G.; Borgo, L.; Creste, S.; Landell, M.G.A.; Mazzafera, P.; Azevedo, R.A. Water Stress Reveals Differential Antioxidant Responses of Tolerant and Non-Tolerant Sugarcane Genotypes. Plant Physiol. Biochem. 2014, 74, 165–175. [Google Scholar] [CrossRef] [PubMed]
- Beauchamp, C.; Fridovich, I. Superoxide Dismutase: Improved Assays and an Assay Applicable to Acrylamide Gels. Anal. Biochem. 1971, 44, 276–287. [Google Scholar] [CrossRef] [PubMed]
- Madamanchi, N.R.; Donahue, J.L.; Cramer, C.L.; Alscher, R.G.; Pedersen, K. Differential Response of Cu, Zn Superoxide Dismutases in Two Pea Cultivars during a Short-Term Exposure to Sulfur Dioxide. Plant Mol. Biol. 1994, 26, 95–103. [Google Scholar] [CrossRef] [PubMed]
- Castillo, F.J.; Penel, C.; Greppin, H. Peroxidase Release Induced by Ozone in Sedum Album Leaves. Plant Physiol. 1984, 74, 846–851. [Google Scholar] [CrossRef]
- Heath, R.L.; Packer, L. Photoperoxidation in Isolated Chloroplasts. I. Kinetics and Stoichiometry of Fatty Acid Peroxidation. Arch. Biochem. Biophys. 1968, 125, 189–198. [Google Scholar] [CrossRef]
- Sayyad-Amin, P.; Jahansooz, M.R.; Borzouei, A.; Ajili, F. Changes in Photosynthetic Pigments and Chlorophyll-a Fluorescence Attributes of Sweet-Forage and Grain Sorghum Cultivars under Salt Stress. J. Biol. Phys. 2016, 42, 601–620. [Google Scholar] [CrossRef]
- Soltys-Kalina, D.; Plich, J.; Strzelczyk-Żyta, D.; Śliwka, J.; Marczewski, W. The Effect of Drought Stress on the Leaf Relative Water Content and Tuber Yield of a Half-Sib Family of ‘Katahdin’-Derived Potato Cultivars. Breed. Sci. 2016, 66, 328–331. [Google Scholar] [CrossRef] [PubMed]
- Tanentzap, F.M.; Stempel, A.; Ryser, P. Reliability of Leaf Relative Water Content (RWC) Measurements after Storage: Consequences for in Situ Measurements. Botany 2015, 93, 535–541. [Google Scholar] [CrossRef]
- Hayat, S.; Hayat, Q.; Alyemeni, M.N.; Wani, A.S.; Pichtel, J.; Ahmad, A. Role of Proline under Changing Environments: A Review. Plant Signal. Behav. 2012, 7, 1456–1466. [Google Scholar] [CrossRef]
- Hebat-Allah, A.H.; Shifaa, O.A.; Sahar, K.M.K.; Fatma, M.E.; Ali, B. Grain-Priming with L-Arginine Improves the Growth Drought Stress. Plants 2022, 11, 1219. [Google Scholar] [CrossRef]
- Kosová, K.; Prášil, I.T.; Vítámvás, P. Protein Contribution to Plant Salinity Response and Tolerance Acquisition. Int. J. Mol. Sci. 2013, 14, 6757–6789. [Google Scholar] [CrossRef] [PubMed]
- Bowler, C.; Van Montagu, M.; Inzé, D. Superoxide Dismutase and Stress Tolerance. Annu. Rev. Plant Physiol. Plant Mol. Biol. 1992, 43, 83–116. [Google Scholar] [CrossRef]
- Alscher, R.G.; Erturk, N.; Heath, L.S. Role of Superoxide Dismutases (SODs) in Controlling Oxidative Stress in Plants. J. Exp. Bot. 2002, 53, 1331–1341. [Google Scholar] [CrossRef]
- Huseynova, I.M.; Aliyeva, D.R.; Aliyev, J.A. Plant Responses to Stresses: Role of Ascorbate Peroxidase in the Antioxidant Protection. Genet. Mol. Biol. 2013, 4, 142–158. [Google Scholar] [CrossRef]
- Attia, M.S.; Hashem, A.H.; Badawy, A.A.; Abdelaziz, A.M. Biocontrol of Early Blight Disease of Eggplant Using Endophytic Aspergillus Terreus: Improving Plant Immunological, Physiological and Antifungal Activities. Bot. Stud. 2022, 63, 26. [Google Scholar] [CrossRef] [PubMed]
- Repetto, M.; Semprine, J.; Boveris, A. Lipid Peroxidation: Chemical Mechanism, Biological Implications and Analytical Determination. In Lipid Peroxidation; Catala, A., Ed.; IntechOpen: London, UK, 2012; pp. 3–30. ISBN 978-953-51-0716-3. [Google Scholar]
- Gandonou, C.B.; Errabii, T.; Abrini, J.; Idaomar, M.; Senhaji, N.S. Selection of Callus Cultures of Sugarcane (Saccharum sp.) Tolerant to NaCl and Their Response to Salt Stress. Plant Cell Tissue Organ Cult. 2006, 87, 9–16. [Google Scholar] [CrossRef]
- Chiconato, D.A.; da Silveira Sousa Junior, G.; dos Santos, D.M.M.; Munns, R. Adaptation of Sugarcane Plants to Saline Soil. Environ. Exp. Bot. 2019, 162, 201–211. [Google Scholar] [CrossRef]
- Xu, Q.; Krishnan, S.; Merewitz, E.; Xu, J.; Huang, B. Gibberellin-Regulation and Genetic Variations in Leaf Elongation for Tall Fescue in Association with Differential Gene Expression Controlling Cell Expansion. Sci. Rep. 2016, 6, 30258. [Google Scholar] [CrossRef]
- Neves-Piestun, B.G.; Bernstein, N. Salinity-Induced Inhibition of Leaf Elongation in Maize Is Not Mediated by Changes in Cell Wall Acidification Capacity. Plant Physiol. 2001, 125, 1419–1428. [Google Scholar] [CrossRef]
- Fricke, W.; Peters, W.S. The Biophysics of Leaf Growth in Salt-Stressed Barley. A Study at the Cell Level. Plant Physiol. 2002, 129, 374–388. [Google Scholar] [CrossRef]
- Jaleel, C.A.; Sankar, B.; Sridharan, R.; Panneerselvam, R. Soil Salinity Alters Growth, Chlorophyll Content, and Secondary Metabolite Accumulation in Catharanthus Roseus. Turk. J. Biol. 2008, 32, 79–83. [Google Scholar]
- Cramer, G.R. Differential Effects of Salinity on Leaf Elongation Kinetics of Three Grass Species. Plant Soil 2003, 253, 233–244. [Google Scholar] [CrossRef]
- Cramer, G.R.; Bowman, D.C. Short-Term Leaf Elongation Kinetics of Maize in Response to Salinity Are Independent of the Root. Plant Physiol. 1991, 253, 233–244. [Google Scholar] [CrossRef] [PubMed]
- Bernstein, N.; Silk, W.; Lauchli, A. Growth and Development of Sorghum Leaves under Conditions of NaCl Stress. Planta 1993, 191, 433–439. [Google Scholar] [CrossRef]
- Jamil, M.; Rehman, S.U.; Kui, J.L.; Jeong, M.K.; Kim, H.S.; Eui, S.R. Salinity Reduced Growth PS2 Photochemistry and Chlorophyll Content in Radish. Sci. Agric. 2007, 62, 111–118. [Google Scholar] [CrossRef]
- Rodrıguez, A.A.; Grunberg, K.A.; Taleisnik, E.L. Reactive Oxygen Species in the Elongation Zone of Maize Leaves Are Necessary for Leaf Extension. J. Plant Physiol. 2002, 129, 1627–1632. [Google Scholar] [CrossRef]
- Rodrıguez, A.A.; Lascano, H.R.; Bustos, D.; Taleisnik, E. Salinity-Induced Decrease in NADPH Oxidase Activity in the Maize Leaf Blade Elongation Zone. J. Plant Physiol. 2007, 164, 223–230. [Google Scholar] [CrossRef]
- Ortega, L.; Fry, S.C.; Taleisnik, E. Why Are Chloris Gayana Leaves Shorter in Salt-Affected Plants? Analyses in the Elongation Zone. J. Exp. Bot. 2006, 57, 3945–3952. [Google Scholar] [CrossRef]
- de Lacerda, C.F.; Cambraia, J.; Oliva, M.A.; Ruiz, H.A.; Prisco, J.T. Solute Accumulation and Distribution during Shoot and Leaf Development in Two Sorghum Genotypes under Salt Stress. Environ. Exp. Bot. 2003, 49, 107–120. [Google Scholar] [CrossRef]
- Azooz, M.M.; Shaddad, M.A.; Abdel-Latef, A.A. Leaf Growth and K+/NA+ Ratio as an Indication of the Salt Tolerance of Three Sorghum Cultivars Grown under Salinity Stress and Iaa Treatment. Acta Agron. Hung. 2004, 52, 287–296. [Google Scholar] [CrossRef]
- Munns, R.; Tester, M. Mechanisms of Salinity Tolerance. Annu. Rev. Plant Biol. 2008, 59, 651–681. [Google Scholar] [CrossRef] [PubMed]
- Maathuis, F.J. Physiological Functions of Mineral Macronutrients. Curr. Opin. Plant Biol. 2009, 12, 250–258. [Google Scholar] [CrossRef] [PubMed]
- Wakeel, A. Potassium-Sodium Interactions in Soil and Plant under Saline-Sodic Conditions. J. Plant Nutr. Soil Sci. 2013, 176, 344–354. [Google Scholar] [CrossRef]
- Almeida, D.M.; Margarida Oliveira, M.; Saibo, N.J.M. Regulation of Na+and K+homeostasis in Plants: Towards Improved Salt Stress Tolerance in Crop Plants. Genet. Mol. Biol. 2017, 40, 326–345. [Google Scholar] [CrossRef]
- Cabot, C.; Sibole, J.V.; Barceló, J.; Poschenrieder, C. Lessons from Crop Plants Struggling with Salinity. Plant Sci. 2014, 226, 2–13. [Google Scholar] [CrossRef]
- Assaha, D.V.M.; Ueda, A.; Saneoka, H.; Al-Yahyai, R.; Yaish, M.W. The Role of Na+ and K+ Transporters in Salt Stress Adaptation in Glycophytes. Front. Physiol. 2017, 8, 509. [Google Scholar] [CrossRef]
- Muhammed, S.; Akbar, M.; Neue, H.U. Effect of Na/Ca and Na/K Ratios in Saline Culture Solution on the Growth and Mineral Nutrition of Rice (Oryza sativa L.). Plant Soil 1987, 104, 57–62. [Google Scholar] [CrossRef]
- Chhipa, B.R.; Lal, P. Na/K Ratios as the Basis of Salt Tolerance in Wheat. Aust. J. Agric. Res. 1995, 46, 533–539. [Google Scholar] [CrossRef]
- Lee, K.S.; Choi, W.Y.; Ko, J.C.; Kim, T.S.; Gregorio, G.B. Salinity Tolerance of Japonica and Indica Rice (Oryza sativa L.) at the Seedling Stage. Planta 2003, 216, 1043–1046. [Google Scholar] [CrossRef]
- Khan, M.A.; Gemenet, D.C.; Villordon, A. Root System Architecture and Abiotic Stress Tolerance: Current Knowledge in Root and Tuber Crops. Front. Plant Sci. 2016, 7, 1584. [Google Scholar] [CrossRef]
- Lynch, J. Root Architecture and Plant Productivity. Plant Physiol. 1995, 109, 7–13. [Google Scholar] [CrossRef] [PubMed]
- Rahneshan, Z.; Nasibi, F.; Moghadam, A.A. Effects of Salinity Stress on Some Growth, Physiological, Biochemical Parameters and Nutrients in Two Pistachio (Pistacia vera L.) Rootstocks. J. Plant Interact. 2018, 13, 73–82. [Google Scholar] [CrossRef]
- Hoque, M.I.U.; Uddin, M.N.; Fakir, M.S.A.; Rasel, M. Drought and Salinity Affect Leaf and Root Anatomical Structures in Three Maize Genotypes. J. Bangladesh Agric. Univ. 2018, 16, 47–55. [Google Scholar] [CrossRef]
- Demmig-Adams, B.; Gilmore, A.M.; Adams, W.W. In Vivo Functions of Carotenoids in Higher Plants. FASEB J. 1996, 10, 403–412. [Google Scholar] [CrossRef]
- Ladygin, V.G.; Shirshikova, G.N. The Current Concepts of Functional Role of Carotenoids in the Eukaryotic Chloroplasts. Zh. Obshch. Biol. 2006, 67, 163–189. [Google Scholar] [PubMed]
- Ivanov, L.A.; Ivanova, L.A.; Ronzhina, D.A.; Yudina, P.K. Changes in the Chlorophyll and Carotenoid Contents in the Leaves of Steppe Plants along a Latitudinal Gradient in South Ural. Russ. J. Plant Physiol. 2013, 60, 812–820. [Google Scholar] [CrossRef]
- Hardo Panintingjati Brotosudarmo, T.; Limantara, L.; Dwi Chandra, R. Heriyanto Chloroplast Pigments: Structure, Function, Assembly and Characterization. In Plant Growth and Regulation-Alterations to Sustain Unfavorable Conditions; Ratnadewi, D., Hamim, Eds.; IntechOpen: London, UK, 2018; pp. 1–111. ISBN 978-1-83881-786-2. [Google Scholar]
- Badawy, A.A.; Alotaibi, M.O.; Abdelaziz, A.M.; Osman, M.S.; Khalil, A.M.A.; Saleh, A.M.; Mohammed, A.E.; Hashem, A.H. Enhancement of Seawater Stress Tolerance in Barley by the Endophytic Fungus Aspergillus Ochraceus. Metabolites 2021, 11, 428. [Google Scholar] [CrossRef]
- Taïbi, K.; Taïbi, F.; Ait Abderrahim, L.; Ennajah, A.; Belkhodja, M.; Mulet, J.M. Effect of Salt Stress on Growth, Chlorophyll Content, Lipid Peroxidation and Antioxidant Defence Systems in Phaseolus vulgaris L. S. Afr. J. Bot. 2016, 105, 306–312. [Google Scholar] [CrossRef]
- Santos, C.V. Regulation of Chlorophyll Biosynthesis and Degradation by Salt Stress in Sunflower Leaves. Sci. Hortic. 2004, 103, 93–99. [Google Scholar] [CrossRef]
- Grzeszczuk, M.; Salachna, P.; Meller, E. Changes in Photosynthetic Pigments, Total Phenolic Content, and Antioxidant Activity of Salvia Coccinea Buc’hoz Ex Etl. Induced by Exogenous Salicylic Acid and Soil Salinity. Molecules 2018, 23, 1296. [Google Scholar] [CrossRef]
- Akcin, A.; Yalcin, E. Effect of Salinity Stress on Chlorophyll, Carotenoid Content, and Proline in Salicornia Prostrata Pall. and Suaeda prostrata Pall. subsp. Prostrata (Amaranthaceae). Rev. Bras. Bot. 2016, 39, 101–106. [Google Scholar] [CrossRef]
- Molazem, D.; Qurbanov, E.M.; Dunyamaliyev, S.A. Role of Proline, Na and Chlorophyll Content in Salt Tolerance of Corn (Zea mays L.). Am. J. Agric. Environ. Sci. 2010, 9, 319–324. [Google Scholar]
- Barrs, H.; Weatherley, P. A Re-Examination of the Relative Turgidity Technique for Estimating Water Deficits in Leaves. Aust. J. Biol. Sci. 1962, 15, 413–428. [Google Scholar] [CrossRef]
- Hand, M.J.; Taffouo, V.D.; Nouck, A.E.; Nyemene, K.P.J.; Tonfack, L.B.; Meguekam, T.L.; Youmbi, E. Effects of Salt Stress on Plant Growth, Nutrient Partitioning, Chlorophyll Content, Leaf Relative Water Content, Accumulation of Osmolytes and Antioxidant Compounds in Pepper (Capsicum annuum L.) Cultivars. Not. Bot. Horti Agrobot. Cluj-Napoca 2017, 45, 481–490. [Google Scholar] [CrossRef]
- Liang, X.; Zhang, L.; Natarajan, S.K.; Becker, D.F. Proline Mechanisms of Stress Survival. Antioxid. Redox Signal. 2013, 19, 998–1011. [Google Scholar] [CrossRef]
- Osman, M.S.; Badawy, A.A.; Osman, A.I.; Abdel Latef, A.A.H. Ameliorative Impact of an Extract of the Halophyte Arthrocnemum Macrostachyum on Growth and Biochemical Parameters of Soybean Under Salinity Stress. J. Plant Growth Regul. 2021, 40, 1245–1256. [Google Scholar] [CrossRef]
- Ashraf, M.; Foolad, M.R. Roles of Glycine Betaine and Proline in Improving Plant Abiotic Stress Resistance. Environ. Exp. Bot. 2007, 59, 206–216. [Google Scholar] [CrossRef]
- Gharsallah, C.; Fakhfakh, H.; Grubb, D.; Gorsane, F. Effect of Salt Stress on Ion Concentration, Proline Content, Antioxidant Enzyme Activities and Gene Expression in Tomato Cultivars. AoB Plants 2016, 8, plw055. [Google Scholar] [CrossRef]
- Ashraf, M.; Harris, P.J.C. Potential Biochemical Indicators of Salinity Tolerance in Plants. Plant Sci. 2004, 166, 3–16. [Google Scholar] [CrossRef]
- Amini, F.; Ehsanpour, A.A. Soluble Proteins, Proline, Carbohydrates and Na+/K+ Changes in Two Tomato (Lycopersicon Esculentum Mill) Cultivars under in Vitro Salt Stress. Am. J. Biochem. Biotechnol. 2005, 1, 212–216. [Google Scholar] [CrossRef]
- Arefian, M.; Vessal, S.; Malekzadeh-Shafaroudi, S.; Siddique, K.H.M.; Bagheri, A. Comparative Proteomics and Gene Expression Analyses Revealed Responsive Proteins and Mechanisms for Salt Tolerance in Chickpea Genotypes. BMC Plant Biol. 2019, 19, 300. [Google Scholar] [CrossRef] [PubMed]
- Xiong, J.; Sun, Y.; Yang, Q.; Tian, H.; Zhang, H.; Liu, Y.; Chen, M. Proteomic Analysis of Early Salt Stress Responsive Proteins in Alfalfa Roots and Shoots. Proteome Sci. 2017, 15, 19. [Google Scholar] [CrossRef] [PubMed]
- Passamani, L.Z.; Barbosa, R.R.; Reis, R.S.; Heringer, A.S.; Rangel, P.L.; Santa-Catarina, C.; Grativol, C.; Veiga, C.F.M.; Souza-Filho, G.A.; Silveira, V. Salt Stress Induces Changes in the Proteomic Profile of Micropropagated Sugarcane Shoots. PLoS ONE 2017, 12, e0176076. [Google Scholar] [CrossRef] [PubMed]
- Aghaei, K.; Ehsanpour, A.A.; Komatsu, S. Proteome Analysis of Potato under Salt Stress. J. Proteome Res. 2008, 7, 4858–4868. [Google Scholar] [CrossRef] [PubMed]
- Ayala-Astorga, G.I.; Alcaraz-Meléndez, L. Salinity Effects on Protein Content, Lipid Peroxidation, Pigments, and Proline in Paulownia Imperialis (Siebold & Zuccarini) and Paulownia Fortunei (Seemann & Hemsley) Grown in Vitro. Electron. J. Biotechnol. 2010, 13, 13–14. [Google Scholar] [CrossRef]
- Amin, M.A.; Ismail, M.A.; Badawy, A.A.; Awad, M.A. Improve the Growth Performance of Helianthus annuus L. and Control of Cutworm Agrotis Ipsilon. Catalysts 2021, 11, 1551. [Google Scholar] [CrossRef]
- Lascano, H.R.; Antonicelli, G.E.; Luna, C.M.; Melchiorre, M.N.; Gómez, L.D.; Racca, R.W.; Trippi, V.S.; Casano, L.M. Antioxidant System Response of Different Wheat Cultivars under Drought: Field and in Vitro Studies. Aust. J. Plant Physiol. 2001, 28, 1095–1102. [Google Scholar] [CrossRef]
- Wu, G.Q.; Zhang, L.N.; Wang, Y.Y. Response of Growth and Antioxidant Enzymes to Osmotic Stress in Two Different Wheat (Triticum Aestivum L.) Cultivars Seedlings. Plant Soil Environ. 2012, 58, 534–539. [Google Scholar] [CrossRef]
- Leonowicz, G.; Trzebuniak, K.F.; Zimak-Piekarczyk, P.; Ślesak, I.; Mysliwa-Kurdziel, B. The Activity of Superoxide Dismutases (SODs) at the Early Stages of Wheat Deetiolation. PLoS ONE 2018, 13, e0194678. [Google Scholar] [CrossRef]
- Meloni, D.A.; Oliva, M.A.; Martinez, C.A.; Cambraia, J. Photosynthesis and Activity of Superoxide Dismutase, Peroxidase and Glutathione Reductase in Cotton under Salt Stress. Environ. Exp. Bot. 2003, 49, 69–76. [Google Scholar] [CrossRef]
- Song, F.; Yang, C.; Liu, X.; Li, G. Effect of Salt Stress on Activity of Superoxide Dismutase (SOD) in Ulmus pumila L. J. For. Res. 2006, 17, 13–16. [Google Scholar] [CrossRef]
- Eyidoǧan, F.; Öktem, H.A.; Yücel, M. Superoxide Dismutase Activity in Salt Stressed Wheat Seedlings. Acta Physiol. Plant. 2003, 25, 263–269. [Google Scholar] [CrossRef]
- Kim, K.Y.; Huh, G.H.; Lee, H.S.; Kwon, S.Y.; Hur, Y.; Kwak, S.S. Molecular Characterization of CDNAs for Two Anionic Peroxidases from Suspension Cultures of Sweet Potato. Mol. Gen. Genet. 1999, 261, 941–947. [Google Scholar] [CrossRef]
- Almagro, L.; Gómez Ros, L.V.; Belchi-Navarro, S.; Bru, R.; Ros Barceló, A.; Pedreño, M.A. Class III Peroxidases in Plant Defence Reactions. J. Exp. Bot. 2009, 60, 377–390. [Google Scholar] [CrossRef] [PubMed]
- Asada, K. Ascorbate Peroxidase—A Hydrogen Peroxide-scavenging Enzyme in Plants. Physiol. Plant. 1992, 85, 235–241. [Google Scholar] [CrossRef]
- Pujari, D.S.; Chanda, S.V. Effect of Salinity Stress on Growth, Peroxidase and IAA Oxidase Activities in Vigna Seedlings. Acta Physiol. Plant. 2002, 24, 435–439. [Google Scholar] [CrossRef]
- Jakovljević, D.Z.; Topuzović, M.D.; Stanković, M.S.; Bojović, B.M. Changes in Antioxidant Enzyme Activity in Response to Salinity-Induced Oxidative Stress during Early Growth of Sweet Basil. Hortic. Environ. Biotechnol. 2017, 58, 240–246. [Google Scholar] [CrossRef]
- Ayala, A.; Muñoz, M.F.; Argüelles, S. Lipid Peroxidation: Production, Metabolism, and Signaling Mechanisms of Malondialdehyde and 4-Hydroxy-2-Nonenal. Oxid. Med. Cell. Longev. 2014, 2014, 360438. [Google Scholar] [CrossRef]
- Schmidt, A.; Kunert, K.J. Lipid Peroxidation in Higher Plants. Plant Physiol. 1986, 82, 700–702. [Google Scholar] [CrossRef]
- De Azevedo Neto, A.D.; Prisco, J.T.; Enéas-Filho, J.; De Abreu, C.E.B.; Gomes-Filho, E. Effect of Salt Stress on Antioxidative Enzymes and Lipid Peroxidation in Leaves and Roots of Salt-Tolerant and Salt-Sensitive Maize Genotypes. Environ. Exp. Bot. 2006, 56, 87–94. [Google Scholar] [CrossRef]













| S. No | Clone | Altitude (ft) | Location | District/State |
|---|---|---|---|---|
| 1 | IND99-880 | 10 | Eswaramangalam | Malappuram, Kerala, India |
| 2. | IND99-884 | 0 | Ponnani | Malappuram, Kerala, India |
| 3. | IND99-895 | 0 | Kottapuram | Ernakulam, Kerala, India |
| 4. | IND99-907 | 10 | PadiyattuKadavu | Ernakulam, Kerala, India |
| 5. | IND01-1134 | 15 | Tarapur | Jagatsingpur, Orissa, India |
| 6. | IND01-1136 | 15 | Kandadhru | Jagatsingpur, Orissa, India |
| 7. | IND03-1253 | 5 | CFO NalleBetapur | Middle Andaman, India |
| 8. | IND03-1255 | 20 | Phoolathala | Middle Andaman, India |
| 9. | IND03-1262 | 10 | Tirur | South Andaman, India |
| 10 | IND02-1208 | 10 | Yethakota | East Godavari, Andhra Pradesh, India |
| 11. | IND02-1260 | 20 | MilderaKatchal | Nicobar Islands, India |
| 12. | IND10-1591 | - | South 24 Paraganas | West Bengal, India |
| 13 | EaLakshadweep | Lakshadweep | Lakshadweep, Islands | |
| 14 | Ea A & N | Andaman & Nicobar | Andaman & Nicobar | |
| 15 | IK76-48 | |||
| 16 | Bethuadahari | Bethuadahari | Nadia, West Bengal, India |
| Entries | Roots Status | Shoot Length | Leaf Length | Root Number | Root Length | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | S | R (%) | C | S | R (%) | C | S | R (%) | C | S | R (%) | ||
| IND99-880 | Dead | 4.93 | 2.93 | 40.61 | 19.75 | 12.50 | 36.71 | 8.33 | 0.00 | 100 | 15.25 | 0.00 | 100 |
| IND99-884 | Dead | 3.50 | 1.25 | 64.29 | 14.25 | 7.75 | 45.61 | 5.67 | 1.00 | 82.35 | 35.33 | 2.33 | 93.40 |
| IND99-907 | Alive | 6.00 | 4.00 | 29.17 | 29.67 | 15.17 | 48.88 | 8.33 | 6.00 | 28.00 | 32.00 | 11.50 | 64.06 |
| IND01-1134 | Alive | 4.25 | 4.00 | 5.88 | 19.50 | 14.92 | 23.50 | 9.00 | 6.33 | 29.63 | 33.33 | 16.67 | 50.00 |
| IND01-1136 | Alive | 5.00 | 3.50 | 30.00 | 22.67 | 11.42 | 49.63 | 8.50 | 2.67 | 68.63 | 27.33 | 23.33 | 14.63 |
| IND02-1208 | Dead | 5.50 | 3.25 | 40.91 | 28.33 | 17.25 | 39.12 | 6.67 | 4.33 | 35.00 | 34.00 | 21.00 | 38.24 |
| IND02-1253 | Dead | 6.75 | 4.00 | 40.74 | 21.67 | 9.09 | 58.07 | 9.67 | 1.00 | 89.66 | 24.33 | 14.67 | 39.73 |
| IND02-1255 | Dead | 9.33 | 2.50 | 73.21 | 31.00 | 15.58 | 49.73 | 8.33 | 0.00 | 100.00 | 24.67 | 0.00 | 100 |
| IND10-1591 | Dead | 7.50 | 6.00 | 20.00 | 30.67 | 18.09 | 41.03 | 10.00 | 1.67 | 83.33 | 31.00 | 2.00 | 93.55 |
| Bethuadahari | Alive | 6.33 | 4.66 | 26.38 | 10.33 | 8.73 | 15.49 | 11.66 | 8.00 | 31.39 | 12.66 | 12.00 | 5.21 |
| IK76-48 | Alive | 4.25 | 3.25 | 23.53 | 16.67 | 9.84 | 40.99 | 6.67 | 1.00 | 85.00 | 29.33 | 13.67 | 53.41 |
| Co 97010 | Dead | 10.13 | 4.63 | 54.32 | 21.75 | 6.75 | 68.97 | 24.00 | 3.67 | 84.70 | 29.00 | 6.00 | 79.31 |
| CD at 5% significance for genotypes | 1.979 | 0.705 | 1.073 | 0.53 | |||||||||
| CD at 5% significance for treatments | 0.808 | 0.288 | 0.438 | 0.28 | |||||||||
| Genotypes | Na (mg/g of Dry Sample) | K (mg/g of Dry Sample) | Na/K Ratio (mg/g of Dry Sample) | ||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| C | T1 | T2 | T3 | T4 | C | T1 | T2 | T3 | T4 | C | T1 | T2 | T3 | T4 | |
| IK76-48 | 5.59 | 8.86 | 14.37 | 15.19 | 15.74 | 13.67 | 16.44 | 16.71 | 16.09 | 14.34 | 0.41 | 0.54 | 0.86 | 0.94 | 1.10 |
| Bethuadahari | 9.74 | 16.44 | 14.20 | 20.54 | 22.04 | 14.78 | 22.54 | 9.32 | 11.67 | 10.47 | 0.66 | 0.73 | 1.52 | 1.76 | 2.11 |
| IND01-1136 | 4.78 | 10.35 | 16.60 | 15.91 | 12.87 | 12.99 | 23.76 | 16.31 | 17.99 | 8.51 | 0.37 | 0.44 | 1.02 | 0.88 | 1.51 |
| IND99-907 | 8.76 | 8.82 | 16.95 | 25.04 | 19.06 | 41.79 | 34.05 | 31.63 | 23.72 | 18.79 | 0.21 | 0.26 | 0.54 | 1.06 | 1.01 |
| IND01-1134 | 9.31 | 8.96 | 14.44 | 17.13 | 17.54 | 13.67 | 13.35 | 14.24 | 9.68 | 10.66 | 0.68 | 0.67 | 1.01 | 1.77 | 1.65 |
| Co 86032 | 6.42 | 12.46 | 16.94 | 19.64 | 19.77 | 19.54 | 18.14 | 16.14 | 13.14 | 3.79 | 0.33 | 0.69 | 1.05 | 1.49 | 5.22 |
| Co 97010 | 10.01 | 15.08 | 23.96 | 18.74 | 18.94 | 26.47 | 21.33 | 17.91 | 11.76 | 7.88 | 0.38 | 0.71 | 1.34 | 1.59 | 2.40 |
| CD (5% level of significance) | G | S | G × S | G | S | G × S | G | S | G × S | ||||||
| 1.72 | 1.30 | 3.43 | 3.27 | ns | 6.54 | 0.34 | ns | 0.68 | |||||||
| Genotypes | Sodium | Calcium | Magnesium | Potassium | ||||
|---|---|---|---|---|---|---|---|---|
| S | C | S | C | S | C | S | C | |
| (i) Elemental composition (mg g−1 of dry samples estimated by ICP-OES) | ||||||||
| Co 97010 | 21.05 | 7.52 | 12.44 | 9.22 | 2.18 | 2.03 | 2.76 | 4.00 |
| IND01-1136 | 22.52 | 8.23 | 15.40 | 12.20 | 4.31 | 3.47 | 3.76 | 4.18 |
| IND99-907 | 21.07 | 14.97 | 10.12 | 9.04 | 2.26 | 3.54 | 9.65 | 19.96 |
| (ii) Cation/K ratio by ICP-OES method | ||||||||
| Co 97010 | 7.64 | 1.88 | 4.52 | 2.30 | 0.79 | 0.51 | ||
| IND01-1136 | 5.99 | 1.97 | 4.10 | 2.92 | 1.15 | 0.83 | ||
| IND99-907 | 2.18 | 0.75 | 1.05 | 0.45 | 0.23 | 0.18 | ||
| (iii) Cation/K ration by SEM-EDX method | ||||||||
| Co 97010 | 1.5 | 0.8 | 3.24 | 1.06 | 0.33 | 0.71 | ||
| IND01-1136 | 1.21 | 0.79 | 1.19 | 1.23 | 0.52 | 0.58 | ||
| IND99-907 | 0.64 | 1.03 | 0.00 | 0.87 | 0.18 | 1.17 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Mahadevaiah, C.; Vignesh, P.; Appunu, C.; Valarmathi, R.; Dhansu, P.; Kumar, A.; Dharshini, S.; Padmanabhan, T.S.S.; Narayan, J.A.; Selvamuthu, K.; et al. Physiological Characterization of Tripidium arundinaceum and Sugarcane (Saccharum spp.) Germplasm for Salinity Stress Tolerance at the Formative Stage. Sustainability 2023, 15, 6962. https://doi.org/10.3390/su15086962
Mahadevaiah C, Vignesh P, Appunu C, Valarmathi R, Dhansu P, Kumar A, Dharshini S, Padmanabhan TSS, Narayan JA, Selvamuthu K, et al. Physiological Characterization of Tripidium arundinaceum and Sugarcane (Saccharum spp.) Germplasm for Salinity Stress Tolerance at the Formative Stage. Sustainability. 2023; 15(8):6962. https://doi.org/10.3390/su15086962
Chicago/Turabian StyleMahadevaiah, Channappa, Palanisamy Vignesh, Chinnasamy Appunu, Ramanathan Valarmathi, Pooja Dhansu, Ashwani Kumar, Selvarajan Dharshini, Thelakat Sasikumar Sarath Padmanabhan, Jayanarayanan Ashwin Narayan, Kannan Selvamuthu, and et al. 2023. "Physiological Characterization of Tripidium arundinaceum and Sugarcane (Saccharum spp.) Germplasm for Salinity Stress Tolerance at the Formative Stage" Sustainability 15, no. 8: 6962. https://doi.org/10.3390/su15086962






