Carbazole Degradation and Genetic Analyses of Sphingobium sp. Strain BS19 Isolated from Antarctic Soil
Abstract
:1. Introduction
2. Materials and Methods
2.1. Collection of Sample and Isolation of Strain
2.2. Bacterial Cell Enumeration
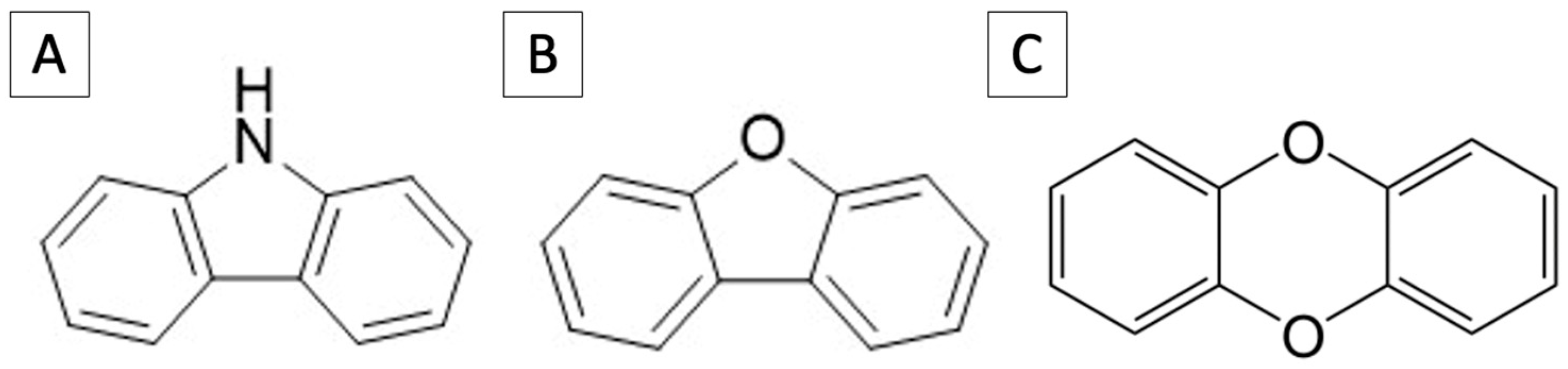
2.3. Carbazole Degradation Analysis
2.4. PAHs and Heterocyclic Compound Utilization Test
2.5. DNA Sequencing Analysis
2.5.1. Total DNA Extraction
2.5.2. 16S ribosomal RNA Gene Sequence Analysis
2.6. Whole Genome Sequencing, Assembly and Analyses
2.7. Gene Expression Detection for Genes Involved in Carbazole Degradation
3. Results
3.1. Isolation and Identification of Carbazole-Degrading Bacterium from Antarctic Soil
3.2. Degradation of Carbazole and Utilization of Similar Compounds
3.3. Whole Genome Sequencing of Strain BS19
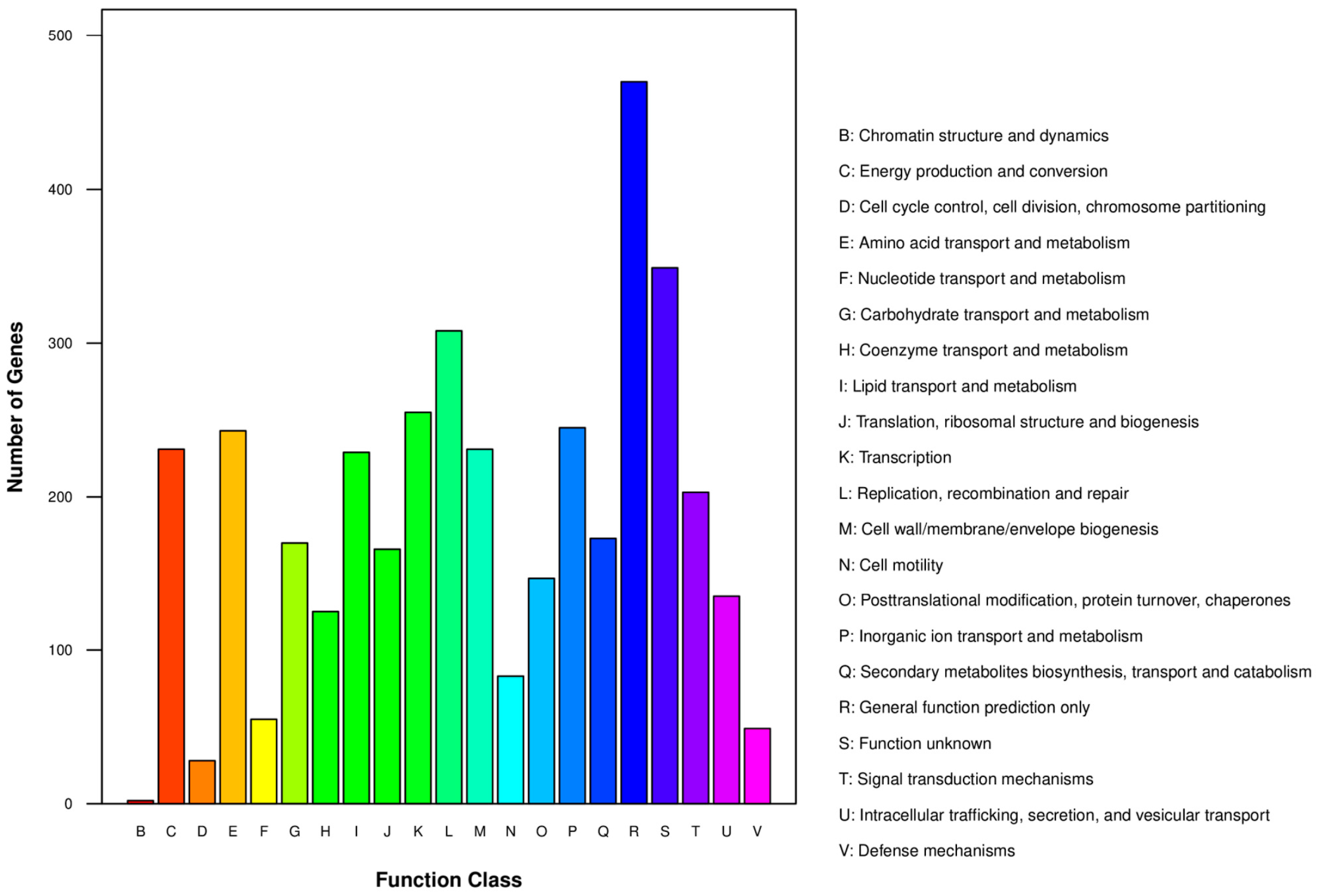
3.4. Gene Function Annotation
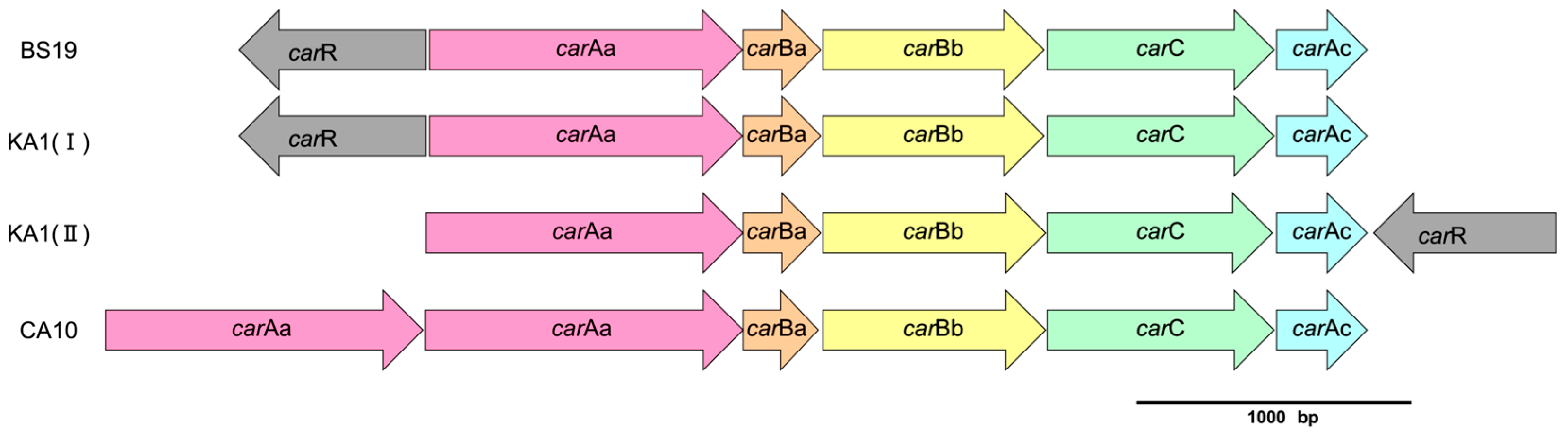
3.5. Identification of Gene Clusters Involved in Carbazole Degradation Pathway
3.6. Detection of Car Gene and Cat Gene Cluster Expression
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bícego, M.C.; Zanardi-Lamardo, E.; Taniguchi, S.; Martins, C.C.; da Silva, D.A.M.; Sasaki, S.T.; Albergaria-Barbosa, A.C.R.; Paolo, F.S.; Weber, R.R.; Montone, R.C. Results from a 15-Year Study on Hydrocarbon Concentrations in Water and Sediment from Admiralty Bay, King George Island, Antarctica. Antarct. Sci. 2009, 21, 209–220. [Google Scholar] [CrossRef]
- Martins, C.C.; Bícego, M.C.; Taniguchi, S.; Montone, R.C. Aliphatic and Polycyclic Aromatic Hydrocarbons in Surface Sediments in Admiralty Bay, King George Island, Antarctica. Antarct. Sci. 2004, 16, 117–122. [Google Scholar] [CrossRef]
- Cripps, G.C.; Priddle, J. Hydrocarbon Content of an Antarctic Infaunal Bivalve—Historical Record or Life Cycle Changes? Antarct. Sci. 1995, 7, 127–136. [Google Scholar] [CrossRef]
- Sims, G.K.; O’Loughlin, E.J.; Crawford, R.L. Degradation of Pyridines in the Environment. Crit. Rev. Environ. Control 1989, 19, 309–340. [Google Scholar] [CrossRef]
- Padoley, K.V.; Mudliar, S.N.; Pandey, R.A. Heterocyclic Nitrogenous Pollutants in the Environment and Their Treatment Options—An Overview. Bioresour. Technol. 2008, 99, 4029–4043. [Google Scholar] [CrossRef]
- Eisentraeger, A.; Brinkmann, C.; Hollert, H.; Sagner, A.; Tiehm, A.; Neuwoehner, J. Heterocyclic compounds: Toxic effects using algae, daphnids, and the salmonella/microsome test taking methodical quantitative aspects into account. Environ. Toxicol. Chem. 2008, 27, 1590. [Google Scholar] [CrossRef]
- Abdulrasheed, M.; Zakaria, N.N.; Roslee, A.F.A.; Shukor, M.Y.; Zulkharnain, A.; Napis, S.; Convey, P.; Alias, S.A.; Gonzalez-Rocha, G.; Ahmad, S.A. Biodegradation of Diesel Oil by Cold-Adapted Bacterial Strains of Arthrobacter spp. from Antarctica. Antarct. Sci. 2020, 32, 5. [Google Scholar] [CrossRef]
- Maliki, I.M.; Abdul-Manas, N.H.; Ahmad, S.A.; Fuse, H.; Ramírez-Moreno, N.; Zulkharnain, A. Removal of Heterocyclic Compound Carbazole Using Cell Immobilization of Thalassospira Profundimaris Strain M02. Rev. Mex. Ing. Quim. 2020, 20, 413–422. [Google Scholar] [CrossRef]
- Zahri, K.N.M.; Zulkharnain, A.; Gomez-Fuentes, C.; Sabri, S.; Ahmad, S.A. Study of Growth Kinetics of Antarctic Bacterial Community for Biodegradation of Waste Canola Oil. Desalination Water Treat 2021, 213, 128–138. [Google Scholar] [CrossRef]
- Lee, G.L.Y.; Ahmad, S.A.; Yasid, N.A.; Zulkharnain, A.; Convey, P.; Wan Johari, W.L.; Alias, S.A.; Gonzalez-Rocha, G.; Shukor, M.Y. Biodegradation of Phenol by Cold-Adapted Bacteria from Antarctic Soils. Polar. Biol. 2018, 41, 553–562. [Google Scholar] [CrossRef]
- Roslee, A.F.A.; Ahmad, S.A.; Gomez-Fuentes, C.; Shaharuddin, N.A.; Khalil, K.A.; Zulkharnain, A. Scientometric Analysis of Diesel Pollutions in Antarctic Territories: A Review of Causes and Potential Bioremediation Approaches. Sustainability 2021, 13, 7064. [Google Scholar] [CrossRef]
- Lim, Z.S.; Wong, R.R.; Wong, C.-Y.; Zulkharnain, A.; Shaharuddin, N.A.; Ahmad, S.A. Bibliometric Analysis of Research on Diesel Pollution in Antarctica and a Review on Remediation Techniques. Appl. Sci. 2021, 11, 1123. [Google Scholar] [CrossRef]
- Verasoundarapandian, G.; Wong, C.-Y.; Shaharuddin, N.A.; Gomez-Fuentes, C.; Zulkharnain, A.; Ahmad, S.A. A Review and Bibliometric Analysis on Applications of Microbial Degradation of Hydrocarbon Contaminants in Arctic Marine Environment at Metagenomic and Enzymatic Levels. Int. J. Environ. Res. Public. Health 2021, 18, 1671. [Google Scholar] [CrossRef] [PubMed]
- Gentile, G.; Bonsignore, M.; Santisi, S.; Catalfamo, M.; Giuliano, L.; Genovese, L.; Yakimov, M.M.; Denaro, R.; Genovese, M.; Cappello, S. Biodegradation Potentiality of Psychrophilic Bacterial Strain Oleispira Antarctica RB-8 T. Mar. Pollut. Bull. 2016, 105, 125–130. [Google Scholar] [CrossRef]
- Aislabie, J.; Foght, J.; Saul, D. Aromatic Hydrocarbon-Degrading Bacteria from Soil near Scott Base, Antarctica. Polar. Biol. 2000, 23, 183–188. [Google Scholar] [CrossRef]
- Lee, G.L.Y.; Zakaria, N.N.; Futamata, H.; Suzuki, K.; Zulkharnain, A.; Shaharuddin, N.A.; Convey, P.; Zahri, K.N.M.; Ahmad, S.A. Metabolic Pathway of Phenol Degradation of a Cold-Adapted Antarctic Bacteria, Arthrobacter sp. Catalysts 2022, 12, 1422. [Google Scholar] [CrossRef]
- Sung, J.Y.; Hwang, Y.; Shin, M.H.; Park, M.S.; Lee, S.H.; Yong, D.; Lee, K. Utility of Conventional Culture and MALDI-TOF MS for Identification of Microbial Communities in Bronchoalveolar Lavage Fluid in Comparison with the GS Junior Next Generation Sequencing System. Ann. Lab. Med. 2018, 38, 110–118. [Google Scholar] [CrossRef] [PubMed]
- Altschul, S.F.; Gish, W.; Miller, W.; Myers, E.W.; Lipman, D.J. Basic Local Alignment Search Tool. J. Mol. Biol. 1990, 215, 403–410. [Google Scholar] [CrossRef]
- Tatusov, R.L.; Koonin, E.V.; Lipman, D.J. A Genomic Perspective on Protein Families. Science (1979) 1997, 278, 631–637. [Google Scholar] [CrossRef]
- Makarova, K.; Wolf, Y.; Koonin, E. Archaeal Clusters of Orthologous Genes (ArCOGs): An Update and Application for Analysis of Shared Features between Thermococcales, Methanococcales, and Methanobacteriales. Life 2015, 5, 818–840. [Google Scholar] [CrossRef]
- Untergasser, A.; Cutcutache, I.; Koressaar, T.; Ye, J.; Faircloth, B.C.; Remm, M.; Rozen, S.G. Primer3—New Capabilities and Interfaces. Nucleic. Acids Res. 2012, 40, e115. [Google Scholar] [CrossRef] [PubMed]
- Yoon, S.-H.; Ha, S.-M.; Kwon, S.; Lim, J.; Kim, Y.; Seo, H.; Chun, J. Introducing EzBioCloud: A Taxonomically United Database of 16S RRNA Gene Sequences and Whole-Genome Assemblies. Int. J. Syst. Evol. Microbiol. 2017, 67, 1613–1617. [Google Scholar] [CrossRef] [PubMed]
- Kasai, D.; Masai, E.; Katayama, Y.; Fukuda, M. Degradation of 3-O-Methylgallate in Sphingomonas Paucimobilis SYK-6 by Pathways Involving Protocatechuate 4,5-Dioxygenase. FEMS Microbiol. Lett. 2007, 274, 323–328. [Google Scholar] [CrossRef] [PubMed]
- Han, S.-R.; Jang, S.-M.; Chi, Y.M.; Kim, B.; Jung, S.-H.; Lee, Y.M.; Uetake, J.; Lee, J.H.; Park, H.; Oh, T.-J. Complete Genome Sequence of Sphingobium sp. Strain PAMC 28499 Reveals a Potential for Degrading Pectin with Comparative Genomics Approach. Genes Genom. 2020, 42, 1087–1096. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Hong, Q.; Li, Q.; Li, C.; Cao, L.; Sun, J.-Q.; Yan, X.; Li, S.-P. Characterization of Isoproturon Biodegradation Pathway in Sphingobium sp. YBL2. Int. Biodeterior. Biodegrad. 2012, 70, 8–13. [Google Scholar] [CrossRef]
- Kwon, S.J.; Lee, P.C. Complete Genome Sequence of Yellow Pigment-Producing Sphingobium sp. Strain HAL-16. Microbiol Resour. Announc. 2020, 9, 41. [Google Scholar] [CrossRef]
- Tabata, M.; Ohhata, S.; Kawasumi, T.; Nikawadori, Y.; Kishida, K.; Sato, T.; Ohtsubo, Y.; Tsuda, M.; Nagata, Y. Complete Genome Sequence of a γ-Hexachlorocyclohexane Degrader, Sphingobium sp. Strain TKS, Isolated from a γ-Hexachlorocyclohexane-Degrading Microbial Community. Genome Announc. 2016, 4, 2. [Google Scholar] [CrossRef]
- Jiang, W.; Zhang, M.; Gao, S.; Zhu, Q.; Qiu, J.; Yan, X.; Xin, F.; Jiang, M.; Hong, Q. Comparative Genomic Analysis of Carbofuran-Degrading Sphingomonads Reveals the Carbofuran Catabolism Mechanism in Sphingobium sp. Strain CFD-1. Appl. Environ. Microbiol. 2022, 88, 22. [Google Scholar] [CrossRef]
- Jiang, W.; Gao, Q.; Zhang, L.; Wang, H.; Zhang, M.; Liu, X.; Zhou, Y.; Ke, Z.; Wu, C.; Qiu, J.; et al. Identification of the Key Amino Acid Sites of the Carbofuran Hydrolase CehA from a Newly Isolated Carbofuran-Degrading Strain Sphingbium sp. CFD-1. Ecotoxicol. Environ. Saf. 2020, 189, 109938. [Google Scholar] [CrossRef]
- Rangu, S.S.; Muralidharan, B.; Tripathi, S.C.; Apte, S.K. Tributyl Phosphate Biodegradation to Butanol and Phosphate and Utilization by a Novel Bacterial Isolate, Sphingobium sp. Strain RSMS. Appl. Microbiol. Biotechnol. 2014, 98, 2289–2296. [Google Scholar] [CrossRef]
- Li, T.; Xu, J.; Brower, A.L.; Xu, Z.-J.; Xu, Y.; Spain, J.C.; Zhou, N.-Y. Molecular Basis and Evolutionary Origin of 1-Nitronaphthalene Catabolism in Sphingobium sp. Strain JS3065. Appl. Environ. Microbiol. 2023, 89, 1. [Google Scholar] [CrossRef] [PubMed]
- Tabata, M.; Ohhata, S.; Nikawadori, Y.; Sato, T.; Kishida, K.; Ohtsubo, Y.; Tsuda, M.; Nagata, Y. Complete Genome Sequence of a γ-Hexachlorocyclohexane-Degrading Bacterium, Sphingobium sp. Strain MI1205. Genome Announc. 2016, 4, 2. [Google Scholar] [CrossRef] [PubMed]
- Fixen, K.R.; Starkenburg, S.R.; Hovde, B.T.; Johnson, S.L.; Deodato, C.R.; Daligault, H.E.; Davenport, K.W.; Harwood, C.S.; Cattolico, R.A. Genome Sequences of Eight Bacterial Species Found in Coculture with the Haptophyte Chrysochromulina Tobin. Genome Announc. 2016, 4, 6. [Google Scholar] [CrossRef] [PubMed]
- Ma, X.; Liang, B.; Qi, M.; Yun, H.; Shi, K.; Li, Z.; Guo, Y.; Yan, P.; Liu, S.-J.; Wang, A. Novel Pathway for Chloramphenicol Catabolism in the Activated Sludge Bacterial Isolate Sphingobium sp. CAP-1. Environ. Sci. Technol. 2020, 54, 7591–7600. [Google Scholar] [CrossRef] [PubMed]
- Nojiri, H.; Nam, J.-W.; Kosaka, M.; Morii, K.-I.; Takemura, T.; Furihata, K.; Yamane, H.; Omori, T. Diverse Oxygenations Catalyzed by Carbazole 1,9a-Dioxygenase from Pseudomonas sp. Strain CA10. J. Bacteriol. 1999, 181, 3105–3113. [Google Scholar] [CrossRef]
- vander Schaaf, N.; Cunningham, A.; Cluff, B.; Kraemer, C.; Reeves, C.; Riester, C.; Slater, L.; Madigan, M.; Sattley, W. Cold-Active, Heterotrophic Bacteria from the Highly Oligotrophic Waters of Lake Vanda, Antarctica. Microorganisms 2015, 3, 391–406. [Google Scholar] [CrossRef]
- Gounot, A.-M. Psychrophilic and Psychrotrophic Microorganisms. Experientia 1986, 42, 1192–1197. [Google Scholar] [CrossRef]
- Inoue, K.; Habe, H.; Yamane, H.; Omori, T.; Nojiri, H. Diversity of Carbazole-Degrading Bacteria Having the Car Gene Cluster: Isolation of a Novel Gram-Positive Carbazole-Degrading Bacterium. FEMS Microbiol. Lett. 2005, 245, 145–153. [Google Scholar] [CrossRef]
- Ouchiyama, N.; Zhang, Y.; Omori, T.; Kodama, T. Biodegradation of Carbazole by Pseudomonas Spp. CA06 and CA10. Biosci. Biotechnol. Biochem. 1993, 57, 455–460. [Google Scholar] [CrossRef]
- Zhao, C.; Zhang, Y.; Li, X.; Wen, D.; Tang, X. Biodegradation of Carbazole by the Seven Pseudomonas sp. Strains and Their Denitrification Potential. J. Hazard Mater. 2011, 190, 253–259. [Google Scholar] [CrossRef]
- Nojiri, H.; Sekiguchi, H.; Maeda, K.; Urata, M.; Nakai, S.I.; Yoshida, T.; Habe, H.; Omori, T. Genetic Characterization and Evolutionary Implications of a Car Gene Cluster in the Carbazole Degrader Pseudomonas sp. Strain CA10. J. Bacteriol. 2001, 183, 12. [Google Scholar] [CrossRef]
- Maeda, K.; Nojiri, H.; Shintani, M.; Yoshida, T.; Habe, H.; Omori, T. Complete Nucleotide Sequence of Carbazole/Dioxin-Degrading Plasmid PCAR1 in Pseudomonas Resinovorans Strain CA10 Indicates Its Mosaicity and the Presence of Large Catabolic Transposon Tn4676. J. Mol. Biol. 2003, 326, 21–33. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M.; Urata, M.; Inoue, K.; Eto, K.; Habe, H.; Omori, T.; Yamane, H.; Nojiri, H. The Sphingomonas Plasmid PCAR3 Is Involved in Complete Mineralization of Carbazole. J. Bacteriol. 2007, 189, 2007–2020. [Google Scholar] [CrossRef] [PubMed]
- Shintani, M.; Fukushima, N.; Tezuka, M.; Yamane, H.; Nojiri, H. Conjugative Transfer of the IncP-7 Carbazole Degradative Plasmid, PCAR1, in River Water Samples. Biotechnol. Lett. 2008, 30, 117–122. [Google Scholar] [CrossRef] [PubMed]
- Assinder, S.J.; Williams, P.A. The TOL Plasmids: Determinants of the Catabolism of Toluene and the Xylenes. Adv. Microb. Physiol. 1990, 31, 1–69. [Google Scholar] [CrossRef] [PubMed]
- Adriaenssens, E.M.; Guerrero, L.D.; Makhalanyane, T.P.; Aislabie, J.M.; Cowan, D.A. Draft Genome Sequence of the Aromatic Hydrocarbon-Degrading Bacterium Sphingobium sp. Strain Ant17, Isolated from Antarctic Soil. Genome Announc. 2014, 2, 2. [Google Scholar] [CrossRef] [PubMed]
- Nagashima, H.; Zulkharnain, A.B.; Maeda, R.; Fuse, H.; Iwata, K.; Omori, T. Cloning and Nucleotide Sequences of Carbazole Degradation Genes from Marine Bacterium Neptuniibacter sp. Strain CAR-SF. Curr. Microbiol. 2010, 61, 50–56. [Google Scholar] [CrossRef] [PubMed]



| Strain | Similarity (%) | Accession No. |
|---|---|---|
| Sphingobium algorifonticola TLA-22T | 97.945 | RZUL01000025 |
| Sphingobium subterraneum II-13T | 97.449 | FJ796422 |
| Sphingobium boeckii 469T | 97.236 | JN591315 |
| Sphingobium aromaticiconvertens DSM 12677T | 97.210 | AM181012 |
| Sphingobium phenoxybenzoativorans SC_3T | 97.165 | MINO01000024 |
| Feature | Count/Value |
|---|---|
| Sequenced genome size (bp) | 4,773,423 |
| GC content (%) | 60.51% |
| Number of contigs | 96 |
| Length of longest contig (bp) | 490,141 |
| Number of protein coding genes | 4690 |
| Number of rRNA | 4 |
| Number of tRNA | 47 |
| Number of non-coding RNA | 66 |
| Strain | Genome Size (Mb) | GC (%) | Number of Scaffolds | Number of CDS | Source | Reference |
|---|---|---|---|---|---|---|
| Sphingobium sp. BS19 | 4.77342 | 60.5 | 96 | 4690 | Antarctic soil | This study |
| Sphingobium sp. SYK-6 | 4.34813 | 65.6 | 2 | 3996 | wastewater | [23] |
| Sphingobium sp. PAMC28499 | 4.88061 | 64.5 | 1 | 4458 | glacier | [24] |
| Sphingobium sp. YBL2 | 5.42742 | 64.4 | 7 | 5064 | soil | [25] |
| Sphingobium sp. KCTC 72723 | 4.42942 | 62.7 | 2 | 4191 | Antarctic soil | [26] |
| Sphingobium sp. TKS | 6.22888 | 63.1 | 11 | 5876 | soil | [27] |
| Sphingobium sp. CFD-2 | 6.02042 | 64.2 | 8 | 5673 | activated sludge | [28] |
| Sphingobium sp. CFD-1 | 4.26258 | 62.2 | 7 | 4171 | activated sludge | [29] |
| Sphingobium sp. RSMS | 5.14875 | 64.6 | 5 | 4758 | waste storage tank | [30] |
| Sphingobium sp. JS3065 | 5.11255 | 63.1 | 5 | 4740 | chemical plant | [31] |
| Sphingobium sp. MI1205 | 4.61937 | 62.3 | 6 | 4471 | soil | [32] |
| Sphingobium sp. RAC03 | 4.36975 | 62.9 | 5 | 4073 | algal phycosphere | [33] |
| Sphingobium sp. CAP-1 | 4.62247 | 64.2 | 5 | 4247 | activated sludge | [34] |
| Sphingobium sp. EP60837 | 4.26170 | 61.9 | 4 | 3971 | soil | * |
| Sphingobium sp. LF-16 | 4.57255 | 64.6 | 1 | 4203 | soil | * |
| Sphingobium sp. YG1 | 5.56389 | 63.1 | 4 | 5066 | marine sediment | * |
| NCBI Locus_Tag | Start | Stop | Strand | Length | Predicted Gene |
|---|---|---|---|---|---|
| Sbs19_23320 | 51,058 | 52,314 | + | 1257 | antA |
| Sbs19_23330 | 52,311 | 52,817 | + | 507 | antB |
| Sbs19_38690 | 45,253 | 44,720 | - | 534 | catR |
| Sbs19_38700 | 45,352 | 46,056 | + | 705 | catB_2 |
| Sbs19_38710 | 46,044 | 46,508 | + | 561 | catB_1 |
| Sbs19_38720 | 46,511 | 46,801 | + | 291 | catC |
| Sbs19_38730 | 46,840 | 47,769 | + | 930 | catA |
| Sbs19_38790 | 54,661 | 54,332 | - | 330 | carAc |
| Sbs19_38800 | 55,526 | 54,702 | - | 825 | carC |
| Sbs19_38810 | 56,372 | 55,569 | - | 804 | carBb |
| Sbs19_38820 | 56,646 | 56,365 | - | 282 | carBa |
| Sbs19_38830 | 57,782 | 56,646 | - | 1137 | carAa |
| Sbs19_38840 | 57,885 | 58,565 | + | 681 | carR |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sato, K.; Take, S.; Ahmad, S.A.; Gomez-Fuentes, C.; Zulkharnain, A. Carbazole Degradation and Genetic Analyses of Sphingobium sp. Strain BS19 Isolated from Antarctic Soil. Sustainability 2023, 15, 7197. https://doi.org/10.3390/su15097197
Sato K, Take S, Ahmad SA, Gomez-Fuentes C, Zulkharnain A. Carbazole Degradation and Genetic Analyses of Sphingobium sp. Strain BS19 Isolated from Antarctic Soil. Sustainability. 2023; 15(9):7197. https://doi.org/10.3390/su15097197
Chicago/Turabian StyleSato, Kenta, Seiryu Take, Siti Aqlima Ahmad, Claudio Gomez-Fuentes, and Azham Zulkharnain. 2023. "Carbazole Degradation and Genetic Analyses of Sphingobium sp. Strain BS19 Isolated from Antarctic Soil" Sustainability 15, no. 9: 7197. https://doi.org/10.3390/su15097197
APA StyleSato, K., Take, S., Ahmad, S. A., Gomez-Fuentes, C., & Zulkharnain, A. (2023). Carbazole Degradation and Genetic Analyses of Sphingobium sp. Strain BS19 Isolated from Antarctic Soil. Sustainability, 15(9), 7197. https://doi.org/10.3390/su15097197








