Abstract
Interest in soil health is growing, though the speed and effectiveness of management practices in improving it are uncertain. We measured biological, chemical, and physical indicators of soil health within a working farm zero, five, and nine years after transitioning from regular applications of inorganic fertilizers and pesticides to cover cropping, compost additions, organic amendments, and rotational grazing. We quantified microbial biomass and composition, soil organic matter (SOM), nutrient availabilities, and water stable aggregates in an avocado orchard, a citrus orchard, a pasture, and a vegetable garden. We found substantial and consistent increases in SOM, water stable aggregates, and microbial biomass, especially during the first five years, whereas nutrient availabilities showed no consistent change. Fungal and bacterial communities shifted but not fungal–bacterial biomass ratios or richness. However, fungal guilds responded differently to shifts in management. The biomass of arbuscular mycorrhizal fungi increased in most crops, and fungal saprotroph relative abundance and richness generally increased, whereas putative fungal pathogens showed the opposite response. Overall, we found substantial and rapid increases in indicators associated with improved soil health following the transition from conventional to regenerative management.
1. Introduction
Mechanical innovation, crop breeding, and the advent of synthetic fertilizers and pesticides have fueled a phenomenal increase in crop productivity during the last century, but there are signs that this increase is slowing [1]. This may, at least partly, be due to environmental degradation, biodiversity loss, and accelerated erosion and carbon loss associated with high-input, conventional agriculture [2,3,4,5]. In response, there has been a growing interest in sustainable food production, including the importance of soil health [6,7]. To what extent, and at what rate, soil health can be rebuilt remains unclear, which affects decisions and expectations associated with management.
The US Department of Agriculture defines soil health as “the continued capacity of soil to function as a vital living ecosystem that sustains plants, animals, and humans” (https://www.nrcs.usda.gov/wps/portal/nrcs/main/soils/health/ (accessed on 15 April 2024)). This definition recognizes soil’s role in producing food and providing essential regulating and supporting services, such as soil, water, and climate protection and biodiversity conservation [8]. But assessing a soil as “healthy” or having “high quality” is complex, site-specific, and sometimes hotly debated [9,10]. Soil organic matter (SOM) is commonly used as a primary indicator of soil health, as increases in SOM can enhance aggregate stability, improve water and nutrient holding capacity, and boost soil fertility and microbial biomass [6,11]. However, the total pool of SOM is large, and changes in SOM are challenging to detect and may not appear until irreversible damage occurs [12]. Soil quality assessment and monitoring tools have been emerging since the 1990s [12], incorporating dynamic indicators such as aggregate stability [13] and microbial biomass and composition [14]. However, the focus on short-term, varied responses across different crops and management practices has hindered our understanding of indicator relationships and their capacity to track the effects of long-term management [13].
USDA-NRCS highlights four approaches that can optimize soil health: (1) minimize disturbance; (2) maximize biodiversity, (3) soil cover, and (4) living roots (https://www.nrcs.usda.gov/conservation-basics/natural-resource-concerns/soils/soil-health (accessed on 15 April 2024)). These broadly align with the objectives of regenerative agriculture, which—while remaining poorly defined [15]—has soil health and building SOM as core themes [16]. Regenerative practices include crop rotation, cover cropping, livestock integration, and using manure and compost instead of chemical inputs, while reducing or eliminating synthetic pesticides [17]. These practices contribute to soil health in complementary ways. For example, crop rotations and cover crops add organic matter and protect soil from erosion. They may also support disease suppression by increasing temporal and spatial crop diversity, which often promotes microbial abundance and diversity [18,19,20]. Compost not only increases SOM and structure but may also suppress diseases through increased microbial activity [21,22,23]. Likewise, well-managed, short-duration grazing followed by longer recovery periods encourages plant growth and root development, promotes microbial biomass, and facilitates nutrient cycling via feces and urine deposition [24]. Our understanding of the rate these practices alter soil properties is limited, as is the consistency of changes across crops.
Soil biota are integral to biogeochemical cycles, redox reactions, disease suppression, and soil structure [23,25,26,27]. The biomass of soil biota is substantial, and one hectare of soil can contain over a metric tonne, dominated by the microbial groups of fungi and bacteria [28]. Soil biota also represent one of the largest reservoirs of biodiversity on Earth, estimated to be home to 59 ± 15% of all species [29]. Advances in molecular methods allow us to characterize this diversity and facilitate the study of non-culturable soil microbes. Using microbial composition or function as soil health indicators promises to offer faster, less expensive, and more informative measurements of composition and function than gross measurements, such as microbial biomass and respiration [14]. Management practices such as no-till and organic production systems promote soil biodiversity [8], which could increase resilience and multifunctionality [30]. Responses may also involve shifts in functional guilds, as illustrated by the increase in saprotrophic fungi with compost additions [31] and lack of responses or even suppression of symbiotic mycorrhizal fungi [32]. Regenerative practices that promote fungi over bacteria may increase SOM, soil structural and ecological stability, and nutrient retention [33,34]. Also, if shifts in management affect the availability of nutrients [8], this may change fungal mutualist to pathogen ratios [35]. Thus, responses by soil biota may be multifaceted and only fully understood by extending measurements beyond microbial biomass.
Here, we sampled a working farm in California zero, five, and nine years following a transition from conventional use of inorganic fertilizers and pesticides to regenerative practices that include compost additions, grazing, and cover cropping. We asked whether this shift would (1) change microbial community richness, composition, biomass, and potential function consistently across crops; (2) promote SOM accumulation and soil aggregate stability; and (3) affect nutrient availability. Our goal was to determine if, and how quickly, soils respond to shifts in management and whether responses were evident in a range of soil health indicators.
2. Methods
2.1. Site and Sampling Methods
Apricot Lane Farm, located in Moorpark, CA (34°18′47.81″ N, 118°55′22.58 W; elevation 239 m), is surrounded by mixed agricultural lands, with a historic mean annual precipitation of 333 mm (http://www.ncdc.noaa.gov (accessed on 10 October 2023)). The soil is a fine to gravely loamy mixture of Mollisols and Alfisols. The farm spans 87 hectares of more than 200 varieties of fruit and vegetable crops, and held organic (CCOF), biodynamic (Demeter), and regenerative organic (ROC) certifications during the study. The farm was managed using conventional methods prior to 2011 (see Supplemental Material for farm history). We sampled established Persea americana (hereafter avocado), established Citrus sp. (citrus), a high-diversity pasture (Avena faua, Bromus caharticus, Cynodon dactylon, Dactylis glomerata, Chenopodium album, Chicorium intybus; pasture), and a vegetable garden (garden), for a total of 4 crop types. Compost, organic amendments, and compost tea applications are applied annually (see Supplemental Material for inputs). Chickens, ducks, sheep, and cattle were rotated through orchards and pastures, but not the garden (Figure 1).

Figure 1.
Contrasting practices with a citrus orchard under conventional management at an adjacent farm with spaces between trees sprayed with glyphosate three times per year to maintain bare soil with representative soil sample (a), compared to the regenerative farm in this study 10 years after implementation of cover cropping, rotational grazing, and compost amendments with representative soil sample (b).
We sampled soil in mid-September 2012, 2017, and 2021, representing 0, 5, and 9 years after transition to regenerative management. Within each crop, we randomly selected 10 locations (>5 m apart) and collected two soil cores per location (0–10 cm depth × 10 cm diameter), which were combined and passed through a 2 mm sieve. We use the term “crop” hereon, although we cannot separate effects of crops and fields as we only accessed one field per crop. We avoided areas of disturbance, compaction, feces, and edges. Surface debris was moved aside before sampling. Orchard soil samples were collected ~80 cm from trunks and 90° apart. We sampled two beds of five garden plants. Samples were transported at 4 °C and processed within 48 h. A subsample (15 mL) was freeze dried for quantification of microbial abundance using phospholipid fatty acids (PLFA) and neutral lipid fatty acids (NLFA), as well as bacterial and fungal community characterizations using amplicon sequencing (described below). The rest was air dried and measured for water stable aggregates (WSA, described below) and sent to Ward Laboratories (Kearney, NE, USA) for measurements of SOM (LOI% at 360 °C for 2 h, which will not affect inorganic carbon [36]), pH (water), PMerlich, K, S, Zn, Fe, Mn, Cu, Ca, Mg, Na, and cation exchange capacity (CEC).
2.2. Water Stable Aggregate and Microbial Analyses
We used a modified version of the wet aggregate stability method [37]. We added 4 g soil to a cup filled with DI H2O for 5 min, followed by 4 min of agitation using a sieve [37]. After drying and weighing the soil, we dispersed the remaining sample with 0.5% sodium hexametaphosphate to correct for sand, washed it through a 250 um sieve, then redried and reweighed it. Water stable aggregates were determined via this formula:
%WSA = (soil mass after aggregate disruption − sand mass)/initial soil mass × 100
We extracted PLFA [38] to quantify bacterial and fungal biomass [39] and NLFA to quantify AMF biomass [40]. We extracted ~2 g of freeze-dried soil with a chloroform–methanol–citrate buffer (1:2:0.8 v/v/v). We loaded extracted lipids onto silica gel columns (Bond Elut LRC, SI 100 mg; Varian, Palo Alto, CA, USA) and eluted lipid fractions with chloroform, acetone, and methanol. The chloroform fraction (NLFA) and the methanol fraction (PLFA) were then subjected to mild alkaline methanolysis to form fatty acid methyl esters (FAMEs). Samples were analyzed on an Agilent 6890N GC and Agilent 5973 MSD (Agilent Technologies, Palo Alto, CA, USA) equipped with a flame ionization detector. The 25 m long column was an Agilent HP-1 (dimethyl-polysiloxane; Agilent Technologies). The PLFA 18:2ω6,9 was used as an indicator for fungi; PLFA i15:0, a15:0, 15:0, i16:0, 16:1ω9, 17:1ω7t, i17:0, a17:0, 18:1w7, cy19:0 were used to indicate bacteria; and NLFA 16:1ω5 was used to indicate AMF, with size and retention times related to the internal standard 19:0 [39]. Fatty acid concentrations were converted to bacterial and fungal biomass using conversions in Frostegård and Bååth [39]. Finally, we divided microbial biomass with SOM to assess potential shifts in SOM quality with the assumption that quality is higher if more microbial biomass is detected per unit of SOM.
To determine if soils harbored distinct microbial communities, we extracted and sequenced bacterial and fungal DNA. Two-step PCR amplification for bacteria and fungi was performed as described elsewhere [40,41]. Briefly, we extracted genomic DNA from 250 mg of freeze-dried soil per sample with a DNeasy PowerSoil Pro DNA isolation kit (Qiagen, Germantown, MD, USA). For bacteria, we amplified the V4 region of the 16S SSU rRNA using the forward primer 515F-Y and reverse primer 806R [42,43]. For fungi, we amplified the internal transcribed spacer 2 (ITS2) region using a 1:1 combination of the forward primers ITS7 [44] and ITS7o [45] and the reverse primer ITS4 [46]. We attached unique barcodes and Nextera flowcell adapters using 15 cycles and 10× diluted PCR 1 products. Amplicons from PCR 2 were purified using AMPure XP beads (Beckman Coulter Genomics, Brae, CA, USA) and pooled based on band strength during gel electrophoresis. Amplicon libraries were sequenced using a MiSeq v2 kit (500 cycles) on a MiSeq sequencing platform (Illumina Inc., San Diego, CA, USA) at the University of Montana Genomics Core Facility.
Raw sequence data were processed using “Quantitative insights into microbial ecology 2” (QIIME2 version 2022.2; https://qiime2.org/ (accessed on 15 May 2023)); [47]. For fungi, we first extracted the ITS2 region from all sequences using the ITSxpress plugin [48]. Bacterial and fungal sequences were quality filtered and dereplicated using the q2-dada2 plugin [49], which uses nucleotide quality scores to produce amplicon sequence variants (ASVs) with 100% similarity representing the true biological variation within each sample. For bacteria, forward and reverse reads were quality-trimmed at 220 and 200 nucleotides before pairing. All ASVs were assigned taxonomy using the QIIME2 q2-feature-classifier-plugin (https://github.com/qiime2/q2-feature-classifier (accessed on 15 May 2023)) [50]. For fungal classification, we used the UNITE fungal database (version 8.3; Nilsson et al., [51]) with a 94% confidence level. For bacterial classification, we used the Genome Taxonomy Database (GTDB [52]) and a 70% confidence level [50]. Raw sequence files were submitted to the Sequence Read Archive (PRJNA1066881). Fungal and bacterial data were rarefied to depths of 2930 and 5100 sequences per sample, respectively.
To explore shifts in fungal guilds over time and to assess constancy across crops, we assigned putative function to ASVs identified to at least the genus-level using the FungalTraits database [53]. In all, an average of 65.2% of sequences per sample were assigned to guilds. Following the method of Schmidt et al. [54], we analyzed the relative abundance of sequences associated with saprotrophic lifestyles (soil, litter, dung, leaf, seed, fruit, wood, and unspecified) and plant pathogens. While this may not reflect absolute abundance due to differences in gene copy numbers among taxa and primer bias [45,55], sequence numbers correlate reasonably well with abundant taxa [56], and discrepancies affect samples equally.
2.3. Statistics
We used R software v.4.2.3 [57] for statistical analysis. We employed linear models with crops and year as fixed effects to examine differences in soil nutrient and structural data, as well as the microbial biomass, richness, and relative abundance of fungal guilds. The DHARMa package version 0.4.6 was used for model diagnostics [58]. Non-conforming data were log-transformed. The functions emmeans in the emmeans package version 1.4.6 [59] and cld in the multcomp package 1.4-25 [60] were used to assess pairwise comparisons using Tukey’s method for multiple comparisons. Relationships between WSA and SOM, between WSA and AMF biomass, and between microbial biomass and SOM were analyzed with Pearson correlation (α = 0.05 for all statistical analyses). To quantify differences in soil nutrients among samples, we performed a PCA using the prcomp function (Figure S1) and PERMANOVA using the vegan package and adonis2 function [61]. The first axis, SPC1, primarily represented gradients in sodium, phosphorus, sulfur, and potassium, while SPC2 represented gradients in zinc, copper, and manganese.
To evaluate differences in fungal and bacterial community composition, we performed PERMANOVA on Bray–Curtis distances of Hellinger-transformed, rarefied sequence abundances using 1000 permutations with the adonis2 function in the vegan package. Main effects included year, crops, and their interaction. To visualize differences in microbial composition among crops and years, we performed Nonmetric Multidimensional Scaling (NMDS) using the metaMDS function in the vegan package and plotted results using the ggplot2 package [62]. To assess relationships between microbial composition, soil nutrients (SPC1 and SPC2), CEC, SOM, pH, and WSA, we used the envfit function to fit significantly correlated (p < 0.001) characteristics onto the ordinations. To identify individual taxa that may be driving shifts in guilds, we used ANCOM-BC2 [63,64], which controls for false discovery rate and potential biases associated with compositional data [65]. We collapsed fungal ASVs to genus-level and removed taxa present in fewer than 33% of samples within each crop. We did not use relative sequence abundance to assess AMF biomass because AMF sometimes do not amplify well with general fungal primers, especially in soil [66]. Instead, we used 16:1ω5 NLFA as this fatty acid reliably tracks AMF biomass [67]. We also did not assess potential shifts in bacterial function based on 16S rRNA because current databases are limited in coverage of bacterial genomes from soil [68].
3. Results
3.1. Shifts in Biological Soil Health Indicators with Management
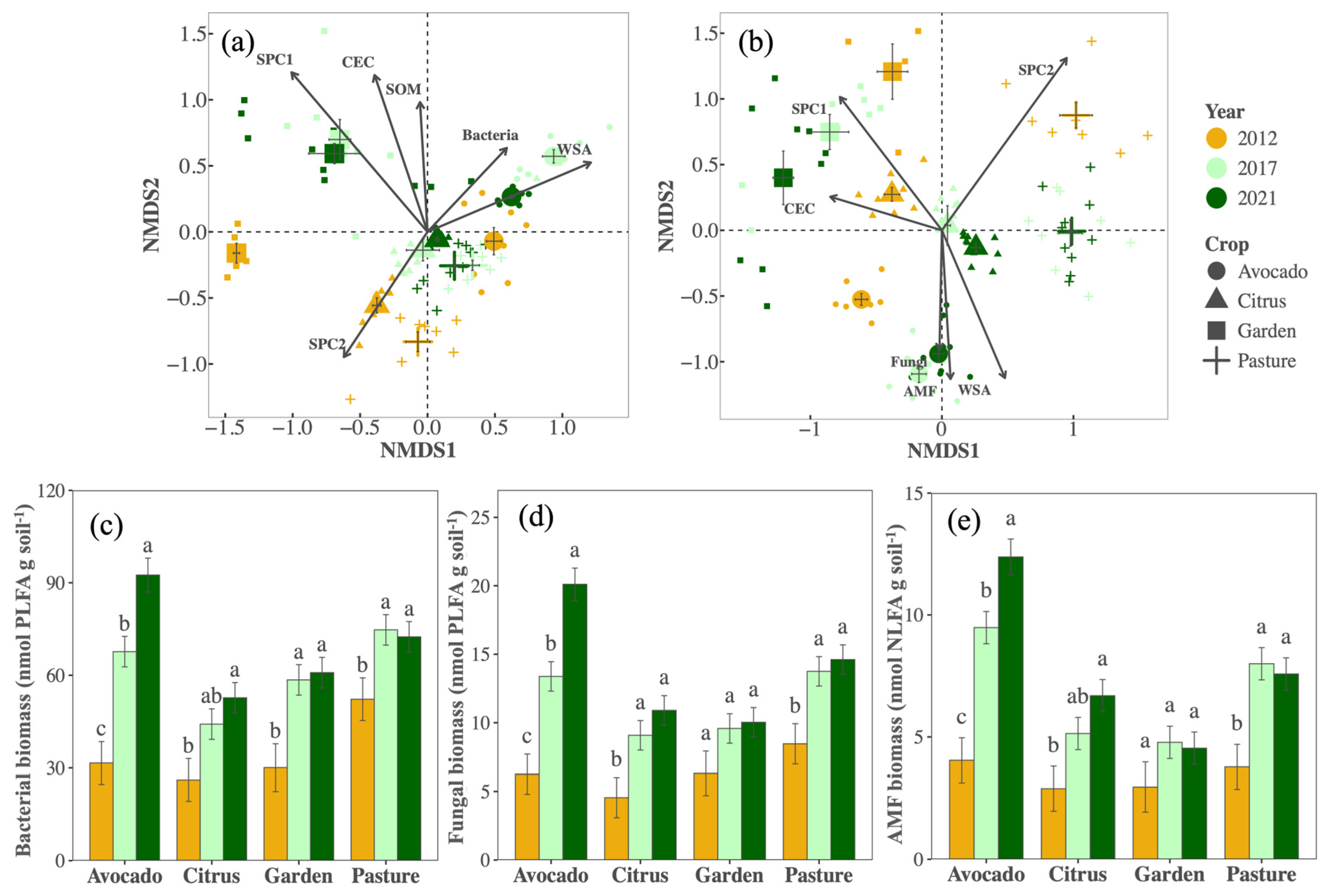
Contrary to expectations, bacterial and overall fungal richness did not change with management (Table S1). However, bacterial and fungal community composition shifted. For bacteria, responses varied somewhat by crop and year (PCrop×Year < 0.001, F = 2.81, R2 = 0.060, Figure 2a), with the bacterial community in the vegetable garden plots being more dissimilar and also changing more over time compared to other crops/fields. Generally, directional shifts in bacterial communities were consistent among crops, were more substantial during the first five years, and correlated with changes in bacterial biomass, WSA, SOM, CEC, and nutrient availability (SPC1 and SPC2). Fungal community composition also responded to changes in management, and responses differed among crops (PCrop×Year < 0.001, F = 3.62, R2 = 0.075) although fungal communities clustered more distinctly by crop/field than bacterial communities (Figure 2b). Fungal composition changes correlated with fungal and AMF biomass, WSA, CEC, and nutrients. Biomass for both bacteria and fungi generally increased over time across all crops, as did AMF biomass (Figure 2c–e). Despite these changes, there were no shifts in the fungi–bacteria ratios (PYear = 0.44).
Figure 2.
Community composition of bacterial (a) and fungal (b) communities, as well as biomass of bacteria (c), fungi (d), and arbuscular mycorrhizal fungi (e), in avocado (circles), citrus (triangles), garden (squares), and pasture (cross) soil samples collected in 2012 (orange), 2017 (light green), and 2021 (dark green). Large symbols in the ordinations represent averages, error bars represent standard errors, and vectors indicate significant (p < 0.001) variables, where CEC represents cation exchange capacity, SOM represents soil organic matter, WSA represents water stable aggregate, fungi, and bacteria, AMF represents biomass, and SPC1 and SPC2 represent the first and second axis in the PCA of soil available nutrients (Figure S1), where SPC1 primarily reflects P, K, S, and Na, and SPC2 primarily reflects Mn, Cu, and Zn. Different letters in figures (c–e) reflect significant (p < 0.05) differences in means within a crop.
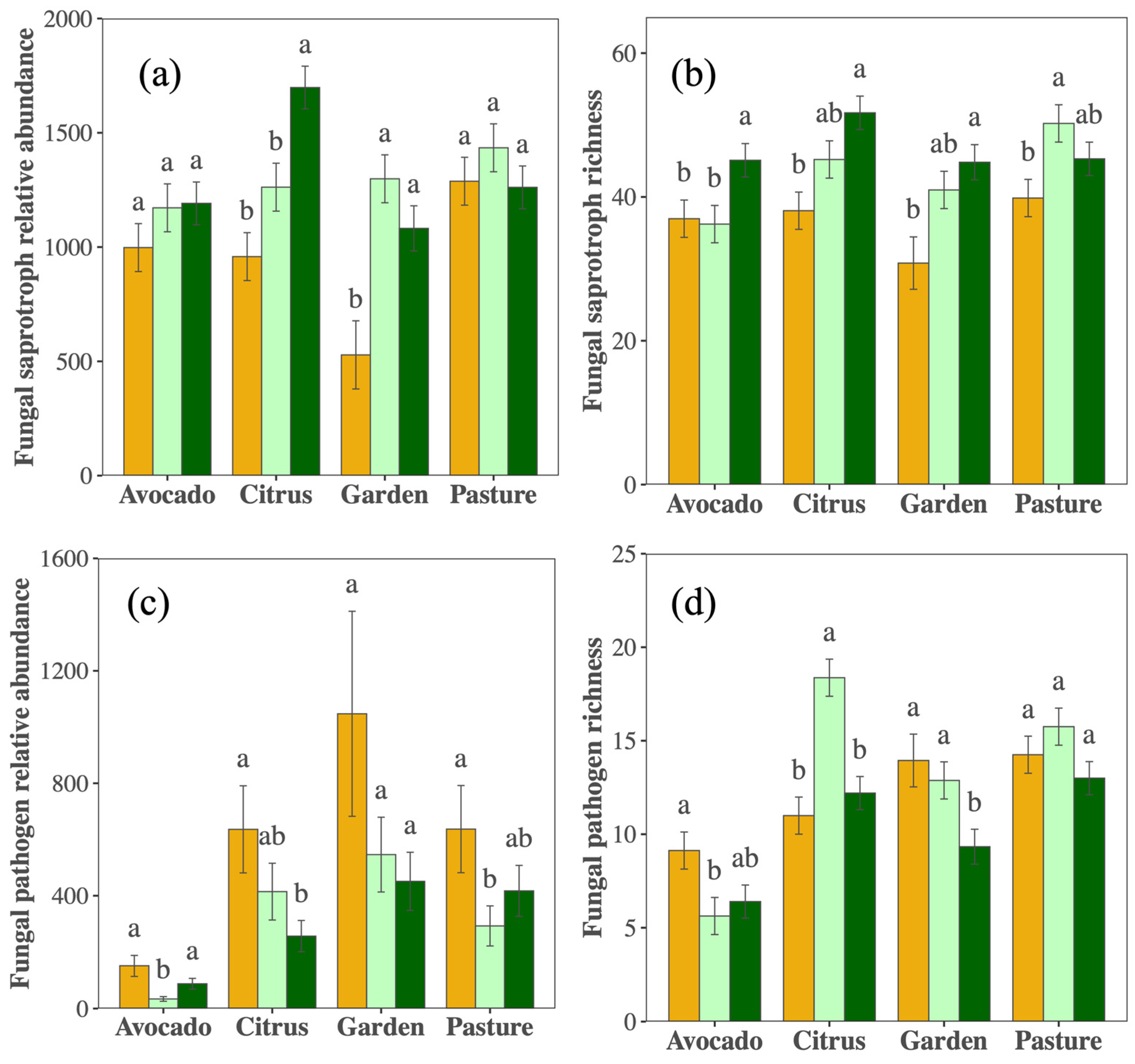
Although overall fungal richness remained stable over time, there were contrasting responses among fungal guilds. Putative saprotroph richness increased across crops, and with increased relative abundance in the citrus orchard and vegetable garden (Figure 3a,b, Table S2). Conversely, the relative abundance of putative pathogens decreased over the sampling period (PYear < 0.001), with significant reductions in pathogen richness in the vegetable garden and a trend in the avocado orchard (Figure 3c,d, Table S2). Indicator species driving these responses are presented in Table S3.
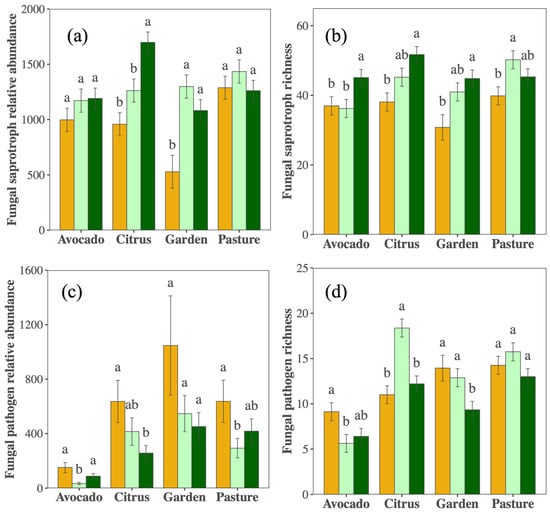
Figure 3.
Relative sequence abundance and ASV richness of fungal saprobes (a,b) and pathogens (c,d) in 2012 (orange), 2017 (light green), and 2021 (dark green) in the citrus, avocado, pasture, and garden. Different letters indicate differences (p < 0.05) within cropping types among years.
3.2. Shifts in Chemical and Physical Soil Health Indicators with Management
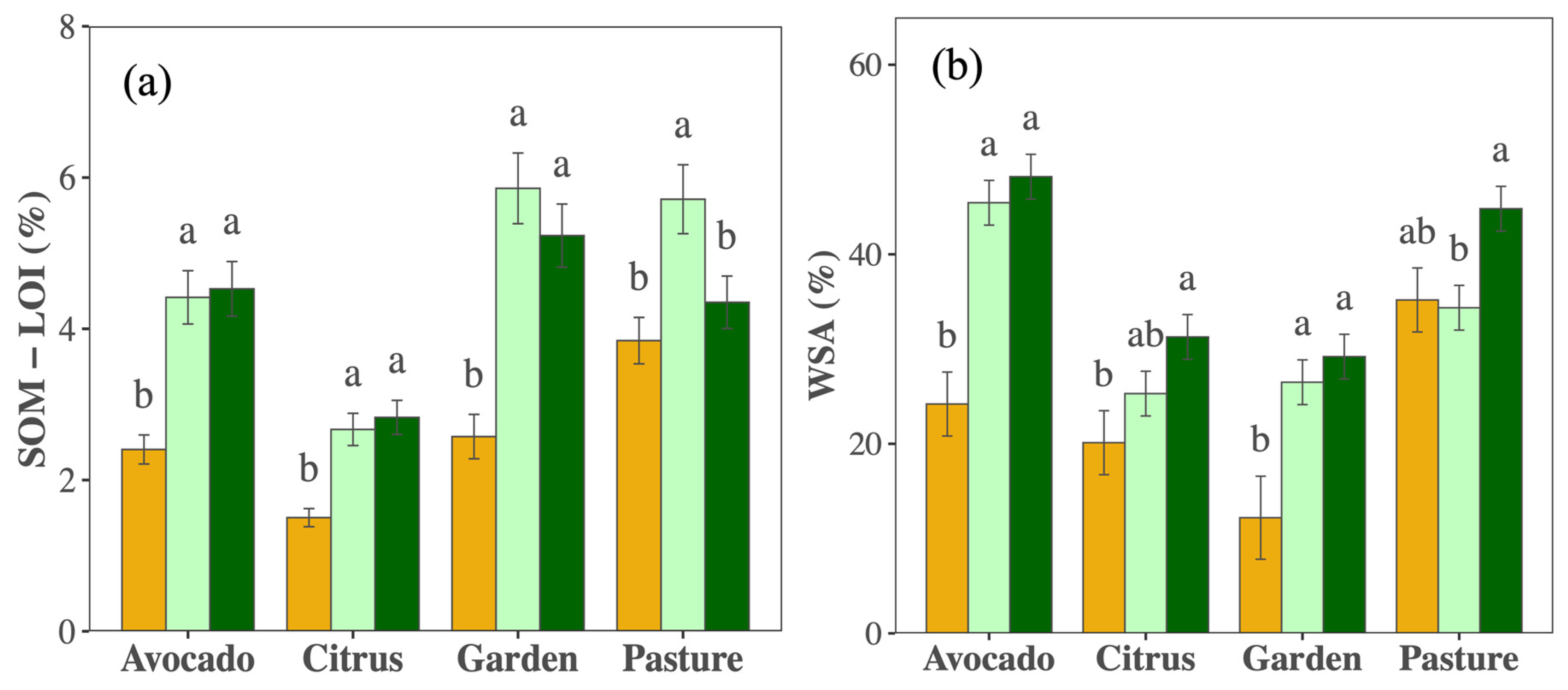
Management changes promoted SOM; though the extent varied by crop and year (PCrop×Year < 0.010, Figure 4a, Table S2), it increased substantially in all crops except in the pasture field, with the most dramatic increases in the first five years. Nutrient availabilities differed greatly among crops/fields (Table S2) but did not change consistently over time (Figure S1). Soil pH remained stable except in the vegetable garden, where it decreased (Table S2). Cation exchange capacity increased in all crops except the citrus orchard (Table S2). Shifts in WSA largely mirrored changes in SOM and depended on crop and year (PCrop×Year = 0.012; Figure 4b).
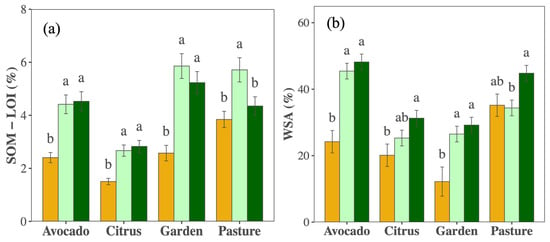
Figure 4.
Mean (±SE) soil organic matter as measured by percentage loss on ignition (SOM—LOI % (a)) and water stable aggregates (WSA % (b)) in the top 0–10 cm soil sampled in 2012 (orange), 2017 (light green), and 2021 (dark green) in citrus, avocado, pasture, and garden plots. Different letters indicate differences among years within crop types (p < 0.05).
3.3. Relationships among Indicators
Many indicators demonstrated interrelated changes. For example, the combined biomass of fungi and bacteria correlated positively with SOM (p < 0.001, R = 0.42), likely due to increased substrate availability. Microbial biomass per unit of SOM generally increased over time, notably in the avocado orchard (Table S2, Figure S2), suggesting an increased substrate quality with management changes. Aggregate stability also correlated positively with SOM (p < 0.001, R = 0.36), overall microbial biomass (p < 0.001, R = 0.58), and AMF biomass (p < 0.001, R = 0.39). These modest R-values likely reflect the complex nature of soil systems, where multiple chemical and biological factors interact to influence soil physical properties.
4. Discussion
Increasingly, conventional agricultural practices are recognized for depleting SOM and reducing microbial diversity, potentially impairing sustainable plant productivity and ecosystem services [69]. Over nine years, we assessed the extent and rate that shifts from conventional to regenerative management changed soil properties within a working farm in California, USA. Surprisingly, estimates of bacterial and fungal diversity did not change. However, microbial biomass, SOM, and aggregate stability—some of the most common biological, chemical, and physical indicators of soil health [12]—increased across most crops, whereas changes in nutrient availabilities were less consistent. We also found shifts in bacterial communities and fungal guilds, with a general increase in mycorrhizal biomass and saprotroph relative abundances and a reduction of putative plant pathogens, potentially suppressing disease. Changes were more pronounced in the first five years than in the subsequent period, suggesting rapid responses to management changes that likely benefit soil health.
4.1. Shifts in Management Changed Biological Indicators in Ways That Appear Beneficial
We observed a 38–178% increase in overall microbial biomass across crops and years. Contrary to expectations, however, we did not observe an increased fungal dominance with shifts from conventional to more regenerative practices [33] as both bacterial and fungal biomass rose. This stability in fungal–bacterial ratios suggests trait overlap between the groups, challenging the reliability of this ratio as a soil health metric [70,71]. What specifically caused the increase in overall microbial biomass is unknown as several management practices were bundled. However, organic amendments have promoted microbial biomass across a range of agricultural studies previously [22], and the substantial compost additions likely played a primary role. Sustaining higher microbial biomass might require frequent amendments, as effects may be fleeting [21]. However, even small quantities of amendments can yield significant benefits [22]. Beyond quantity, SOM quality could change. In this study, management shifts coincided with increased microbial biomass per unit SOM in most crops, suggesting improved SOM quality. Since many studies measure microbial biomass and SOM, incorporating this ratio could provide a valuable additional metric to assess changes in soil health.
Maximizing biodiversity is often highlighted as one key aspect of promoting soil health because higher microbial richness is associated with increased functional diversity and resilience [30]. While richness can be higher in organic than in conventional systems [8,72], we found no change in bacterial and fungal richness despite substantial increases in biomass and altered composition. At least for fungi, this may be due to opposite responses among guilds because saprotroph richness increased while pathogen richness generally declined. This, incidentally, indicates that aggregate richness estimates can obscure guild-level responses and therefore provide limited information related to soil health. The increase in saprotroph richness coincided with increased relative abundance, which was likely driven by the increased substrate availability and complexity from the compost additions and cover crops [31]. The reduced pathogen relative abundance and richness may result from improved plant health or antagonistic interactions with other soil biota [23]. AMF biomass also increased, except in the vegetable garden, where high soil P and frequent tillage may have suppressed abundance [73] (Table S2). Although AMF, as obligate symbionts, do not derive carbon from compost, additions can promote AMF abundance [74], possibly because AMF scavenge nutrients from organic-rich patches [75]. Overall, the increase in mutualist abundance relative to putative pathogens observed in most crops here aligns with predictions [76] and previous studies reporting reduced disease incidence following the implementation of regenerative practices [77,78]. Given that synthetic pesticides are seldom used in regenerative production systems [17], disease suppression, though underlying mechanisms are not fully understood [77], could be an important component of soil health.
4.2. Rapid and Consistent Shifts in Chemical and Physical Soil Health Indicators
Changes in SOM pools are predicted to be hard to detect [12], yet we recorded a 13–100% increase across crops within five years, suggesting that changes can be substantial and rapid. The smallest increase was in the pasture, where initial SOM levels were highest. This increase in SOM likely promoted nutrient holding capacity and aggregate stability directly, as well as indirectly by feeding soil biota [6,11]. As an increase in SOM from 1% to 3% can double plant water availability and retention [78], this represents an increasingly important ecosystem service given California’s water restrictions and erratic weather associated with climate change.
The combination of several management practices on this farm prevents us from assessing their relative importance. However, the doubling of SOM in the garden, where cover crops were rare and grazers absent, suggests the large compost additions were especially important. This is supported by findings from seven California farms, where similar compost applications resulted in soil carbon values three times higher than controls and increased microbial activity and water holding capacity [79]. SOM cannot increase in perpetuity, however, and depending on the quantity and quality of inputs, as well as soil structure and texture [80,81], the soil will become saturated. The slower accumulation and even slight declines between 2017 and 2021 across all crops may indicate saturation, though SOM could still increase in deeper layers, as observed in no-till agriculture [82]. Because soil contains twice as much carbon as the earth’s atmosphere [83], soil carbon sequestration could be an important regulating service. Whether the increase in SOM seen here represents sequestration requires knowledge about long-term stability, which depends on the input, aggregate stability, level of disturbance, and microbial composition and activity among other things [84]. Nonetheless, organic amendments applied to the soil surface were incorporated and coincided with rapid shifts in other parameters.
Increased aggregate stability likely improved the soil environment for roots and soil biota and increased infiltration. This will reduce crusting, ponding, and surface runoff during large storm events, leading to increased resilience to drought. The effect of organic matter additions on aggregate stability depends on the chemical composition, with short-term benefits likely deriving from decomposition of labile litter and microbial binding agents, whereas longer-term effects likely involve humified compounds [85]. The positive correlation between AMF abundance and aggregate stability suggests a potential role of these fungi as well, as documented previously [27].
Converting from inorganic fertilizer additions to systems reliant on organic matter turnover may alter nutrient availability, although previous work is inconsistent. For example, Ca, K, Mg, and Mn were higher with organic amendments compared with synthetic fertilizers in on-farm trials in Maryland and Virginia [86], and Ca, S, N, and P were higher in regenerative almond orchards in California [87]. But organic production systems had lower soil fertility than conventional production systems in experiments in Switzerland [8]. We observed differences in nutrient availabilities among crops, likely reflecting crop-specific needs, with no consistent overall shift over time. This aligns with a Norwegian study, which found that nutrient management in regenerative systems needs to be tailored to crops to optimize nutrient availability [88]. We also observed no directional shift in soil pH—arguably the most important driver of biogeochemical processes and microbial communities in soil [25,89]—as it remained stable except for the vegetable garden where it decreased. CEC, however, increased over time, which was likely driven by the increase in organic matter [90]. Overall, the promotion of SOM and aggregate stability without dramatic changes in nutrient availabilities is encouraging, as balancing nutrient supply and demand becomes challenging with non-synthetic fertilizers [91].
4.3. Limitations and Future Directions
This study was conducted on a working farm, which lends realism and limitations. First, the inclusion of multiple crops helped assess the generality of responses, but crops were not replicated and were spatially confounded, which means comparisons among crops should be interpreted cautiously. Second, without control areas maintaining conventional practices, we could not directly compare the effects of management changes on soil function, leaving open the possibility that observed shifts might be temporal rather than management-related. However, there is no reason to expect SOM to increase solely over time given that the avocado and citrus orchards were more than 20 years old. Finally, our focus was on soil health and thus we do not have information about yield responses. Yields are known to decrease, at least initially, when converting from conventional to regenerative practices, although the increased market value of crops may offset losses in income [8].
5. Conclusions
The importance of soil health is increasingly being recognized, but to what extent and at what rate soil health can be rebuilt remains uncertain. We show that conversion from conventional to regenerative practices can result in rapid and sustained shifts in many abiotic and biotic soil properties important for soil health, including soil organic matter, aggregate stability, and microbial biomass. Our results also highlight the potential of molecular tools—still underrepresented in soil health assessments [7,12]—to detect potential shifts among fungal guilds that could influence soil function.
Supplementary Materials
The following supporting information can be downloaded at https://www.mdpi.com/article/10.3390/su16135509/s1: Supplementary text: Farm history and inputs; Table S1: Fertilizer and amendments to the various crops over the study period; Table S2: Means (±SE) and statistics of abiotic and biotic properties in the four crops over time; Table S3: Indicator species analysis of fungal genera and lifestyles in the four crops; Figure S1: PCA of soil nutrient availabilities in the four crops over time; Figure S2: Microbial biomass per unit SOM in the four crops over time.
Author Contributions
Y.L. and P.W.R. conceived of the study; Y.L., M.E.D., M.M. and G.D.L.R. collected and processed the samples; M.M., M.E.D. and L.S.B. analyzed data; Y.L. wrote the first draft; and Y.L., M.M., L.S.B., M.E.D., S.G., J.R., P.W.R. and G.D.L.R. provided input on earlier drafts. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by MPG Ranch.
Institutional Review Board Statement
Not applicable.
Informed Consent Statement
Not applicable.
Data Availability Statement
All data required to repeat analyses are either already available (sequencing data) or will be uploaded to publicly available sites upon acceptance.
Acknowledgments
Y.L., M.M., L.S.B., M.E.D., G.D.L.R., S.G., and P.W.R. are thankful to MPG Ranch for funding, and J.R. thanks the Knut and Alice Wallenberg Foundation for support (KAW 2022.0175).
Conflicts of Interest
Authors Ylva Lekberg, Morgan McLeod, Lorinda S. Bullington, Mary Ellyn DuPre and Philip W. Ramsey were employed by the company MPG Ranch. Authors Gabriela De La Roca and Shawn Greenbaum were employed by the company Apricot Lane Farms. The remaining authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Pardey, P.G.; Alston, J.M. Unpacking the Agricultural Black Box: The Rise and Fall of American Farm Productivity Growth. J. Econ. Hist. 2021, 81, 114–155. [Google Scholar] [CrossRef]
- Broussard, W.; Turner, R.E. A Century of Changing Land-Use and Water-Quality Relationships in the Continental US. Front. Ecol. Environ. 2009, 7, 302–307. [Google Scholar] [CrossRef]
- Tibbett, M.; Fraser, T.D.; Duddigan, S. Identifying Potential Threats to Soil Biodiversity. PeerJ 2020, 8, e9271. [Google Scholar] [CrossRef] [PubMed]
- Galloway, J.N.; Cowling, E.B. Reactive Nitrogen and the World: 200 Years of Change. In Ambio; Royal Swedish Academy of Sciences: Stockholm, Sweden, 2002; Volume 31, pp. 64–71. [Google Scholar] [CrossRef]
- Montgomery, D.R. Soil Erosion and Agricultural Sustainability. Proc. Natl. Acad. Sci. USA 2007, 104, 13268–13272. [Google Scholar] [CrossRef] [PubMed]
- Karlen, D.L.; Veum, K.S.; Sudduth, K.A.; Obrycki, J.F.; Nunes, M.R. Soil Health Assessment: Past Accomplishments, Current Activities, and Future Opportunities. Soil Tillage Res. 2019, 195, 104365. [Google Scholar] [CrossRef]
- Lehmann, J.; Bossio, D.A.; Kögel-Knabner, I.; Rillig, M.C. The Concept and Future Prospects of Soil Health. In Nature Reviews Earth and Environment; Springer Nature: Berlin/Heidelberg, Germany, 2020; pp. 544–553. [Google Scholar] [CrossRef]
- Wittwer, R.A.; Bender, S.F.; Hartman, K.; Hydbom, S.; Lima, R.A.A.; Loaiza, V.; Nemecek, T.; Oehl, F.; Olsson, P.A.; Petchey, O.; et al. Organic and Conservation Agriculture Promote Ecosystem Multifunctionality. Sci. Adv. 2021, 7, eabg6995. [Google Scholar] [CrossRef]
- Letey, J.; Sojka, R.E.; Upchurch, D.R.; Cassel, D.K.; Olson, K.R.; Payne, W.A.; Petrie, S.E.; Price, G.H.; Reginato, R.J.; Scott, H.D.; et al. Deficiencies in the Soil Quality Concept and Its Application. J. Soil Water Conserv. 2003, 58, 180–187. [Google Scholar]
- Ritz, K.; Black, H.I.J.; Campbell, C.D.; Harris, J.A.; Wood, C. Selecting Biological Indicators for Monitoring Soils: A Framework for Balancing Scientific and Technical Opinion to Assist Policy Development. Ecol. Indic. 2009, 9, 1212–1221. [Google Scholar] [CrossRef]
- Lehman, R.M.; Cambardella, C.A.; Stott, D.E.; Acosta-Martinez, V.; Manter, D.K.; Buyer, J.S.; Maul, J.E.; Smith, J.L.; Collins, H.P.; Halvorson, J.J.; et al. Understanding and Enhancing Soil Biological Health: The Solution for Reversing Soil Degradation. Sustainability 2015, 7, 988–1027. [Google Scholar] [CrossRef]
- Bünemann, E.K.; Bongiorno, G.; Bai, Z.; Creamer, R.E.; De Deyn, G.; de Goede, R.; Fleskens, L.; Geissen, V.; Kuyper, T.W.; Mäder, P.; et al. Soil Quality—A Critical Review. Soil Biol. Biochem. 2018, 120, 105–125. [Google Scholar] [CrossRef]
- Stewart, R.D.; Jian, J.; Gyawali, A.J.; Thomason, W.E.; Badgley, B.D.; Reiter, M.S.; Strickland, M.S. What We Talk about When We Talk about Soil Health. Agric. Environ. Lett. 2018, 3, 180033. [Google Scholar] [CrossRef]
- Sahu, P.K.; Singh, D.P.; Prabha, R.; Meena, K.K.; Abhilash, P.C. Connecting Microbial Capabilities with the Soil and Plant Health: Options for Agricultural Sustainability. Ecol. Indic. 2019, 105, 601–612. [Google Scholar] [CrossRef]
- Giller, K.E.; Hijbeek, R.; Andersson, J.A.; Sumberg, J. Regenerative Agriculture: An Agronomic Perspective. Outlook Agric. 2021, 50, 13–25. [Google Scholar] [CrossRef] [PubMed]
- Schreefel, L.; Schulte, R.P.O.; de Boer, I.J.M.; Schrijver, A.P.; van Zanten, H.H.E. Regenerative Agriculture—The Soil Is the Base. Glob. Food Sec. 2020, 26, 100404. [Google Scholar] [CrossRef]
- Khangura, R.; Ferris, D.; Wagg, C.; Bowyer, J. Regenerative Agriculture—A Literature Review on the Practices and Mechanisms Used to Improve Soil Health. Sustainability 2023, 15, 2338. [Google Scholar] [CrossRef]
- Shah, K.K.; Modi, B.; Pandey, H.P.; Subedi, A.; Aryal, G.; Pandey, M.; Shrestha, J. Diversified Crop Rotation: An Approach for Sustainable Agriculture Production. Adv. Agric. 2021, 2021, 8924087. [Google Scholar] [CrossRef]
- Sharma, P.; Singh, A.; Kahlon, C.S.; Brar, A.S.; Grover, K.K.; Dia, M.; Steiner, R.L. The Role of Cover Crops towards Sustainable Soil Health and Agriculture—A Review Paper. Am. J. Plant Sci. 2018, 9, 1935–1951. [Google Scholar] [CrossRef]
- McDaniel, M.D.; Tiemann, L.K.; Grandy, A.S. Does Agricultural Crop Diversity Enhance Soil Microbial Biomass and Organic Matter Dynamics? A Meta-Analysis. Ecol. Appl. 2014, 24, 560–570. [Google Scholar] [CrossRef] [PubMed]
- Siedt, M.; Schäffer, A.; Smith, K.E.C.; Nabel, M.; Roß-Nickoll, M.; van Dongen, J.T. Comparing Straw, Compost, and Biochar Regarding Their Suitability as Agricultural Soil Amendments to Affect Soil Structure, Nutrient Leaching, Microbial Communities, and the Fate of Pesticides. Sci. Total Environ. 2021, 751, 141607. [Google Scholar] [CrossRef]
- Kallenbach, C.; Grandy, A.S. Controls over Soil Microbial Biomass Responses to Carbon Amendments in Agricultural Systems: A Meta-Analysis. Agric. Ecosyst. Environ. 2011, 144, 241–252. [Google Scholar] [CrossRef]
- Bonanomi, G.; Lorito, M.; Vinale, F.; Woo, S.L. Organic Amendments, Beneficial Microbes, and Soil Microbiota: Toward a Unified Framework for Disease Suppression. Annu. Rev. Phytopathol. Annu. Rev. Phytopathol. 2018, 56, 1–20. [Google Scholar] [CrossRef]
- Teague, R.; Kreuter, U. Managing Grazing to Restore Soil Health, Ecosystem Function, and Ecosystem Services. Front. Sustain. Food Syst. 2020, 4, 534187. [Google Scholar] [CrossRef]
- Fierer, N. Embracing the Unknown: Disentangling the Complexities of the Soil Microbiome. Nat. Rev. Microbiol. 2017, 15, 579–590. [Google Scholar] [CrossRef] [PubMed]
- Delavaux, C.S.; Smith-Ramesh, L.M.; Kuebbing, S.E. Beyond Nutrients: A Meta-Analysis of the Diverse Effects of Arbuscular Mycorrhizal Fungi on Plants and Soils. Ecology 2017, 98, 2111–2119. [Google Scholar] [CrossRef]
- Rillig, M.C.; Mummey, D.L. Mycorrhizas and Soil Structure. New Phytol. 2006, 171, 41–53. [Google Scholar] [CrossRef] [PubMed]
- He, L.; Mazza Rodrigues, J.L.; Soudzilovskaia, N.A.; Barceló, M.; Olsson, P.A.; Song, C.; Tedersoo, L.; Yuan, F.; Yuan, F.; Lipson, D.A.; et al. Global Biogeography of Fungal and Bacterial Biomass Carbon in Topsoil. Soil. Biol. Biochem. 2020, 151, 108024. [Google Scholar] [CrossRef]
- Anthony, M.A.; Bender, S.F.; van der Heijden, M.G.A. Enumerating Soil Biodiversity. Proc. Natl. Acad. Sci. USA 2023, 120, e2304663120. [Google Scholar] [CrossRef]
- Wagg, C.; Bender, S.F.; Widmer, F.; Van Der Heijden, M.G.A. Soil Biodiversity and Soil Community Composition Determine Ecosystem Multifunctionality. Proc. Natl. Acad. Sci. USA 2014, 111, 5266–5270. [Google Scholar] [CrossRef] [PubMed]
- Dang, Q.; Wang, Y.; Xiong, S.; Yu, H.; Zhao, X.; Tan, W.; Cui, D.; Xi, B. Untangling the Response of Fungal Community Structure, Composition and Function in Soil Aggregate Fractions to Food Waste Compost Addition. Sci. Total Environ. 2021, 769, 145248. [Google Scholar] [CrossRef]
- Cavagnaro, T.R. Impacts of Compost Application on the Formation and Functioning of Arbuscular Mycorrhizas. Soil Biol. Biochem. 2014, 78, 38–44. [Google Scholar] [CrossRef]
- Six, J.; Frey, S.D.; Thiet, R.K.; Batten, K.M. Bacterial and Fungal Contributions to Carbon Sequestration in Agroecosystems. Soil Sci. Soc. Am. J. 2006, 70, 555–569. [Google Scholar] [CrossRef]
- Wardle, D.A.; Bardgett, R.D.; Klironomos, J.N.; Setälä, H.; Wim, H.; Der Putten, V.; Wall, D.H. Ecological Linkages between Aboveground and Belowground Biota. Science 2004, 304, 1629–1633. [Google Scholar] [CrossRef] [PubMed]
- Lekberg, Y.; Arnillas, C.A.; Borer, E.T.; Bullington, L.S.; Fierer, N.; Kennedy, P.G.; Leff, J.W.; Luis, A.D.; Seabloom, E.W.; Henning, J.A. Nitrogen and Phosphorus Fertilization Consistently Favor Pathogenic over Mutualistic Fungi in Grassland Soils. Nat. Commun. 2021, 12, 3484. [Google Scholar] [CrossRef] [PubMed]
- Wang, Q.; Li, Y.; Wang, Y. Optimizing the Weight Loss-on-Ignition Methodology to Quantify Organic and Carbonate Carbon of Sediments from Diverse Sources. Environ. Monit. Assess. 2011, 174, 241–257. [Google Scholar] [CrossRef]
- Bourget, J.; Kemp, J.G. Wet Sieving Apparatus for Stability Analysis of Soil Aggregates. Can. J. Soil Sci. 1957, 37, 60–61. [Google Scholar] [CrossRef]
- Frostegård, Å.; Tunlid, A.; Bååth, E. Phospholipid Fatty Acid Composition, Biomass, and Activity of Microbial Communities from Two Soil Types Experimentally Exposed to Different Heavy Metals. Appl. Environ. Microbiol. 1993, 59, 3605–3617. [Google Scholar] [CrossRef] [PubMed]
- Frostegård, A.; Bååth, E. The Use of Phospholipid Fatty Acid Analysis to Estimate Bacterial and Fungal Biomass in Soil; Springer: Berlin/Heidelberg, Germany, 1996; Volume 22. [Google Scholar]
- McTee, M.R.; Lekberg, Y.; Mummey, D.; Rummel, A.; Ramsey, P.W. Do Invasive Plants Structure Microbial Communities to Accelerate Decomposition in Intermountain Grasslands? Ecol. Evol. 2017, 7, 11227–11235. [Google Scholar] [CrossRef] [PubMed]
- Bullington, L.S.; Lekberg, Y.; Sniezko, R.; Larkin, B. The Influence of Genetics, Defensive Chemistry and the Fungal Microbiome on Disease Outcome in Whitebark Pine Trees. Mol. Plant Pathol. 2018, 19, 1847–1858. [Google Scholar] [CrossRef] [PubMed]
- Apprill, A.; Mcnally, S.; Parsons, R.; Weber, L. Minor Revision to V4 Region SSU RRNA 806R Gene Primer Greatly Increases Detection of SAR11 Bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every Base Matters: Assessing Small Subunit RRNA Primers for Marine Microbiomes with Mock Communities, Time Series and Global Field Samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Ihrmark, K.; Bödeker, I.T.M.; Cruz-Martinez, K.; Friberg, H.; Kubartova, A.; Schenck, J.; Strid, Y.; Stenlid, J.; Brandström-Durling, M.; Clemmensen, K.E.; et al. New Primers to Amplify the Fungal ITS2 Region—Evaluation by 454-Sequencing of Artificial and Natural Communities. FEMS Microbiol. Ecol. 2012, 82, 666–677. [Google Scholar] [CrossRef] [PubMed]
- Kohout, P.; Sudová, R.; Janoušková, M.; Čtvrtlíková, M.; Hejda, M.; Pánková, H.; Slavíková, R.; Štajerová, K.; Vosátka, M.; Sýkorová, Z. Comparison of Commonly Used Primer Sets for Evaluating Arbuscular Mycorrhizal Fungal Communities: Is There a Universal Solution? Soil Biol. Biochem. 2014, 68, 482–493. [Google Scholar] [CrossRef]
- White, T.; Bruns, T.D.; Lee, S.; Taylor, J. Amplification and Direct Sequencing of Fungal Ribosomal RNA Genes for Phylogenetics. In PCR Protocols: A Guide to Methods and Applications; Innis, M., Gelfand, D., Shinsky, J., White, T., Eds.; Academic Press: London, UK, 1990; pp. 315–322. [Google Scholar]
- Bolyen, E.; Rideout, J.R.; Dillon, M.R.; Bokulich, N.A.; Abnet, C.C.; Al-Ghalith, G.A.; Alexander, H.; Alm, E.J.; Arumugam, M.; Asnicar, F.; et al. Reproducible, Interactive, Scalable and Extensible Microbiome Data Science Using QIIME 2. Nat. Biotechnol. 2019, 37, 852–857. [Google Scholar] [CrossRef] [PubMed]
- Rivers, A.R.; Weber, K.C.; Gardner, T.G.; Liu, S.; Armstrong, S.D. ITSxpress: Software to Rapidly Trim Internally Transcribed Spacer Sequences with Quality Scores for Marker Gene Analysis. F1000Research 2018, 7, 1418. [Google Scholar] [CrossRef] [PubMed]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Bokulich, N.A.; Kaehler, B.D.; Rideout, J.R.; Dillon, M.; Bolyen, E.; Knight, R.; Huttley, G.A.; Gregory Caporaso, J. Optimizing Taxonomic Classification of Marker-Gene Amplicon Sequences with QIIME 2′s Q2-Feature-Classifier Plugin. Microbiome 2018, 6, 90. [Google Scholar] [CrossRef] [PubMed]
- Nilsson, R.H.; Larsson, K.H.; Taylor, A.F.S.; Bengtsson-Palme, J.; Jeppesen, T.S.; Schigel, D.; Kennedy, P.; Picard, K.; Glöckner, F.O.; Tedersoo, L.; et al. The UNITE Database for Molecular Identification of Fungi: Handling Dark Taxa and Parallel Taxonomic Classifications. Nucleic Acids Res. 2019, 47, D259–D264. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Chuvochina, M.; Rinke, C.; Mussig, A.J.; Chaumeil, P.A.; Hugenholtz, P. GTDB: An Ongoing Census of Bacterial and Archaeal Diversity through a Phylogenetically Consistent, Rank Normalized and Complete Genome-Based Taxonomy. Nucleic Acids Res. 2022, 50, D785–D794. [Google Scholar] [CrossRef] [PubMed]
- Põlme, S.; Abarenkov, K.; Henrik Nilsson, R.; Lindahl, B.D.; Clemmensen, K.E.; Kauserud, H.; Nguyen, N.; Kjøller, R.; Bates, S.T.; Baldrian, P.; et al. FungalTraits: A User-Friendly Traits Database of Fungi and Fungus-like Stramenopiles. Fungal Divers 2020, 105, 1–16. [Google Scholar] [CrossRef]
- Schmidt, R.; Mitchell, J.; Scow, K. Cover Cropping and No-till Increase Diversity and Symbiotroph:Saprotroph Ratios of Soil Fungal Communities. Soil Biol. Biochem. 2019, 129, 99–109. [Google Scholar] [CrossRef]
- Corradi, N.; Croll, D.; Colard, A.; Kuhn, G.; Ehinger, M.; Sanders, I.R. Gene Copy Number Polymorphisms in an Arbuscular Mycorrhizal Fungal Population. Appl. Environ. Microbiol. 2007, 73, 366–369. [Google Scholar] [CrossRef] [PubMed]
- Skelton, J.; Cauvin, A.; Hunter, M.E. Environmental DNA Metabarcoding Read Numbers and Their Variability Predict Species Abundance, but Weakly in Non-Dominant Species. Environ. DNA 2023, 5, 1092–1104. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria; Available online: https://www.r-project.org/ (accessed on 15 May 2024).
- Hartig, F. DHARMa: Residual Diagnostics for Hierarchical (Multi-Level/Mixed) Regression Models. R package version 0.4.6. Available online: https://cran.r-project.org/web/packages/DHARMa/vignettes/DHARMa.html (accessed on 15 May 2024).
- Lenth, R.; Singmann, H.; Love, J.; Buerkner, P.; Herve, M. Emmeans: Estimated Marginal Means, Aka Least-Squares Means. Available online: https://CRAN.R-project.org/package=emmeans (accessed on 15 May 2024).
- Hothorn, T.; Bretz, F.; Westfall, P. Simultaneous Inference in General Parametric Models Simultaneous Inference in General Parametric Models. Biom. J. J. Math. Methods Biosci. 2008, 50, 346–363. [Google Scholar]
- Oksanen, J.; Blanchet, F.G.; Friendly, M.; Kindt, R.; Legendre, P.; McGlinn, D.; Minchin, P.R.; O’Hara, R.B.; Simpson, G.L.; Solymos, P.; et al. Vegan: Community Ecology Package. 2020. Available online: https://www.researchgate.net/publication/346579465_vegan_community_ecology_package_version_25-7_November_2020 (accessed on 15 May 2024).
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016. [Google Scholar]
- Lin, H.; Das Peddada, S. Analysis of Compositions of Microbiomes with Bias Correction. Nat. Commun. 2020, 11, 3514. [Google Scholar] [CrossRef] [PubMed]
- Lin, H.; Eggesbø, M.; Das Peddada, S. Linear and Nonlinear Correlation Estimators Unveil Undescribed Taxa Interactions in Microbiome Data. Nat. Commun. 2022, 13, 4946. [Google Scholar] [CrossRef] [PubMed]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and Microbial Differential Abundance Strategies Depend upon Data Characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]
- Tedersoo, L.; Bahram, M.; Põlme, S.; Kõljalg, U.; Yorou, N.S.; Wijesundera, R.; Ruiz, L.V.; Vasco-Palacios, A.M.; Thu, P.Q.; Suija, A.; et al. Global Diversity and Geography of Soil Fungi. Science 2014, 346, 1052–1053. [Google Scholar] [CrossRef] [PubMed]
- Lekberg, Y.; Bååth, E.; Frostegård, Å.; Hammer, E.; Hedlund, K.; Jansa, J.; Kaiser, C.; Ramsey, P.W.; Řezanka, T.; Rousk, J.; et al. Fatty Acid 16:1ω5 as a Proxy for Arbuscular Mycorrhizal Fungal Biomass: Current Challenges and Ways Forward. Biol. Fertil. Soils 2022, 58, 835–842. [Google Scholar] [CrossRef]
- Baldrian, P. The Known and the Unknown in Soil Microbial Ecology. In FEMS Microbiology Ecology; Oxford University Press: Oxford, UK, 2019. [Google Scholar] [CrossRef]
- Bender, S.F.; Wagg, C.; van der Heijden, M.G.A. An Underground Revolution: Biodiversity and Soil Ecological Engineering for Agricultural Sustainability. Trends Ecol. Evol. 2016, 31, 440–452. [Google Scholar] [CrossRef]
- Fierer, N.; Wood, S.A.; Bueno de Mesquita, C.P. How Microbes Can, and Cannot, Be Used to Assess Soil Health. Soil Biol. Biochem. 2021, 153, 108111. [Google Scholar] [CrossRef]
- Strickland, M.S.; Rousk, J. Considering Fungal: Bacterial Dominance in Soils—Methods, Controls, and Ecosystem Implications. Soil Biol. Biochem. 2010, 42, 1385–1395. [Google Scholar] [CrossRef]
- Hartmann, M.; Frey, B.; Mayer, J.; Mäder, P.; Widmer, F. Distinct Soil Microbial Diversity under Long-Term Organic and Conventional Farming. ISME J. 2015, 9, 1177–1194. [Google Scholar] [CrossRef]
- Sheng, M.; Lalande, R.; Hamel, C.; Ziadi, N.; Shi, Y. Growth of Corn Roots and Associated Arbuscular Mycorrhizae Are Affected by Long-Term Tillage and Phosphorus Fertilization. Agron. J. 2012, 104, 1672–1678. [Google Scholar] [CrossRef]
- Hammer, E.C.; Nasr, H.; Wallander, H. Effects of Different Organic Materials and Mineral Nutrients on Arbuscular Mycorrhizal Fungal Growth in a Mediterranean Saline Dryland. Soil Biol. Biochem. 2011, 43, 2332–2337. [Google Scholar] [CrossRef]
- Hodge, A.; Fitter, A.H. Substantial Nitrogen Acquisition by Arbuscular Mycorrhizal Fungi from Organic Material Has Implications for N Cycling. Proc. Natl. Acad. Sci. USA 2010, 107, 13754–13759. [Google Scholar] [CrossRef]
- Hermans, S.M.; Lear, G.; Case, B.S.; Buckley, H.L. IScience The Soil Microbiome: An Essential, but Neglected, Component of Regenerative Agroecosystems. iScience 2023, 26, 106028. [Google Scholar] [CrossRef] [PubMed]
- Jayaraman, S.; Naorem, A.K.; Lal, R.; Dalal, R.C.; Sinha, N.K.; Patra, A.K.; Chaudhari, S.K. Disease-Suppressive Soils-Beyond Food Production: A Critical Review. J. Soil Sci. Plant Nutr. 2021, 21, 1437–1465. [Google Scholar] [CrossRef] [PubMed]
- Hudson, B.D. Soil Organic Matter and Available Water Capacity. J. Soil Water Conserv. 1994, 49, 189194. [Google Scholar]
- Brown, S.; Cotton, M. Changes in Soil Properties and Carbon Content Following Compost Application: Results of On-Farm Sampling. Compos. Sci. Util. 2011, 19, 87–96. [Google Scholar] [CrossRef]
- Six, J.; Conant, R.T.; Paul, E.A.; Paustian, K. Stabilization Mechanisms of Soil Organic Matter: Implications for C-Saturation of Soils. Plant Soil 2002, 241, 155–176. [Google Scholar] [CrossRef]
- Kong, A.Y.Y.; Six, J.; Bryant, D.C.; Denison, R.F.; van Kessel, C. The Relationship between Carbon Input, Aggregation, and Soil Organic Carbon Stabilization in Sustainable Cropping Systems. Soil Sci. Soc. Am. J. 2005, 69, 1078–1085. [Google Scholar] [CrossRef]
- Nicoloso, R.S.; Rice, C.W.; Amado, T.J.C.; Costa, C.N.; Akley, E.K. Carbon Saturation and Translocation in a No-till Soil under Organic Amendments. Agric. Ecosyst. Environ. 2018, 264, 73–84. [Google Scholar] [CrossRef]
- Smith, P.; Martino, D.; Cai, Z.; Gwary, D.; Janzen, H.; Kumar, P.; McCarl, B.; Ogle, S.; O’Mara, F.; Rice, C.; et al. Greenhouse Gas Mitigation in Agriculture. In Philosophical Transactions of the Royal Society B: Biological Sciences; Royal Society: London, UK, 2008; pp. 789–813. [Google Scholar] [CrossRef]
- Bailey, V.L.; Pries, C.H.; Lajtha, K. What Do We Know about Soil Carbon Destabilization? Environ. Res. Lett. 2019, 14, 083004. [Google Scholar] [CrossRef]
- Abiven, S.; Menasseri, S.; Chenu, C. The Effects of Organic Inputs over Time on Soil Aggregate Stability—A Literature Analysis. Soil Biol. Biochem. 2009, 41, 1–12. [Google Scholar] [CrossRef]
- Bulluck Iii, L.R.; Brosius, M.; Evanylo, G.K.; Ristaino, J.B.; Edu, J.R. Organic and Synthetic Fertility Amendments Influence Soil Microbial, Physical and Chemical Properties on Organic and Conventional Farms. Appl. Soil Ecol. 2002, 19, 147–160. [Google Scholar] [CrossRef]
- Fenster, T.L.D.; Oikawa, P.Y.; Lundgren, J.G. Regenerative Almond Production Systems Improve Soil Health, Biodiversity, and Profit. Front. Sustain. Food Syst. 2021, 5, 664359. [Google Scholar] [CrossRef]
- Bakken, A.K.; Breland, T.A.; Haraldsen, T.; Aamlid, T.; Sveistrup, T. Soil Fertility in Three Cropping Systems after Conversion from Conventional to Organic Farming. Acta Agric. Scand. B Soil Plant Sci. 2006, 56, 81–90. [Google Scholar] [CrossRef]
- Neina, D. The Role of Soil PH in Plant Nutrition and Soil Remediation. Appl. Environ. Soil Sci. 2019, 2019, 5794869. [Google Scholar] [CrossRef]
- Ramos, F.T.; Dores, E.F.d.C.; Weber, O.L.d.S.; Beber, D.C.; Campelo, J.H.; Maia, J.C.d.S. Soil Organic Matter Doubles the Cation Exchange Capacity of Tropical Soil under No-till Farming in Brazil. J. Sci. Food Agric. 2018, 98, 3595–3602. [Google Scholar] [CrossRef]
- Sacco, D.; Moretti, B.; Monaco, S.; Grignani, C. Six-Year Transition from Conventional to Organic Farming: Effects on Crop Production and Soil Quality. Eur. J. Agron. 2015, 69, 10–20. [Google Scholar] [CrossRef]
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).