Response of Soil Nitrogen-Cycling Genes to the Coupling Effects of Arbuscular Mycorrhizal Fungi Inoculation and Biochar Application in Maize Rhizosphere
Abstract
:1. Introduction
2. Material and Methods
2.1. Soil Characteristics
2.2. Materials of Maize, AM Fungus, and Biochar
2.3. Experiment Design
2.4. Samples Collection
2.5. Soil Microbial DNA Analysis
2.6. Statistical Analysis and Data Processing
3. Results
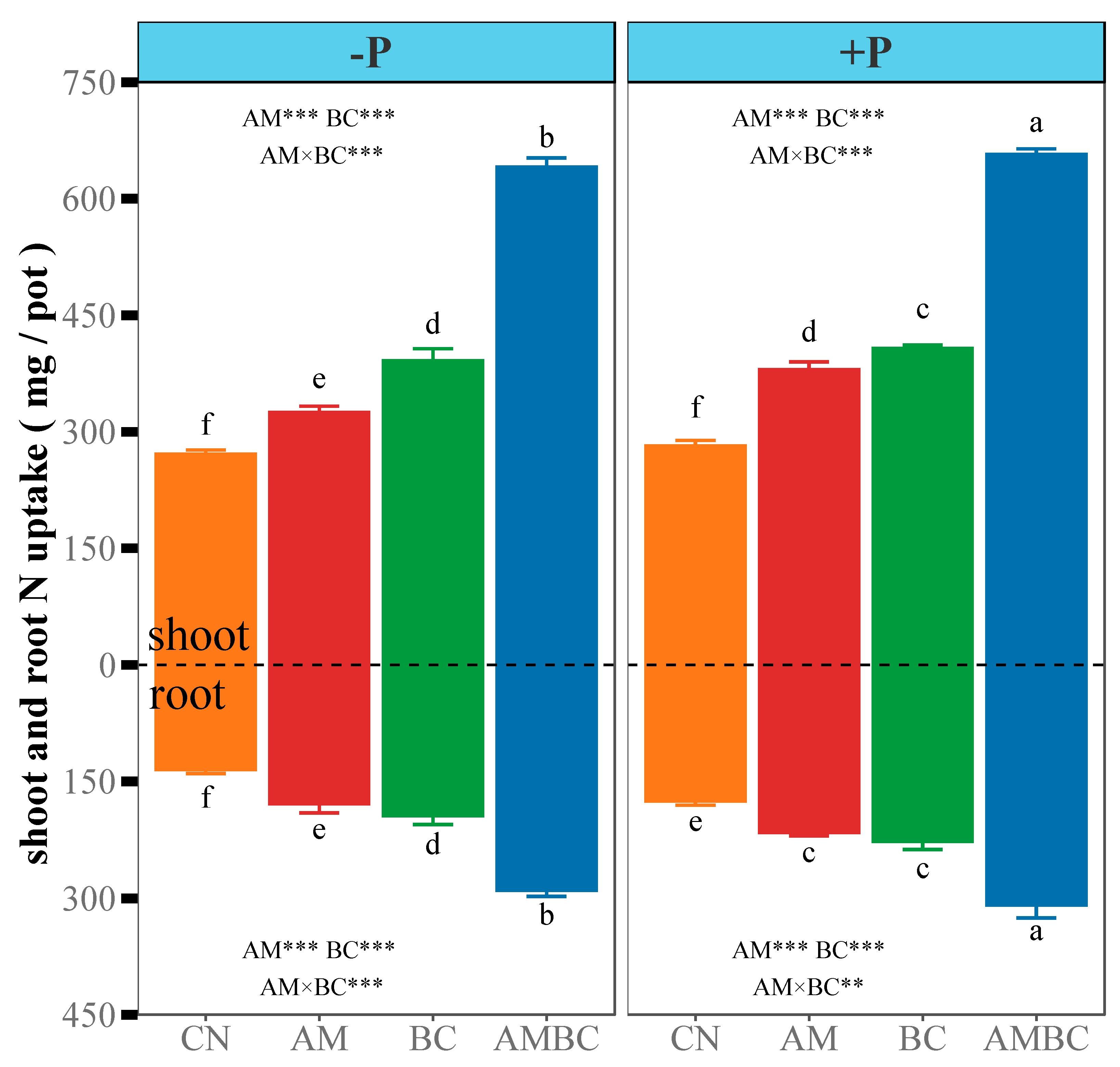
3.1. N Uptake and Root Morphology of Maize
3.2. Soil Microbial Network Analysis
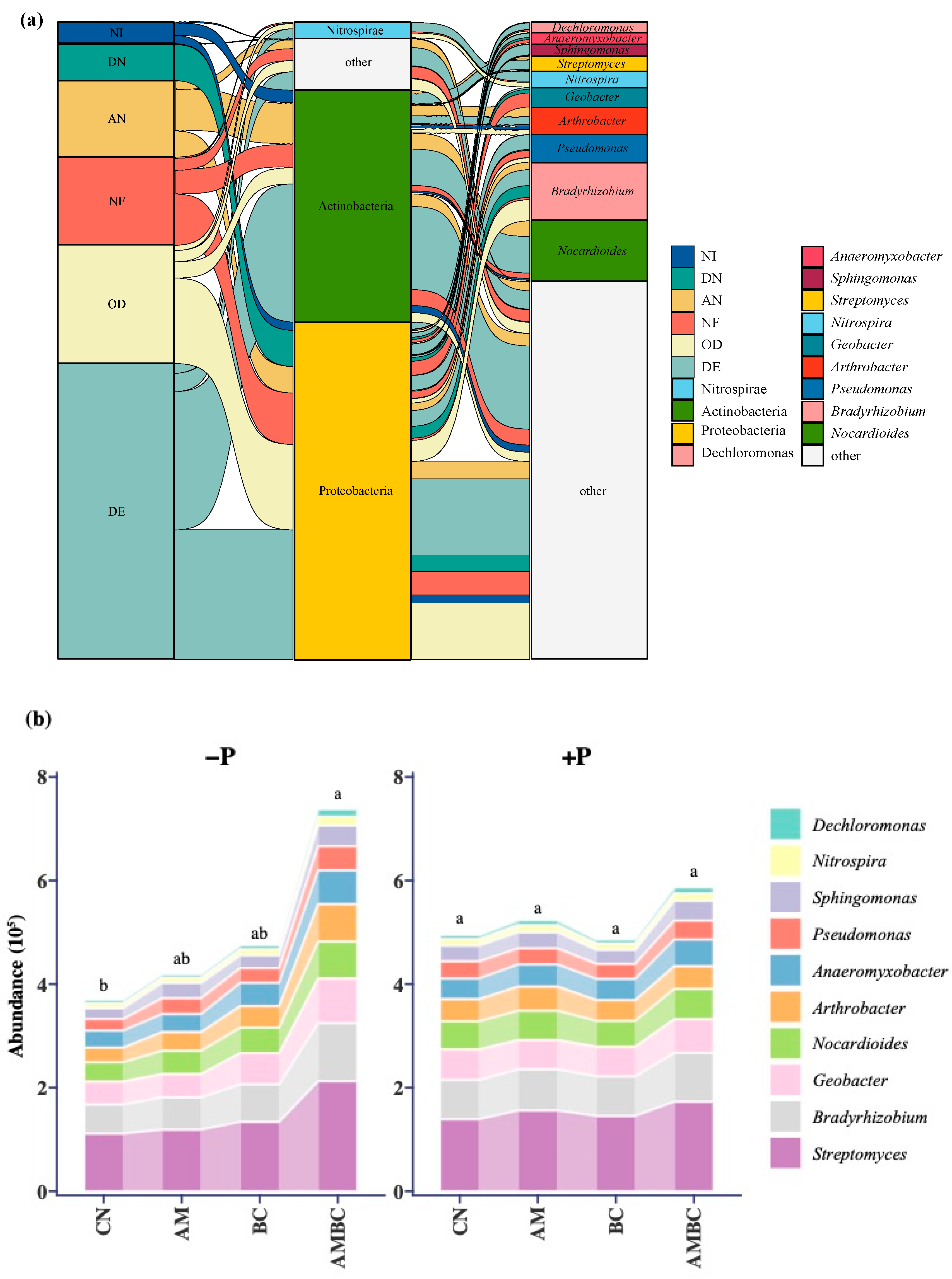
3.3. Abundances and Structures of N-Cycle Genes in Each Process
3.4. Analysis of the Composition of N-Cycle Genes in Soil Microbe
3.5. Correlation Analysis of Soil N Content and N-Cycling Genes
4. Discussion
4.1. Coupling Effects of AMF and Biochar on N Uptake and Root Morphology of Maize
4.2. Coupling Effects of AMF and Biochar on Soil Microbial Community
4.3. Coupling Effects of AMF and Biochar on the Abundance and Structure of N-Cycle Genes
4.4. Taxonomic Analysis of Composition of N-Cycle Genes and Mantel Test
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Zhao, B.; Jia, X.; Yu, N.; Murray, J.D.; Yi, K.; Wang, E. Microbe-dependent and independent nitrogen and phosphate acquisition and regulation in plants. New Phytol. 2023. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.; Zhang, L.; Feng, G.; George, T.S. Arbuscular mycorrhizal fungi have a greater role than root hairs of maize for priming the rhizosphere microbial community and enhancing rhizosphere organic P mineralization. Soil Biol. Biochem. 2022, 171, 108713. [Google Scholar] [CrossRef]
- Meng, L.; Zhang, A.; Wang, F.; Han, X.; Wang, D.; Li, S. Arbuscular mycorrhizal fungi and rhizobium facilitate nitrogen uptake and transfer in soybean/maize intercropping system. Front. Plant Sci. 2015, 6, 339. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Zhou, J.; George, T.S.; Limpens, E.; Feng, G. Arbuscular mycorrhizal fungi conducting the hyphosphere bacterial orchestra. Trends Plant Sci. 2022, 27, 402–411. [Google Scholar] [CrossRef] [PubMed]
- Hestrin, R.; Hammer, E.C.; Mueller, C.W.; Lehmann, J. Synergies between mycorrhizal fungi and soil microbial communities increase plant nitrogen acquisition. Commun. Biol. 2019, 2, 233. [Google Scholar] [CrossRef] [PubMed]
- Zhang, R.; Mu, Y.; Li, X.; Li, S.; Sang, P.; Wang, X.; Wu, H.; Xu, N. Response of the arbuscular mycorrhizal fungi diversity and community in maize and soybean rhizosphere soil and roots to intercropping systems with different nitrogen application rates. Sci. Total Environ. 2020, 740, 139810. [Google Scholar] [CrossRef] [PubMed]
- Zhu, C.; Tian, G.; Luo, G.; Kong, Y.; Guo, J.; Wang, M.; Guo, S.; Ling, N.; Shen, Q. N-fertilizer-driven association between the arbuscular mycorrhizal fungal community and diazotrophic community impacts wheat yield. Agric. Ecosyst. Environ. 2018, 254, 191–201. [Google Scholar] [CrossRef]
- Cheng, Z.; Meng, L.; Yin, T.; Li, Y.; Zhang, Y.; Li, S. Changes in Soil Rhizobia Diversity and Their Effects on the Symbiotic Efficiency of Soybean Intercropped with Maize. Agronomy 2023, 13, 997. [Google Scholar] [CrossRef]
- Sun, R.; Wang, F.; Hu, C.; Liu, B. Metagenomics reveals taxon-specific responses of the nitrogen-cycling microbial community to long-term nitrogen fertilization. Soil Biol. Biochem. 2021, 156, 108214. [Google Scholar] [CrossRef]
- Sun, Y.; Li, Y.; Chang, J.; Tian, L.; Ji, L.; Guo, L.; Gao, Q.; van Veen, J.A.; Tian, C. Chemotaxis mediates nitrogen acquisition of maize under long-term nitrogen input. Soil Biol. Biochem. 2023, 184, 109118. [Google Scholar] [CrossRef]
- Gui, H.; Gao, Y.; Wang, Z.; Shi, L.; Yan, K.; Xu, J. Arbuscular mycorrhizal fungi potentially regulate N2O emissions from agricultural soils via altered expression of denitrification genes. Sci. Total Environ. 2021, 774, 145133. [Google Scholar] [CrossRef]
- Li, J.; Meng, B.; Yang, X.; Cui, N.; Zhao, T.; Chai, H.; Zhang, T.; Sun, W. Suppression of AMF accelerates N(2)O emission by altering soil bacterial community and genes abundance under varied precipitation conditions in a semiarid grassland. Front Microbiol. 2022, 13, 961969. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.; Zhang, Y.; Jiang, S.; Deng, Y.; Christie, P.; Murray, P.J.; Li, X.; Zhang, J. Arbuscular mycorrhizal fungi in soil and roots respond differently to phosphorus inputs in an intensively managed calcareous agricultural soil. Sci. Rep. 2016, 6, 24902. [Google Scholar] [CrossRef]
- Deepika, S.; Kothamasi, D. Soil moisture—A regulator of arbuscular mycorrhizal fungal community assembly and symbiotic phosphorus uptake. Mycorrhiza 2015, 25, 67–75. [Google Scholar] [CrossRef]
- Azcón, R.; Ambrosano, E.; Charest, C. Nutrient acquisition in mycorrhizal lettuce plants under different phosphorus and nitrogen concentration. Plant Sci. 2003, 165, 1137–1145. [Google Scholar] [CrossRef]
- Liao, D.; Sun, C.; Liang, H.; Wang, Y.; Bian, X.; Dong, C.; Niu, X.; Yang, M.; Xu, G.; Chen, A.; et al. SlSPX1-SlPHR complexes mediate the suppression of arbuscular mycorrhizal symbiosis by phosphate repletion in tomato. Plant Cell 2022, 34, 4045–4065. [Google Scholar] [CrossRef]
- Gul, S.; Whalen, J.K.; Thomas, B.W.; Sachdeva, V.; Deng, H.Y. Physico-chemical properties and microbial responses in biochar-amended soils: Mechanisms and future directions. Agric. Ecosyst. Environ. 2015, 206, 46–59. [Google Scholar] [CrossRef]
- Liu, M.; Zhao, Z.; Chen, L.; Wang, L.; Ji, L.; Xiao, Y. Influences of arbuscular mycorrhizae, phosphorus fertiliser and biochar on alfalfa growth, nutrient status and cadmium uptake. Ecotoxicol. Environ. Saf. 2020, 196, 110537. [Google Scholar] [CrossRef] [PubMed]
- Hammer, E.C.; Balogh-Brunstad, Z.; Jakobsen, I.; Olsson, P.A.; Stipp, S.L.S.; Rillig, M.C. A mycorrhizal fungus grows on biochar and captures phosphorus from its surfaces. Soil Biol. Biochem. 2014, 77, 252–260. [Google Scholar] [CrossRef]
- Gao, S.; DeLuca, T.H. Wood biochar impacts soil phosphorus dynamics and microbial communities in organically-managed croplands. Soil Biol. Biochem. 2018, 126, 144–150. [Google Scholar] [CrossRef]
- Van Zwieten, L.; Kimber, S.; Morris, S.; Chan, K.Y.; Downie, A.; Rust, J.; Joseph, S.; Cowie, A. Effects of biochar from slow pyrolysis of papermill waste on agronomic performance and soil fertility. Plant Soil 2009, 327, 235–246. [Google Scholar] [CrossRef]
- Gao, S.; DeLuca, T.H.; Cleveland, C.C. Biochar additions alter phosphorus and nitrogen availability in agricultural ecosystems: A meta-analysis. Sci. Total Environ. 2019, 654, 463–472. [Google Scholar] [CrossRef] [PubMed]
- Xiao, Z.; Rasmann, S.; Yue, L.; Lian, F.; Zou, H.; Wang, Z. The effect of biochar amendment on N-cycling genes in soils: A meta-analysis. Sci. Total Environ. 2019, 696, 133984. [Google Scholar] [CrossRef] [PubMed]
- Zhang, L.; Jing, Y.; Chen, C.; Xiang, Y.; Rezaei Rashti, M.; Li, Y.; Deng, Q.; Zhang, R. Effects of biochar application on soil nitrogen transformation, microbial functional genes, enzyme activity, and plant nitrogen uptake: A meta-analysis of field studies. GCB Bioenergy 2021, 13, 1859–1873. [Google Scholar] [CrossRef]
- Tian, J.; Kuang, X.; Tang, M.; Chen, X.; Huang, F.; Cai, Y.; Cai, K. Biochar application under low phosphorus input promotes soil organic phosphorus mineralization by shifting bacterial phoD gene community composition. Sci. Total Environ. 2021, 779, 146556. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Chi, S.; Lin, C.; Cai, C.; Yang, L.; Peng, K.; Huang, X.; Liu, J. Combination of biochar and AMF promotes phosphorus utilization by stimulating rhizosphere microbial co-occurrence networks and lipid metabolites of Phragmites. Sci. Total Environ. 2022, 845, 157339. [Google Scholar] [CrossRef] [PubMed]
- Kjeldahl, J. Neue Methode zur Bestimmung des Stickstoffs in organischen Körpern. Z. Anal. Chem. 1883, 22, 366–382. [Google Scholar] [CrossRef]
- Nell, M.; Votsch, M.; Vierheilig, H.; Steinkellner, S.; Zitterl-Eglseer, K.; Franz, C.; Novak, J. Effect of phosphorus uptake on growth and secondary metabolites of garden sage (Salvia officinalis L.). J. Sci. Food Agric. 2009, 89, 1090–1096. [Google Scholar] [CrossRef]
- Dudhagara, P.; Bhavsar, S.; Bhagat, C.; Ghelani, A.; Bhatt, S.; Patel, R. Web Resources for Metagenomics Studies. Genom. Proteom. Bioinform. 2015, 13, 296–303. [Google Scholar] [CrossRef]
- Barna, G.; Makó, A.; Takács, T.; Skic, K.; Füzy, A.; Horel, Á. Biochar Alters Soil Physical Characteristics, Arbuscular Mycorrhizal Fungi Colonization, and Glomalin Production. Agronomy 2020, 10, 1933. [Google Scholar] [CrossRef]
- Solaiman, Z.M.; Abbott, L.K.; Murphy, D.V. Biochar phosphorus concentration dictates mycorrhizal colonisation, plant growth and soil phosphorus cycling. Sci. Rep. 2019, 9, 5062. [Google Scholar] [CrossRef] [PubMed]
- Chang, Y.; Rossi, L.; Zotarelli, L.; Gao, B.; Shahid, M.A.; Sarkhosh, A. Biochar improves soil physical characteristics and strengthens root architecture in Muscadine grape (Vitis rotundifolia L.). Chem. Biol. Technol. Agric. 2021, 8, 1–11. [Google Scholar] [CrossRef]
- Jabborova, D.; Annapurna, K.; Azimov, A.; Tyagi, S.; Pengani, K.R.; Sharma, P.; Vikram, K.V.; Poczai, P.; Nasif, O.; Ansari, M.J.; et al. Co-inoculation of biochar and arbuscular mycorrhizae for growth promotion and nutrient fortification in soybean under drought conditions. Front. Plant Sci. 2022, 13, 947547. [Google Scholar] [CrossRef] [PubMed]
- Jabborova, D.; Annapurna, K.; Paul, S.; Kumar, S.; Saad, H.A.; Desouky, S.; Ibrahim, M.F.M.; Elkelish, A. Beneficial Features of Biochar and Arbuscular Mycorrhiza for Improving Spinach Plant Growth, Root Morphological Traits, Physiological Properties, and Soil Enzymatic Activities. J. Fungi 2021, 7, 571. [Google Scholar] [CrossRef] [PubMed]
- Xiang, Y.; Deng, Q.; Duan, H.; Guo, Y. Effects of biochar application on root traits: A meta-analysis. GCB Bioenergy 2017, 9, 1563–1572. [Google Scholar] [CrossRef]
- Kuypers, M.M.M.; Marchant, H.K.; Kartal, B. The microbial nitrogen-cycling network. Nat. Rev. Microbiol. 2018, 16, 263–276. [Google Scholar] [CrossRef]
- Hartman, K.; van der Heijden, M.G.A.; Wittwer, R.A.; Banerjee, S.; Walser, J.C.; Schlaeppi, K. Cropping practices manipulate abundance patterns of root and soil microbiome members paving the way to smart farming. Microbiome 2018, 6, 14. [Google Scholar] [CrossRef]
- Toussaint, J.-P.; St-Arnaud, M.; Charest, C. Nitrogen transfer and assimilation between the arbuscular mycorrhizal fungus Glomus intraradices Schenck & Smith and Ri T-DNA roots of Daucus carota L. in an in vitro compartmented system. Can. J. Microbiol. 2004, 50, 251–260. [Google Scholar] [CrossRef]
- Lehmann, J.; Rillig, M.C.; Thies, J.; Masiello, C.A.; Hockaday, W.C.; Crowley, D. Biochar effects on soil biota—A review. Soil Biol. Biochem. 2011, 43, 1812–1836. [Google Scholar] [CrossRef]
- Caranto, J.D.; Lancaster, K.M. Nitric oxide is an obligate bacterial nitrification intermediate produced by hydroxylamine oxidoreductase. Proc. Natl. Acad. Sci. USA 2017, 114, 8217–8222. [Google Scholar] [CrossRef]
- Yu, H.; Liu, X.; Yang, C.; Peng, Y.; Yu, X.; Gu, H.; Zheng, X.; Wang, C.; Xiao, F.; Shu, L.; et al. Co-symbiosis of arbuscular mycorrhizal fungi (AMF) and diazotrophs promote biological nitrogen fixation in mangrove ecosystems. Soil Biol. Biochem. 2021, 161, 108382. [Google Scholar] [CrossRef]
- Wunsch, P.; Herb, M.; Wieland, H.; Schiek, U.M.; Zumft, W.G. Requirements for Cu(A) and Cu-S center assembly of nitrous oxide reductase deduced from complete periplasmic enzyme maturation in the nondenitrifier Pseudomonas putida. J. Bacteriol. 2003, 185, 887–896. [Google Scholar] [CrossRef] [PubMed]
- Wunsch, P.; Zumft, W.G. Functional domains of NosR, a novel transmembrane iron-sulfur flavoprotein necessary for nitrous oxide respiration. J. Bacteriol. 2005, 187, 1992–2001. [Google Scholar] [CrossRef] [PubMed]
- Lanciano, P.; Vergnes, A.; Grimaldi, S.; Guigliarelli, B.; Magalon, A. Biogenesis of a respiratory complex is orchestrated by a single accessory protein. J. Biol. Chem. 2007, 282, 17468–17474. [Google Scholar] [CrossRef] [PubMed]
- Bartnikas, T.B.; Wang, Y.; Bobo, T.; Veselov, A.; Scholes, C.P.; Shapleigh, J.P. Characterization of a member of the NnrR regulon in Rhodobacter sphaeroides 2.4.3 encoding a haem–copper protein. The GenBank accession number for nnrS is U62403. Microbiology 2002, 148, 825–833. [Google Scholar] [CrossRef]
- Jones, C.M.; Stres, B.; Rosenquist, M.; Hallin, S. Phylogenetic analysis of nitrite, nitric oxide, and nitrous oxide respiratory enzymes reveal a complex evolutionary history for denitrification. Mol. Biol. Evol. 2008, 25, 1955–1966. [Google Scholar] [CrossRef]
- Kits, K.D.; Klotz, M.G.; Stein, L.Y. Methane oxidation coupled to nitrate reduction under hypoxia by the Gammaproteobacterium Methylomonas denitrificans, sp. nov. type strain FJG1. Environ. Microbiol. 2015, 17, 3219–3232. [Google Scholar] [CrossRef]
- Hu, L.T.; Foxall, P.A.; Russell, R.; Mobley, H.L. Purification of recombinant Helicobacter pylori urease apoenzyme encoded by ureA and ureB. Infect. Immun. 1992, 60, 2657–2666. [Google Scholar] [CrossRef] [PubMed]
- Saez, L.P.; Cabello, P.; Ibanez, M.I.; Luque-Almagro, V.M.; Roldan, M.D.; Moreno-Vivian, C. Cyanate Assimilation by the Alkaliphilic Cyanide-Degrading Bacterium Pseudomonas pseudoalcaligenes CECT5344: Mutational Analysis of the cyn Gene Cluster. Int. J. Mol. Sci. 2019, 20, 3008. [Google Scholar] [CrossRef]
- Ensign, S.A.; Hyman, M.R.; Arp, D.J. In vitro activation of ammonia monooxygenase from Nitrosomonas europaea by copper. J. Bacteriol. 1993, 175, 1971–1980. [Google Scholar] [CrossRef]
- Hooper, A.B.; Terry, K.R. Hydroxylamine oxidoreductase of Nitrosomonas. Production of nitric oxide from hydroxylamine. Biochim. Biophys. Acta 1979, 571, 12–20. [Google Scholar] [CrossRef] [PubMed]
- Gardner, P.R.; Gardner, A.M.; Martin, L.A.; Dou, Y.; Li, T.; Olson, J.S.; Zhu, H.; Riggs, A.F. Nitric-oxide dioxygenase activity and function of flavohemoglobins. Sensitivity to nitric oxide and carbon monoxide inhibition. J. Biol. Chem. 2000, 275, 31581–31587. [Google Scholar] [CrossRef] [PubMed]
- Daims, H.; Lucker, S.; Wagner, M. A New Perspective on Microbes Formerly Known as Nitrite-Oxidizing Bacteria. Trends Microbiol. 2016, 24, 699–712. [Google Scholar] [CrossRef] [PubMed]
- Kuang, B.; Xiao, R.; Hu, Y.; Wang, Y.; Zhang, L.; Wei, Z.; Bai, J.; Zhang, K.; Acuna, J.J.; Jorquera, M.A.; et al. Metagenomics reveals biogeochemical processes carried out by sediment microbial communities in a shallow eutrophic freshwater lake. Front Microbiol. 2022, 13, 1112669. [Google Scholar] [CrossRef]
- Bennett, A.B.; Pankievicz, V.C.S.; Ané, J.-M. A Model for Nitrogen Fixation in Cereal Crops. Trends Plant Sci. 2020, 25, 226–235. [Google Scholar] [CrossRef]
- de Lima, D.R.M.; Dos Santos, I.B.; Oliveira, J.T.C.; da Costa, D.P.; de Queiroz, J.V.J.; Romagnoli, E.M.; Andreote, F.D.; Freire, F.J.; Kuklinsky-Sobral, J. Genetic diversity of N-fixing and plant growth-promoting bacterial community in different sugarcane genotypes, association habitat and phenological phase of the crop. Arch. Microbiol. 2021, 203, 1089–1105. [Google Scholar] [CrossRef]
- Yan, Y.; Yang, J.; Dou, Y.; Chen, M.; Ping, S.; Peng, J.; Lu, W.; Zhang, W.; Yao, Z.; Li, H.; et al. Nitrogen fixation island and rhizosphere competence traits in the genome of root-associated Pseudomonas stutzeri A1501. Proc. Natl. Acad. Sci. USA 2008, 105, 7564–7569. [Google Scholar] [CrossRef]
- Sellstedt, A.; Richau, K.H. Aspects of nitrogen-fixing Actinobacteria, in particular free-living and symbiotic Frankia. FEMS Microbiol. Lett. 2013, 342, 179–186. [Google Scholar] [CrossRef] [PubMed]
- Zhang, M.; Pan, L.; Liu, L.; Su, C.; Dou, L.; Su, Z.; He, Z. Phosphorus and nitrogen removal by a novel phosphate-accumulating organism, Arthrobacter sp. HHEP5 capable of heterotrophic nitrification-aerobic denitrification: Safety assessment, removal characterization, mechanism exploration and wastewater treatment. Bioresour. Technol. 2020, 312, 123633. [Google Scholar] [CrossRef]
- Liu, Y.; Zhang, Y.; Huang, Y.; Niu, J.; Huang, J.; Peng, X.; Peng, F. Spatial and temporal conversion of nitrogen using Arthrobacter sp. 24S4-2, a strain obtained from Antarctica. Front. Microbiol. 2023, 14, 1040201. [Google Scholar] [CrossRef]
- Calderoli, P.A.; Collavino, M.M.; Behrends Kraemer, F.; Morras, H.J.M.; Aguilar, O.M. Analysis of nifH-RNA reveals phylotypes related to Geobacter and Cyanobacteria as important funsctional components of the N(2) -fixing community depending on depth and agricultural use of soil. Microbiol. Open 2017, 6, e00502. [Google Scholar] [CrossRef] [PubMed]
- Masuda, Y.; Shiratori, Y.; Ohba, H.; Ishida, T.; Takano, R.; Satoh, S.; Shen, W.; Gao, N.; Itoh, H.; Senoo, K. Enhancement of the nitrogen-fixing activity of paddy soils owing to iron application. Soil Sci. Plant Nutr. 2021, 67, 243–247. [Google Scholar] [CrossRef]
- Liu, Y.; Guo, Z.; Xue, C.; Gao, W.; Wang, G.; Liu, X. Changes in N2-fixation activity, abundance and composition of diazotrophic communities in a wheat field under elevated CO2 and canopy warming. Appl. Soil Ecol. 2021, 165, 104017. [Google Scholar] [CrossRef]
- Ma, J.; Bei, Q.; Wang, X.; Liu, G.; Cadisch, G.; Lin, X.; Zhu, J.; Sun, X.; Xie, Z. Paddy System with a Hybrid Rice Enhances Cyanobacteria Nostoc and Increases N2 Fixation. Pedosphere 2019, 29, 374–387. [Google Scholar] [CrossRef]



| Treatment | Root Weight | Root Length | Root Superficial Area | Root Volume | |
|---|---|---|---|---|---|
| g/Plant | cm | cm 2 | cm 3 | ||
| −P | NC | 21.66 ± 0.38 e | 3896 ± 414 e | 17,260 ± 1262 e | 1148 ± 77 e |
| AM | 24.01 ± 0.72 d | 5759 ± 414 c | 22,392 ± 1297 c | 1869 ± 127 bc | |
| BC | 23.83 ± 0.55 d | 4647 ± 332 d | 19,925 ± 1067 d | 1420 ± 172 de | |
| AMBC | 34.29 ± 0.46 b | 6952 ± 279 ab | 27,442 ± 1157 ab | 2277 ± 167 a | |
| +P | NC | 24.77 ± 0.49 d | 4722 ± 217 d | 19,904 ± 625 d | 1445 ± 150 de |
| AM | 27.47 ± 0.69 c | 6501 ± 232 b | 25,761 ± 880 b | 1915 ± 78 b | |
| BC | 27.83 ± 1.01 c | 4775 ± 220 d | 19,699 ± 610 d | 1552 ± 57 cd | |
| AMBC | 35.47 ± 0.87 a | 7229 ± 274 a | 28,236 ± 1334 a | 2563 ± 435 a | |
| ANOVA (p value) | |||||
| P | *** | ** | ** | * | |
| AM | *** | *** | *** | *** | |
| BC | *** | *** | *** | *** | |
| P × AM | * | NS | NS | NS | |
| P × BC | NS | * | ** | NS | |
| AM × BC | ** | * | * | * | |
| P × AM × BC | * | NS | NS | NS | |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Meng, L.; Cheng, Z.; Li, S. Response of Soil Nitrogen-Cycling Genes to the Coupling Effects of Arbuscular Mycorrhizal Fungi Inoculation and Biochar Application in Maize Rhizosphere. Sustainability 2024, 16, 3349. https://doi.org/10.3390/su16083349
Meng L, Cheng Z, Li S. Response of Soil Nitrogen-Cycling Genes to the Coupling Effects of Arbuscular Mycorrhizal Fungi Inoculation and Biochar Application in Maize Rhizosphere. Sustainability. 2024; 16(8):3349. https://doi.org/10.3390/su16083349
Chicago/Turabian StyleMeng, Lingbo, Zeyu Cheng, and Shumin Li. 2024. "Response of Soil Nitrogen-Cycling Genes to the Coupling Effects of Arbuscular Mycorrhizal Fungi Inoculation and Biochar Application in Maize Rhizosphere" Sustainability 16, no. 8: 3349. https://doi.org/10.3390/su16083349




