Natural Hydrocarbon-Contaminated Springs as a Reservoir of Microorganisms Useful for Bioremediation: Isolation and Multilevel Analysis of Hydrocarbonoclastic Bacteria from the Agri Valley (Southern Italy)
Abstract
:1. Introduction
2. Materials and Methods
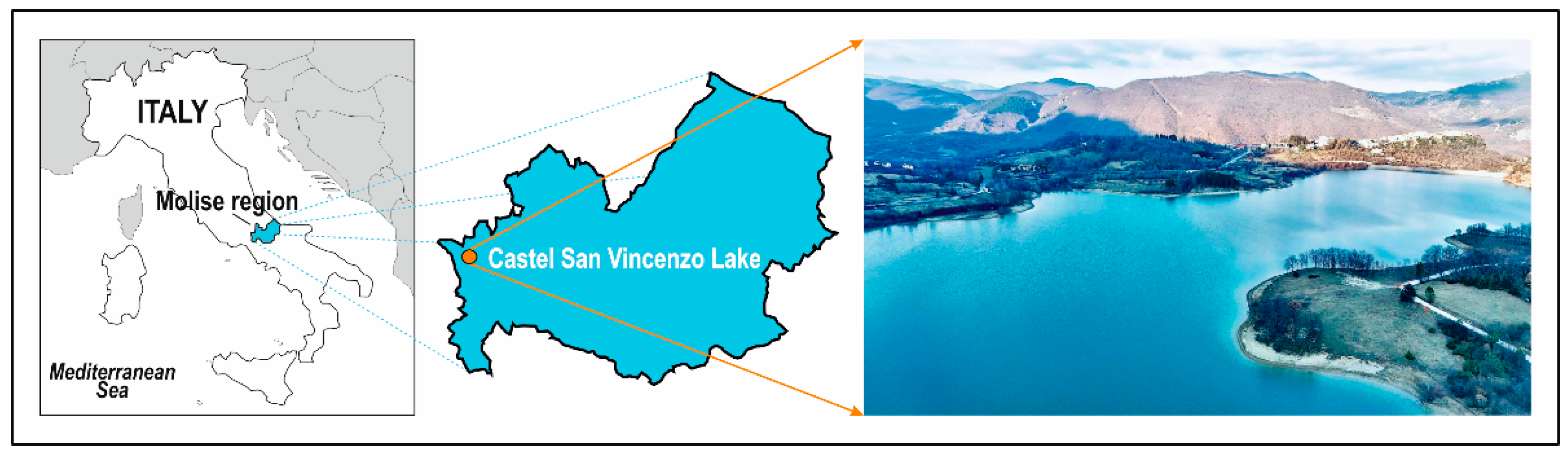
2.1. Study Area and Sample Collection
2.2. Isolation and Genome Sequencing of Hydrocarbon-Oxidising Bacterial Strains
2.3. Comparative Genome Analysis of the Selected Hydrocarbon-Oxidising Bacterial Strains
2.4. Prediction of Genes Involved in Toxic Compound Degradation
2.5. Emulsification Properties and Microbial Adhesion to Hydrocarbons Assays
2.6. Water and Soil DNA Extraction and 16S rDNA Gene Sequencing
2.7. Bacterial Consortium and Mesocosm Experiment Setup
3. Results
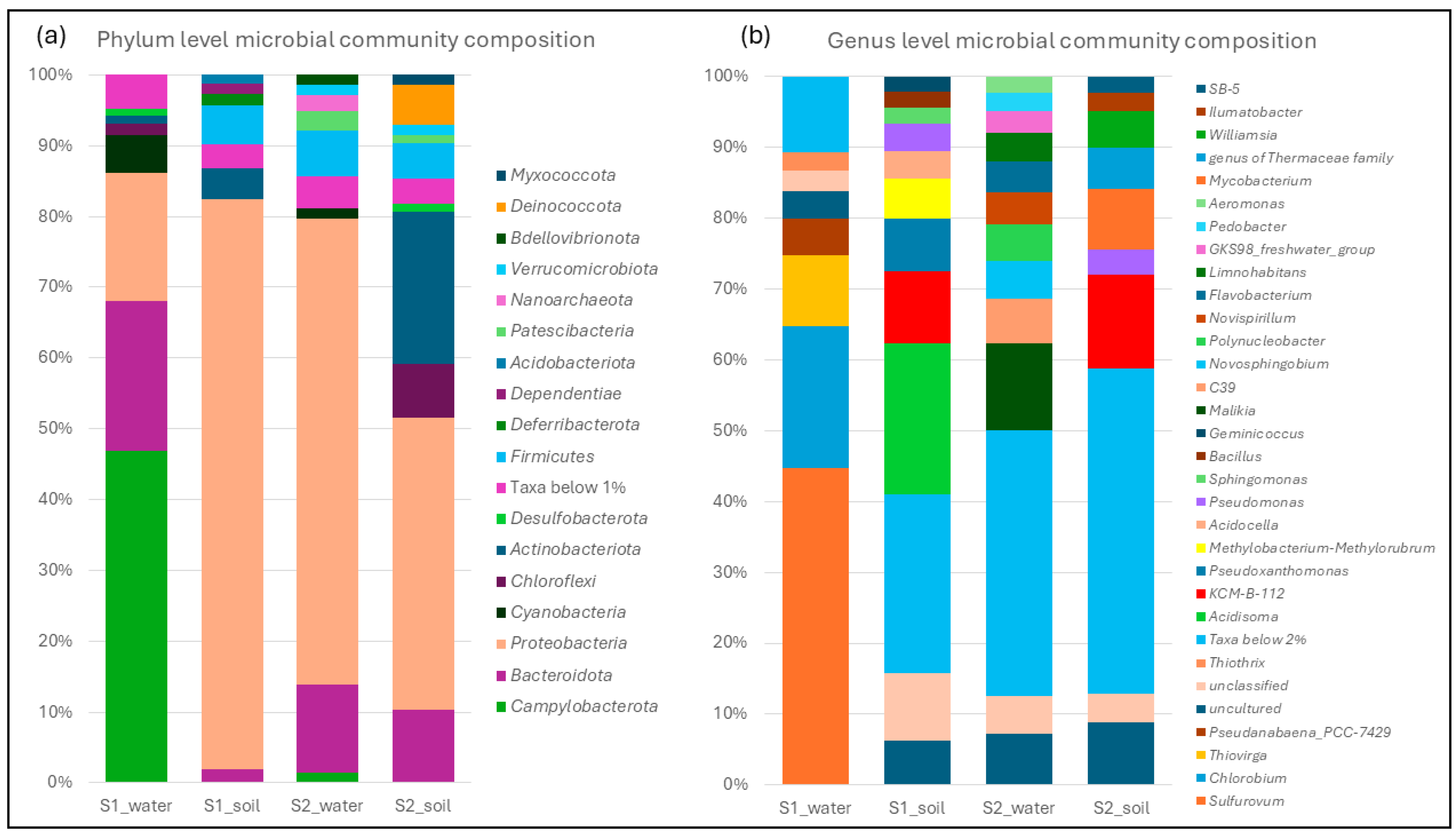
3.1. Isolation, Characterisation, and Genome Sequencing of Hydrocarbonoclastic Bacteria, Along with Microbial Community Analysis of Spring Waters and Surrounding Soils
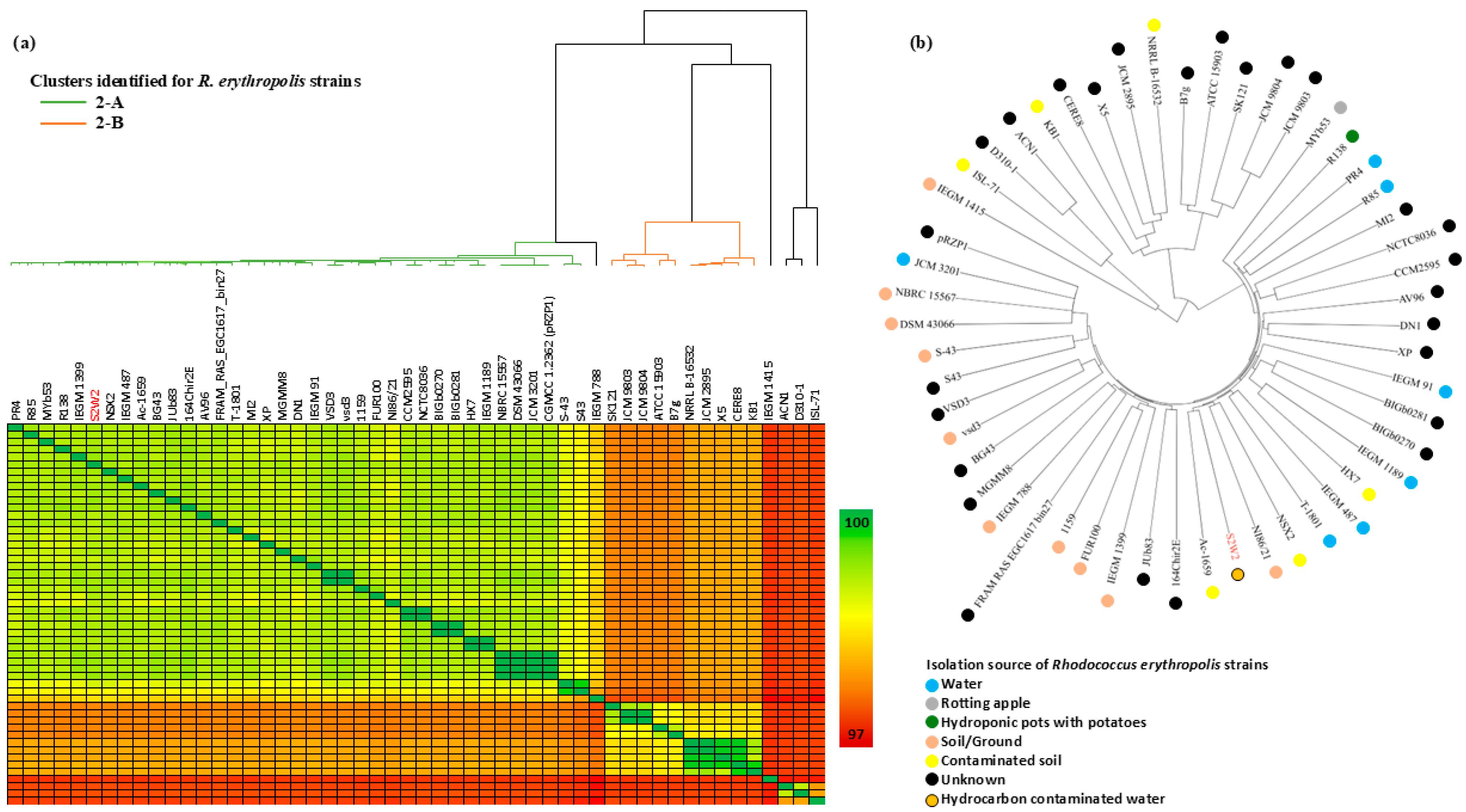
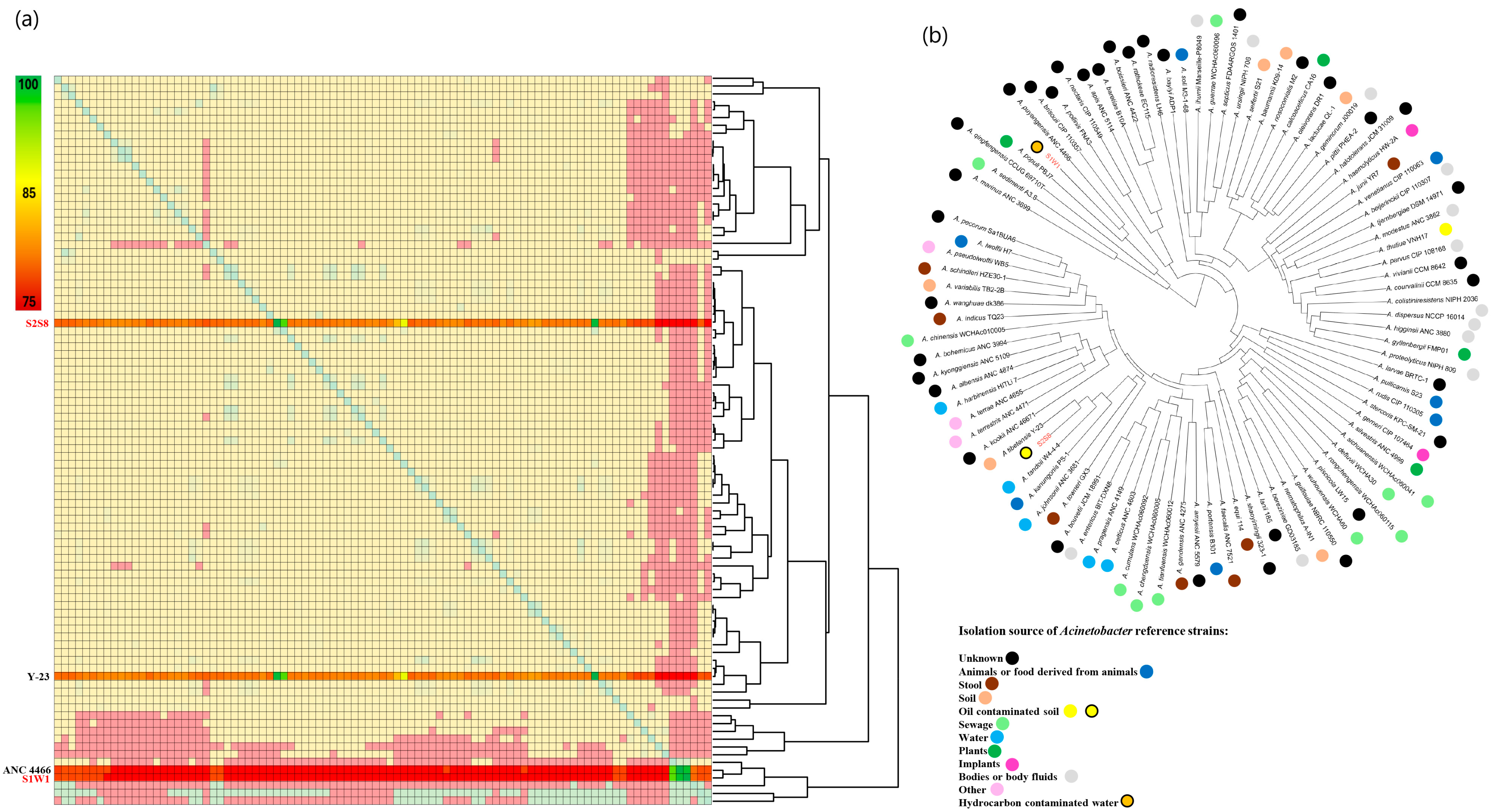
3.2. Comparative Genome Analysis of the Four Isolated Hydrocarbonoclastic Bacterial Strains
3.3. Assessment of the Unique Genetic Repertoire and Prediction of Genes Involved in Toxic Compound Degradation
3.4. Assessment of Emulsification Properties and Microbial Adhesion to Hydrocarbons
3.5. Construction of a Microbial Consortium and Bioaugmentation of Artificially Hydrocarbon-Contaminated Lake Waters to Assess the Degradative Capacity of the Four Hydrocarbonoclastic Strains in a Different Environmental Context
4. Discussion and Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Ossai, I.C.; Ahmed, A.; Hassan, A.; Hamid, F.S. Remediation of Soil and Water Contaminated with Petroleum Hydrocarbon: A Review. Environ. Technol. Innov. 2020, 17, 100526. [Google Scholar]
- Das, N.; Das, A.; Das, S.; Bhatawadekar, V.; Pandey, P.; Choure, K.; Damare, S.; Pandey, P. Petroleum Hydrocarbon Catabolic Pathways as Targets for Metabolic Engineering Strategies for Enhanced Bioremediation of Crude-Oil-Contaminated Environments. Fermentation 2023, 9, 196. [Google Scholar] [CrossRef]
- Della-Flora, I.K.; Clerici, N.J.; Dupont, G.K.; Serafini, C.G.; Daroit, D.J. Remediation of Soil Contaminated with a Commercial Diesel-Biodiesel Blend (B12): A Microcosm Evaluation on the Effects of (in)Organic Amendments. Chemosphere 2022, 287, 132059. [Google Scholar] [CrossRef]
- Wu, M.; Feng, S.; Liu, Z.; Tang, S. Bioremediation of Petroleum-Contaminated Soil Based on Both Toxicity Risk Control and Hydrocarbon Removal—Progress and Prospect. Environ. Sci. Pollut. Res. 2024, 31, 59795–59818. [Google Scholar] [CrossRef]
- Ambaye, T.G.; Chebbi, A.; Formicola, F.; Rosatelli, A.; Prasad, S.; Gomez, F.H.; Sbaffoni, S.; Franzetti, A.; Vaccari, M. Ex-Situ Bioremediation of Petroleum Hydrocarbon Contaminated Soil Using Mixed Stimulants: Response and Dynamics of Bacterial Community and Phytotoxicity. J. Environ. Chem. Eng. 2022, 10, 108814. [Google Scholar] [CrossRef]
- Mekonnen, B.A.; Aragaw, T.A.; Genet, M.B. Bioremediation of Petroleum Hydrocarbon Contaminated Soil: A Review on Principles, Degradation Mechanisms, and Advancements. Front. Environ. Sci. 2024, 12, 1354422. [Google Scholar]
- Mishra, P.; Kiran, N.S.; Romanholo Ferreira, L.F.; Yadav, K.K.; Mulla, S.I. New Insights into the Bioremediation of Petroleum Contaminants: A Systematic Review. Chemosphere 2023, 326, 138391. [Google Scholar] [CrossRef]
- Atakpa, E.O.; Zhou, H.; Jiang, L.; Ma, Y.; Liang, Y.; Li, Y.; Zhang, D.; Zhang, C. Improved Degradation of Petroleum Hydrocarbons by Co-Culture of Fungi and Biosurfactant-Producing Bacteria. Chemosphere 2022, 290, 13333. [Google Scholar] [CrossRef]
- IEA. Oil Market Report—November 2024; IEA: Paris, France, 2024; Available online: https://www.iea.org/reports/oil-market-report-november-2024 (accessed on 24 March 2025).
- Wang, S.; Li, C.; Zhang, L.; Chen, Q.; Wang, S. Assessing the Ecological Impacts of Polycyclic Aromatic Hydrocarbons Petroleum Pollutants Using a Network Toxicity Model. Environ. Res. 2023, 245, 117901. [Google Scholar] [CrossRef]
- Falih, K.T.; Mohd Razali, S.F.; Abdul Maulud, K.N.; Abd Rahman, N.; Abba, S.I.; Yaseen, Z.M. Assessment of Petroleum Contamination in Soil, Water, and Atmosphere: A Comprehensive Review. Int. J. Environ. Sci. Technol. 2024, 21, 8803–8832. [Google Scholar] [CrossRef]
- Baig, Z.T.; Abbasi, S.A.; Memon, A.G.; Naz, A.; Soomro, A.F. Assessment of Degradation Potential of Pseudomonas Species in Bioremediating Soils Contaminated with Petroleum Hydrocarbons. J. Chem. Technol. Biotechnol. 2022, 97, 455–465. [Google Scholar] [CrossRef]
- Goma-Tchimbakala, E.J.C.D.; Pietrini, I.; Goma-Tchimbakala, J.; Corgnati, S.P. Use of Shotgun Metagenomics to Assess the Microbial Diversity and Hydrocarbons Degrading Functions of Auto-Mechanic Workshops Soils Polluted with Gasoline and Diesel Fuel. Microorganisms 2023, 11, 722. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.J.; Upasani, V.N. A New Look on Factors Affecting Microbial Degradation of Petroleum Hydrocarbon Pollutants. Int. Biodeterior. Biodegradation 2017, 120, 71–83. [Google Scholar] [CrossRef]
- Xu, X.; Liu, W.; Tian, S.; Wang, W.; Qi, Q.; Jiang, P.; Gao, X.; Li, F.; Li, H.; Yu, H. Petroleum Hydrocarbon-Degrading Bacteria for the Remediation of Oil Pollution Under Aerobic Conditions: A Perspective Analysis. Front. Microbiol. 2018, 9, 2885. [Google Scholar] [CrossRef] [PubMed]
- Varjani, S.; Upasani, V.N. Influence of Abiotic Factors, Natural Attenuation, Bioaugmentation and Nutrient Supplementation on Bioremediation of Petroleum Crude Contaminated Agricultural Soil. J. Environ. Manag. 2019, 245, 358–366. [Google Scholar] [CrossRef]
- Curiel-Alegre, S.; Velasco-Arroyo, B.; Rumbo, C.; Khan, A.H.A.; Tamayo-Ramos, J.A.; Rad, C.; Gallego, J.L.R.; Barros, R. Evaluation of Biostimulation, Bioaugmentation, and Organic Amendments Application on the Bioremediation of Recalcitrant Hydrocarbons of Soil. Chemosphere 2022, 307, 135638. [Google Scholar] [CrossRef]
- Omenna, E.C.; Omage, K.; Ezaka, E.; Azeke, M.A. Bio-Augmentation and Bio-Stimulation with Kenaf Core Enhanced Bacterial Enzyme Activities during Bio-Degradation of Petroleum Hydrocarbon in Polluted Soil. Sci. Rep. 2024, 14, 8. [Google Scholar] [CrossRef]
- Sayed, K.; Baloo, L.; Sharma, N.K. Bioremediation of Total Petroleum Hydrocarbons (Tph) by Bioaugmentation and Biostimulation in Water with Floating Oil Spill Containment Booms as Bioreactor Basin. Int. J. Environ. Res. Public Health 2021, 18, 2226. [Google Scholar] [CrossRef]
- Varjani, S.J.; Gnansounou, E. Microbial Dynamics in Petroleum Oilfields and Their Relationship with Physiological Properties of Petroleum Oil Reservoirs. Bioresour. Technol. 2017, 245, 1258–1265. [Google Scholar] [CrossRef]
- Sun, S.; Su, Y.; Chen, S.; Cui, W.; Zhao, C.; Liu, Q. Bioremediation of Oil-Contaminated Soil: Exploring the Potential of Endogenous Hydrocarbon Degrader Enterobacter sp. SAVR S-1. Appl. Soil. Ecol. 2022, 173, 104387. [Google Scholar] [CrossRef]
- Dell’Anno, F.; Joaquim van Zyl, L.; Trindade, M.; Buschi, E.; Cannavacciuolo, A.; Pepi, M.; Sansone, C.; Brunet, C.; Ianora, A.; de Pascale, D.; et al. Microbiome Enrichment from Contaminated Marine Sediments Unveils Novel Bacterial Strains for Petroleum Hydrocarbon and Heavy Metal Bioremediation. Environ. Pollut. 2023, 317, 120772. [Google Scholar] [CrossRef]
- Radhakrishnan, A.; Balaganesh, P.; Vasudevan, M.; Natarajan, N.; Chauhan, A.; Arora, J.; Ranjan, A.; Rajput, V.D.; Sushkova, S.; Minkina, T.; et al. Bioremediation of Hydrocarbon Pollutants: Recent Promising Sustainable Approaches, Scope, and Challenges. Sustainability 2023, 15, 5847. [Google Scholar] [CrossRef]
- Dhar, K.; Panneerselvan, L.; Venkateswarlu, K.; Megharaj, M. Efficient Bioremediation of PAHs-Contaminated Soils by a Methylotrophic Enrichment Culture. Biodegradation 2022, 33, 575–591. [Google Scholar] [CrossRef] [PubMed]
- Tyagi, M.; da Fonseca, M.M.R.; de Carvalho, C.C.C.R. Bioaugmentation and Biostimulation Strategies to Improve the Effectiveness of Bioremediation Processes. Biodegradation 2011, 22, 231–241. [Google Scholar] [PubMed]
- Varjani, S.; Pandey, A.; Upasani, V.N. Petroleum Sludge Polluted Soil Remediation: Integrated Approach Involving Novel Bacterial Consortium and Nutrient Application. Sci. Total Environ. 2021, 763, 142934. [Google Scholar] [CrossRef]
- Yuan, L.; Wang, K.; Zhao, Q.; Yang, L.; Wang, G.; Jiang, M.; Li, L. An Overview of in Situ Remediation for Groundwater Co-Contaminated with Heavy Metals and Petroleum Hydrocarbons. J. Environ. Manag. 2024, 349, 119342. [Google Scholar]
- Varjani, S.J. Microbial Degradation of Petroleum Hydrocarbons. Bioresour. Technol. 2017, 223, 277–286. [Google Scholar]
- Rizzo, P.; Malerba, M.; Bucci, A.; Sanangelantoni, A.M.; Remelli, S.; Celico, F. Potential Enhancement of the In-Situ Bioremediation of Contaminated Sites through the Isolation and Screening of Bacterial Strains in Natural Hydrocarbon Springs. Water 2020, 12, 2090. [Google Scholar] [CrossRef]
- Schirripa Spagnolo, G.; Agosta, F.; Aldega, L.; Prosser, G.; Smeraglia, L.; Tavani, S.; Looser, N.; Guillong, M.; Bernasconi, S.M.; Billi, A.; et al. Structural Architecture and Maturity of Val d’Agri Faults, Italy: Inferences from Natural and Induced Seismicity. J. Struct. Geol. 2024, 180, 105084. [Google Scholar] [CrossRef]
- Lugli, G.A.; Fontana, F.; Tarracchini, C.; Milani, C.; Mancabelli, L.; Turroni, F.; Ventura, M. MEGAnnotator2: A Pipeline for the Assembly and Annotation of Microbial Genomes. Microbiome Res. Rep. 2023, 2, 15. [Google Scholar] [CrossRef]
- Seemann, T. Prokka: Rapid Prokaryotic Genome Annotation. Bioinformatics 2014, 30, 2068–2069. [Google Scholar] [CrossRef] [PubMed]
- Ding, W.; Baumdicker, F.; Neher, R.A. PanX: Pan-Genome Analysis and Exploration. Nucleic Acids Res. 2018, 46, E5. [Google Scholar] [CrossRef]
- Letunic, I.; Bork, P. Interactive Tree of Life (ITOL) v5: An Online Tool for Phylogenetic Tree Display and Annotation. Nucleic Acids Res. 2021, 49, W293–W296. [Google Scholar] [CrossRef] [PubMed]
- Caspi, R.; Billington, R.; Keseler, I.M.; Kothari, A.; Krummenacker, M.; Midford, P.E.; Ong, W.K.; Paley, S.; Subhraveti, P.; Karp, P.D. The MetaCyc Database of Metabolic Pathways and Enzymes-a 2019 Update. Nucleic Acids Res 2020, 48, D445–D453. [Google Scholar] [CrossRef]
- Buchfink, B.; Reuter, K.; Drost, H.G. Sensitive Protein Alignments at Tree-of-Life Scale Using DIAMOND. Nat. Methods 2021, 18, 366–368. [Google Scholar] [CrossRef]
- Larkin, M.A.; Blackshields, G.; Brown, N.P.; Chenna, R.; Mcgettigan, P.A.; McWilliam, H.; Valentin, F.; Wallace, I.M.; Wilm, A.; Lopez, R.; et al. Clustal W and Clustal X Version 2.0. Bioinformatics 2007, 23, 2947–2948. [Google Scholar] [CrossRef] [PubMed]
- Rani, M.; Weadge, J.T.; Jabaji, S. Isolation and Characterization of Biosurfactant-Producing Bacteria from Oil Well Batteries with Antimicrobial Activities Against Food-Borne and Plant Pathogens. Front. Microbiol. 2020, 11, 64. [Google Scholar] [CrossRef]
- Choudhary, J.; Dubey, R.C.; Sengar, G.; Dheeman, S. Evaluation of Probiotic Potential and Safety Assessment of Lactobacillus Pentosus MMP4 Isolated from Mare’s Lactation. Probiotics Antimicrob. Proteins 2019, 11, 403–412. [Google Scholar] [CrossRef]
- Caprari, C.; Bucci, A.; Ciotola, A.C.; Del Grosso, C.; Dell’Edera, I.; Di Bartolomeo, S.; Di Pilla, D.; Divino, F.; Fortini, P.; Monaco, P.; et al. Microbial Biocontrol Agents and Natural Products Act as Salt Stress Mitigators in Lactuca sativa L. Plants 2024, 13, 2505. [Google Scholar] [CrossRef]
- Caprari, C.; Bucci, A.; Divino, F.; Giovacchini, S.; Mirone, E.; Monaco, P.; Perrella, G.; Quaranta, L.; Scalabrino, S.; Ranalli, G. Collection Methods of Wild Barn Owl Pellets at Low Environmental Contamination and Proposals of Microbiological and Ecological Investigations. Ann. Microbiol. 2024, 74, 14. [Google Scholar] [CrossRef]
- Takahashi, S.; Tomita, J.; Nishioka, K.; Hisada, T.; Nishijima, M. Development of a Prokaryotic Universal Primer for Simultaneous Analysis of Bacteria and Archaea Using Next-Generation Sequencing. PLoS ONE 2014, 9, e105592. [Google Scholar] [CrossRef]
- Caporaso, J.G.; Kuczynski, J.; Stombaugh, J.; Bittinger, K.; Bushman, F.D.; Costello, E.K.; Fierer, N.; Pẽa, A.G.; Goodrich, J.K.; Gordon, J.I.; et al. QIIME Allows Analysis of High-Throughput Community Sequencing Data. Nat. Methods 2010, 7, 335–336. [Google Scholar]
- Callahan, B.J.; McMurdie, P.J.; Rosen, M.J.; Han, A.W.; Johnson, A.J.A.; Holmes, S.P. DADA2: High-Resolution Sample Inference from Illumina Amplicon Data. Nat. Methods 2016, 13, 581–583. [Google Scholar] [CrossRef] [PubMed]
- Quast, C.; Pruesse, E.; Yilmaz, P.; Gerken, J.; Schweer, T.; Yarza, P.; Peplies, J.; Glöckner, F.O. The SILVA Ribosomal RNA Gene Database Project: Improved Data Processing and Web-Based Tools. Nucleic Acids Res. 2013, 41, D590–D596. [Google Scholar] [CrossRef]
- Introduction to Bioinformatics in Microbiology; Christensen, H., Ed.; Learning Materials in Biosciences; Springer International Publishing: Cham, Switzerland, 2018; ISBN 978-3-319-99279-2. [Google Scholar]
- Yu, T.; Liu, X.; Ai, J.; Wang, J.; Guo, Y.; Liu, X.; He, X.; Deng, Z.; Jiang, Y. Microbial Community Succession during Crude Oil-Degrading Bacterial Enrichment Cultivation and Construction of a Degrading Consortium. Front. Microbiol. 2022, 13, 1044448. [Google Scholar] [CrossRef]
- Chen, W.; Kong, Y.; Li, J.; Sun, Y.; Min, J.; Hu, X. Enhanced Biodegradation of Crude Oil by Constructed Bacterial Consortium Comprising Salt-Tolerant Petroleum Degraders and Biosurfactant Producers. Int. Biodeterior. Biodegrad. 2020, 154, 105047. [Google Scholar] [CrossRef]
- APAT; CNR-IRSA. Metodi Analiti per le Acque; Manuali e Linee Guida 29/2003; APAT: Rome, Italy, 2003; ISBN 88-448-0083-7. [Google Scholar]
- Sayers, E.W.; Bolton, E.E.; Brister, J.R.; Canese, K.; Chan, J.; Comeau, D.C.; Connor, R.; Funk, K.; Kelly, C.; Kim, S.; et al. Database Resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2022, 50, D20–D26. [Google Scholar] [CrossRef]
- Kato, Y.; Asano, Y. Occurrence of a Novel Lyase Catalyzing β-Elimination Reaction toward Threo-3-Chloro-L-Aspartate in Pseudomonas Putida TPU 7151. Biosci. Biotechnol. Biochem. 2001, 65, 435–437. [Google Scholar] [CrossRef] [PubMed]
- Pratt, B.; Riesen, R.; Johnston, C.G. PLFA Analyses of Microbial Communities Associated with PAH-Contaminated Riverbank Sediment. Microb. Ecol. 2012, 64, 680–691. [Google Scholar] [CrossRef]
- Shao, Z.Q.; Seffens, W.; Mulbry, W.; Behki, R.M. Cloning and Expression of the S-Triazine Hydrolase Gene (TrzA) from Rhodococcus Corallinus and Development of Rhodococcus Recombinant Strains Capable of Dealkylating and Dechlorinating the Herbicide Atrazine. J. Bacteriol. 1995, 177, 5748–5755. [Google Scholar] [CrossRef]
- Sumathi, K.; Manian, R. Bioremediation of Polycyclic Aromatic Hydrocarbons Contaminated Soils: Recent Progress, Perspectives and Challenges. Environ. Monit. Assess. 2023, 195, 1441. [Google Scholar]
- Sui, X.; Wang, X.; Li, Y.; Ji, H. Remediation of Petroleum-Contaminated Soils with Microbial and Microbial Combined Methods: Advances, Mechanisms and Challenges. Sustainability 2021, 13, 9267. [Google Scholar] [CrossRef]
- Cazzini, F.F. The History of the Upstream Oil and Gas Industry in Italy. Geol. Soc. Lond. Spéc. Publ. 2018, 465, 243–274. [Google Scholar] [CrossRef]
- Rizzo, P.; Bucci, A.; Sanangelantoni, A.M.; Iacumin, P.; Celico, F. Coupled Microbiological-Isotopic Approach for Studying Hydrodynamics in Deep Reservoirs: The Case of the Val d’agri Oilfield (Southern Italy). Water 2020, 12, 1483. [Google Scholar] [CrossRef]
- Wang, J.; Zheng, Q.; Wang, S.; Zeng, J.; Yuan, Q.; Zhong, Y.; Jiang, L.; Shao, Z. Characterization of Two Novel Chemolithoautotrophic Bacteria of Sulfurovum from Marine Coastal Environments and Further Comparative Genomic Analyses Revealed Species Differentiation among Deep-Sea Hydrothermal Vent and Non-Vent Origins. Front. Mar. Sci. 2023, 10, 1222526. [Google Scholar] [CrossRef]
- Li, W.; Zhang, M.; Kang, D.; Chen, W.; Yu, T.; Xu, D.; Zeng, Z.; Li, Y.; Zheng, P. Mechanisms of Sulfur Selection and Sulfur Secretion in a Biological Sulfide Removal (BISURE) System. Environ. Int. 2020, 137, 105549. [Google Scholar] [CrossRef]
- Kushkevych, I.; Procházka, J.; Gajdács, M.; Rittmann, S.K.M.R.; Vítězová, M. Molecular Physiology of Anaerobic Phototrophic Purple and Green Sulfur Bacteria. Int. J. Mol. Sci. 2021, 22, 6398. [Google Scholar] [CrossRef]
- Frantsuzova, E.; Delegan, Y.; Bogun, A.; Sokolova, D.; Nazina, T. Comparative Genomic Analysis of the Hydrocarbon-Oxidizing Dibenzothiophene-Desulfurizing Gordonia Strains. Microorganisms 2022, 11, 4. [Google Scholar] [CrossRef]
- Ubani, O.; Atagana, H.I. Biotreatment of Crude Oil Waste Sludge Using a Novel Bacterial Formula. Environ. Chall. 2024, 15, 100943. [Google Scholar] [CrossRef]
- Hao, D.H.; Lin, J.Q.; Song, X.; Lin, J.Q.; Su, Y.J.; Qu, Y.B. Isolation, Identification, and Performance Studies of a Novel Paraffin-Degrading Bacterium of Gordonia Amicalis LH3. Biotechnol. Bioprocess Eng. 2008, 13, 61–68. [Google Scholar] [CrossRef]
- Sowani, H.; Kulkarni, M.; Zinjarde, S. Uptake and Detoxification of Diesel Oil by a Tropical Soil Actinomycete Gordonia Amicalis HS-11: Cellular Responses and Degradation Perspectives. Environ. Pollut. 2020, 263, 114538. [Google Scholar] [CrossRef]
- Thi Mo, L.; Irina, P.; Natalia, S.; Irina, N.; Lenar, A.; Andrey, F.; Ekaterina, A.; Sergey, A.; Olga, P. Hydrocarbons Biodegradation by Rhodococcus: Assimilation of Hexadecane in Different Aggregate States. Microorganisms 2022, 10, 1594. [Google Scholar] [CrossRef] [PubMed]
- Huang, L.; Xie, J.; Lv, B.-y.; Shi, X.-f.; Li, G.-q.; Liang, F.-l.; Lian, J. yan Optimization of Nutrient Component for Diesel Oil Degradation by Acinetobacter Beijerinckii ZRS. Mar. Pollut. Bull. 2013, 76, 325–332. [Google Scholar] [CrossRef] [PubMed]
- Mara, K.; Decorosi, F.; Viti, C.; Giovannetti, L.; Papaleo, M.C.; Maida, I.; Perrin, E.; Fondi, M.; Vaneechoutte, M.; Nemec, A.; et al. Molecular and Phenotypic Characterization of Acinetobacter Strains Able to Degrade Diesel Fuel. Res. Microbiol. 2012, 163, 161–172. [Google Scholar] [CrossRef] [PubMed]
- Ho, M.T.; Li, M.S.M.; McDowell, T.; MacDonald, J.; Yuan, Z.C. Characterization and Genomic Analysis of a Diesel-Degrading Bacterium, Acinetobacter Calcoaceticus CA16, Isolated from Canadian Soil. BMC Biotechnol. 2020, 20, 39. [Google Scholar] [CrossRef]
- Espeche, M.E.; Maccormack, W.P.; Fraile, E.R. Factors affecting growth of an n-hexadecane degrader Acinetobacter species isolated from a highly polluted urban river. Int. Biodeterior. Biodegrad. 1994, 33, 187–196. [Google Scholar] [CrossRef]
- Akinde, S.B.; Obire, O. Aerobic Heterotrophic Bacteria and Petroleum-Utilizing Bacteria from Cow Dung and Poultry Manure. World J. Microbiol. Biotechnol. 2008, 24, 1999–2002. [Google Scholar] [CrossRef]
- Matin, M.; Pedregosa, A.; Rios, S.; Laborda, F. Study of Factors Influencing the Degradation of Heating Oil by A Cinetobacter Calcoaceticus MM5. Biodeterior. Biodegrad. 1996, 38, 69–75. [Google Scholar]
- Luo, Q.; Wang, Y.; Chen, Q.; Sun, W.; Zhu, B. Bioremediation of Diesel Oil Polluted Seawater by a Hydrocarbon-Degrading Bacterial Consortium with Oleophilic Nutrients. Reg. Stud. Mar. Sci. 2024, 71. [Google Scholar] [CrossRef]
- Adebusoye, S.A.; Ilori, M.O.; Amund, O.O.; Teniola, O.D.; Olatope, S.O. Microbial Degradation of Petroleum Hydrocarbons in a Polluted Tropical Stream. World J. Microbiol. Biotechnol. 2007, 23, 1149–1159. [Google Scholar] [CrossRef]
- Fischer, R.; Bleichrodt, F.S.; Gerischer, U.C. Aromatic Degradative Pathways in Acinetobacter Baylyi Underlie Carbon Catabolite Repression. Microbiology 2008, 154, 3095–3103. [Google Scholar] [CrossRef] [PubMed]
- Dohare, S.; Rawat, H.K.; Bhargava, Y.; Kango, N. Characterization of Diesel Degrading Indigenous Bacterial Strains, Acinetobacter pittii and Pseudomonas aeruginosa, Isolated from Oil Contaminated Soils. Indian J. Microbiol. 2024, 64, 749–757. [Google Scholar] [CrossRef]
- Kuppan, N.; Padman, M.; Mahadeva, M.; Srinivasan, S.; Devarajan, R. A Comprehensive Review of Sustainable Bioremediation Techniques: Eco Friendly Solutions for Waste and Pollution Management. Waste Manag. Bull. 2024, 2, 154–171. [Google Scholar] [CrossRef]
- Tesfaye, E.L.; Bogale, F.M.; Aragaw, T.A. Biodegradation of Polycyclic Aromatic Hydrocarbons: The Role of Ligninolytic Enzymes and Advances of Biosensors for in-Situ Monitoring. Emerg. Contam. 2025, 11, 100424. [Google Scholar]
- Cabello-Yeves, P.J.; Callieri, C.; Picazo, A.; Schallenberg, L.; Huber, P.; Roda-Garcia, J.J.; Bartosiewicz, M.; Belykh, O.I.; Tikhonova, I.V.; Torcello-Requena, A.; et al. Elucidating the Picocyanobacteria Salinity Divide through Ecogenomics of New Freshwater Isolates. BMC Biol. 2022, 20, 175. [Google Scholar] [CrossRef]
- Cai, H.; Cui, H.; Zeng, Y.; Wang, Y.; Jiang, H. Niveispirillum lacus sp. Nov., Isolated from Cyanobacterial Aggregates in a Eutrophic Lake. Int. J. Syst. Evol. Microbiol. 2018, 68, 507–512. [Google Scholar] [CrossRef]
- Cruz-Hernández, M.A.; Mendoza-Herrera, A.; Bocanegra-García, V.; Rivera, G. Azospirillum Spp. from Plant Growth-Promoting Bacteria to Their Use in Bioremediation. Microorganisms 2022, 10, 1057. [Google Scholar] [CrossRef]
- Xie, C.H.; Yokota, A. Azospirillum oryzae sp. Nov., a Nitrogen-Fixing Bacterium Isolated from the Roots of the Rice Plant Oryza Sativa. Int. J. Syst. Evol. Microbiol. 2005, 55, 1435–1438. [Google Scholar] [CrossRef]
- Dekhil, S.B.; Cahill, M.; Stackebrandt, E.; Sly, L.I. Transfer of Conglomeromonas Largomobilis Subsp. largomobilis to the Genus Azospirillum as Azospirillum Largomobile. Nov., and Elevation of Conglomeromonas Largomobilis. Parooensis to the New Type Species of Conglomeromonas, Conglomeromonas Parooensis sp. Nov. Syst. Appl. Microbiol. 1997, 20, 72–77. [Google Scholar] [CrossRef]
- Lin, S.Y.; Shen, F.T.; Young, L.S.; Zhu, Z.L.; Chen, W.M.; Young, C.C. Azospirillum formosense sp. Nov., a Diazotroph from Agricultural Soil. Int. J. Syst. Evol. Microbiol. 2012, 62, 1185–1190. [Google Scholar] [CrossRef]
- Khammas, K.M.; Ageron, E.; Grimont, P.A.D.; Kaiser, P. Azospirillum irakense sp. nov., a nitrogen-fixing bacterium associated with rice roots and. Res. Microbiol. 1991, 140, 679–693. [Google Scholar] [CrossRef]
- Wu, D.; Zhang, X.J.; Liu, H.C.; Zhou, Y.G.; Wu, X.L.; Nie, Y.; Kang, Y.Q.; Cai, M. Azospirillum oleiclasticum sp. Nov, a Nitrogen-Fixing and Heavy Oil Degrading Bacterium Isolated from an Oil Production Mixture of Yumen Oilfield. Syst. Appl. Microbiol. 2021, 44, 126171. [Google Scholar] [CrossRef]
- Yang, Y.; Zhang, R.; Feng, J.; Wang, C.; Chen, J. Azospirillum griseum sp. Nov., Isolated from Lakewater. Int. J. Syst. Evol. Microbiol. 2019, 69, 3676–3681. [Google Scholar] [CrossRef]
- Lavrinenko, K.; Chernousova, E.; Gridneva, E.; Dubinina, G.; Akimov, V.; Kuever, J.; Lysenko, A.; Grabovich, M. Azospirillum thiophilum sp. Nov., a Diazotrophic Bacterium Isolated from a Sulfide Spring. Int. J. Syst. Evol. Microbiol. 2010, 60, 2832–2837. [Google Scholar] [CrossRef] [PubMed]
- Zhou, S.; Han, L.; Wang, Y.; Yang, G.; Li, Z.; Hu, P. Azospirillum humicireducens sp. Nov., a Nitrogen-Fixing Bacterium Isolated from a Microbial Fuel Cell. Int. J. Syst. Evol. Microbiol. 2013, 63, 2618–2624. [Google Scholar] [CrossRef] [PubMed]
- Lin, S.Y.; Young, C.C.; Hupfer, H.; Siering, C.; Arun, A.B.; Chen, W.M.; Lai, W.A.; Shen, F.T.; Rekha, P.D.; Yassin, A.F. Azospirillum picis sp. Nov., Isolated from Discarded Tar. Int. J. Syst. Evol. Microbiol. 2009, 59, 761–765. [Google Scholar] [CrossRef]
- Young, C.C.; Hupfer, H.; Siering, C.; Ho, M.J.; Arun, A.B.; Lai, W.A.; Rekha, P.D.; Shen, F.T.; Hung, M.H.; Chen, W.M.; et al. Azospirillum rugosum sp. Nov., Isolated from Oil-Contaminated Soil. Int. J. Syst. Evol. Microbiol. 2008, 58, 959–963. [Google Scholar] [CrossRef]




| S2S5 | S2W2 | S2S8 | S1W1 | |
|---|---|---|---|---|
| Sequencing output: | 1,172,742 | 785,928 | 1,115,851 | 1,002,674 |
| High quality reads: | 1,132,596 | 756,097 | 1,085,988 | 970,118 |
| Filtered reads: | 1,132,594 | 756,087 | 1,085,987 | 970,117 |
| Contigs generated at: | K127 | K127 | K127 | K127 |
| 16S rRNA gene identity: | Gordonia amicalis JCM 11271 100.000 | Pseudomonas viridiflava 100.000 | Bacillus mycoides 98.813 | Acinetobacter soli 98.046 |
| ANI screening: | Gordonia amicalis 98.4177 | Rhodococcus erythropolis 98.6463 | Acinetobacter kanungonis 85.9859 | Acinetobacter populi 91.6293 |
| Genome completeness: | 99.76 | 99.94 | 99.73 | 100.00 |
| Genome contamination: | 0.3 | 0.09 | 0.96 | 0 |
| Average coverage: | 110,7166 | 58,0149 | 152,2452 | 120,9935 |
| Number of contigs: | 110 | 32 | 54 | 83 |
| Genome length: | 5,030,312 | 6,366,085 | 3,445,643 | 3,858,766 |
| Number of genes: | 4629 | 5885 | 3269 | 3625 |
| Number of rRNA genes: | 5 | 6 | 5 | 6 |
| Number of tRNA genes: | 49 | 54 | 67 | 59 |
| Certain taxonomical level: | Gordonia amicalis | Rhodococcus erythropolis | Acinetobacter sp. | Acinetobacter sp. |
| Hydrocarbon-Oxidising Bacterial Strains | EC % in BH | EI24 % in BH | EI48 % in BH | EC % in LB | EI24 % in LB | EI48 % in LB | MATH % |
|---|---|---|---|---|---|---|---|
| Acinetobacter puyangensis S1W1 | 50.00 | - | - | 30.00 | 66.67 | 66.67 | |
| Rhodococcus erythropolis S2W2 | 26.67 | - | - | 20.00 | 55.55 | - | 90.62 |
| Gordonia amicalis S2S5 | 53.33 | 53.33 | 46.67 | 83.33 | 100.00 | 90.48 | 76.00 |
| Acinetobacter tibetensis S2S8 | 13.33 | - | - | 40.00 | 93.33 | 20.00 | 11.11 |
| Parameters | Values |
|---|---|
| pH | 7.89 |
| Temperature (°C) | 23.50 |
| Conductivity (µS/cm) | 246.30 |
| Total hydrocarbons (mg/L) | <10.00 |
| Cl− (mg/L) | 2.06 |
| SO42− (mg/L) | 7.34 |
| Na+ (mg/L) | 1.69 |
| K+ (mg/L) | 0.68 |
| Mg2+ (mg/L) | 4.17 |
| Ca2+ (mg/L) | 49.75 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2025 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cavone, C.; Monaco, P.; Fantasma, F.; Rizzo, P.; Tarracchini, C.; Petraro, S.; Ventura, M.; Milani, C.; Celico, F.; Naclerio, G.; et al. Natural Hydrocarbon-Contaminated Springs as a Reservoir of Microorganisms Useful for Bioremediation: Isolation and Multilevel Analysis of Hydrocarbonoclastic Bacteria from the Agri Valley (Southern Italy). Sustainability 2025, 17, 3083. https://doi.org/10.3390/su17073083
Cavone C, Monaco P, Fantasma F, Rizzo P, Tarracchini C, Petraro S, Ventura M, Milani C, Celico F, Naclerio G, et al. Natural Hydrocarbon-Contaminated Springs as a Reservoir of Microorganisms Useful for Bioremediation: Isolation and Multilevel Analysis of Hydrocarbonoclastic Bacteria from the Agri Valley (Southern Italy). Sustainability. 2025; 17(7):3083. https://doi.org/10.3390/su17073083
Chicago/Turabian StyleCavone, Cristina, Pamela Monaco, Francesca Fantasma, Pietro Rizzo, Chiara Tarracchini, Silvia Petraro, Marco Ventura, Christian Milani, Fulvio Celico, Gino Naclerio, and et al. 2025. "Natural Hydrocarbon-Contaminated Springs as a Reservoir of Microorganisms Useful for Bioremediation: Isolation and Multilevel Analysis of Hydrocarbonoclastic Bacteria from the Agri Valley (Southern Italy)" Sustainability 17, no. 7: 3083. https://doi.org/10.3390/su17073083
APA StyleCavone, C., Monaco, P., Fantasma, F., Rizzo, P., Tarracchini, C., Petraro, S., Ventura, M., Milani, C., Celico, F., Naclerio, G., & Bucci, A. (2025). Natural Hydrocarbon-Contaminated Springs as a Reservoir of Microorganisms Useful for Bioremediation: Isolation and Multilevel Analysis of Hydrocarbonoclastic Bacteria from the Agri Valley (Southern Italy). Sustainability, 17(7), 3083. https://doi.org/10.3390/su17073083











