Retrieving Foliar Traits of Quercus garryana var. garryana across a Modified Landscape Using Leaf Spectroscopy and LiDAR
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Site
2.2. Airborne Data Aquistion and Processing
2.3. Leaf Spectroscopy
2.4. Partial Least-Squares Regression
2.5. Principal Component Analysis
2.6. Statistical Analysis
3. Results
3.1. Leaf-Level Spectroscopy
3.2. Model Performance
3.3. Principal Component Analyses
3.3.1. PCA 1: ITV between Agricultural and Meadow Sub-Sites
3.3.2. PCA 2: ITV in Spatial Relation to Anthropogenic Landscape Modifications
3.4. Statistical Analysis
3.4.1. PCA 1: ITV between Agricultural and Meadow Sub-Sites
3.4.2. PCA 2: ITV in Spatial Relation to Anthropogenic Landscape Modifications
4. Discussion
4.1. Remote Sensing of Intraspecific Trait Variation
4.2. Spectral Separation
4.3. Model Performance
4.4. Functional Trait Categories
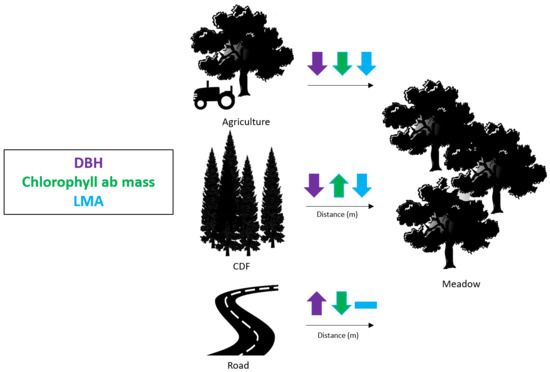
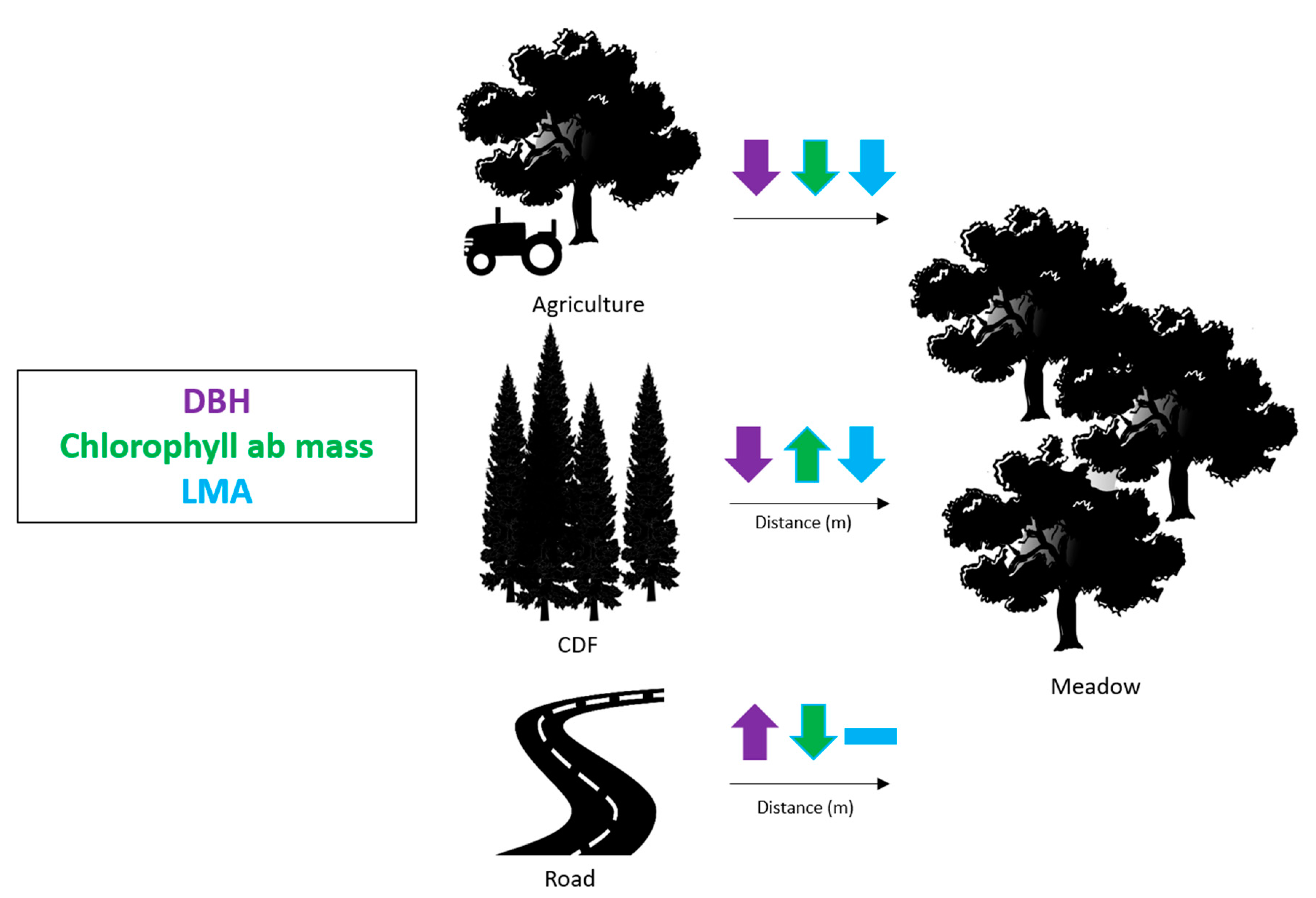
4.5. Functional Trait Variation due to Land Use
4.6. Functional Trait Variation in Relation to Landscape Modifications
4.7. Future Directions
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Steffen, W.; Grinevald, J.; Crutzen, P.; Mcneill, J. The anthropocene: Conceptual and historical perspectives. Philos. Trans. R. Soc. A Math. Phys. Eng. Sci. 2011, 369, 842–867. [Google Scholar] [CrossRef]
- Johnson, C.N.; Balmford, A.; Brook, B.W.; Buettel, J.C.; Galetti, M.; Guangchun, L.; Wilmshurst, J.M. Biodiversity losses and conservation responses in the Anthropocene. Science 2017, 356, 270–275. [Google Scholar] [CrossRef] [PubMed]
- Zeller, U.; Starik, N.; Göttert, T. Biodiversity, land use and ecosystem services—An organismic and comparative approach to different geographical regions. Glob. Ecol. Conserv. 2017, 10, 114–125. [Google Scholar] [CrossRef]
- Kerr, J.T.; Ostrovsky, M. From space to species: Ecological applications for remote sensing. Trends Ecol. Evol. 2003, 18, 299–305. [Google Scholar] [CrossRef]
- Bock, C.H.; Poole, G.H.; Parker, P.E.; Gottwald, T.R. Plant disease severity estimated visually, by digital photography and image analysis, and by hyperspectral imaging. CRC Crit. Rev. Plant Sci. 2010, 29, 59–107. [Google Scholar] [CrossRef]
- Eitel, J.U.H.; Gessler, P.E.; Smith, A.M.S.; Robberecht, R. Suitability of existing and novel spectral indices to remotely detect water stress in Populus spp. For. Ecol. Manag. 2006, 229, 170–182. [Google Scholar] [CrossRef]
- Newbold, T.; Hudson, L.N.; Phillips, H.R.P.; Hill, S.L.L.; Contu, S.; Lysenko, I.; Blandon, A.; Butchart, S.H.M.; Booth, H.L.; Day, J.; et al. A global model of the response of tropical and sub-tropical forest biodiversity to anthropogenic pressures. Proc. R. Soc. B Biol. Sci. 2014, 281, 1–10. [Google Scholar] [CrossRef] [Green Version]
- Van Cleemput, E.; Vanierschot, L.; Fernández-Castilla, B.; Honnay, O.; Somers, B. The functional characterization of grass- and shrubland ecosystems using hyperspectral remote sensing: Trends, accuracy and moderating variables. Remote Sens. Environ. 2018, 209, 747–763. [Google Scholar] [CrossRef]
- Tong, A.; He, Y. Estimating and mapping chlorophyll content for a heterogeneous grassland: Comparing prediction power of a suite of vegetation indices across scales between years. ISPRS J. Photogramm. Remote Sens. 2017, 126, 146–167. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Spectranomics: Emerging science and conservation opportunities at the interface of biodiversity and remote sensing. Glob. Ecol. Conserv. 2016, 8, 212–219. [Google Scholar] [CrossRef] [Green Version]
- Ingh, A.D.S.; Erbin, S.H.P.S.; Eil, B.R.E.M.C.N.; Ingdon, C.L.C.K. Imaging spectroscopy algorithms for mapping canopy foliar chemical and morphological traits and their uncertainties. Ecol. Appl. 2015, 25, 2180–2197. [Google Scholar]
- Asner, G.P.; Martin, R.E.; Knapp, D.E.; Tupayachi, R.; Anderson, C.; Carranza, L.; Martinez, P.; Houcheime, M.; Sinca, F.; Weiss, P. Spectroscopy of canopy chemicals in humid tropical forests. Remote Sens. Environ. 2011, 115, 3587–3598. [Google Scholar] [CrossRef]
- Kitajima, K.; Joseph Wright, S.; Westbrook, J.W. Leaf cellulose density as the key determinant of inter-and intra-specific variation in leaf fracture toughness in a species-rich tropical forest. Interface Focus 2016, 6, 20150100. [Google Scholar] [CrossRef] [PubMed]
- Butler, E.E.; Datta, A.; Flores-Moreno, H.; Chen, M.; Wythers, K.R.; Fazayeli, F.; Banerjee, A.; Atkin, O.K.; Kattge, J.; Amiaud, B.; et al. Mapping local and global variability in plant trait distributions. Proc. Natl. Acad. Sci. USA 2017, 201708984. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Serbin, S.P.; Singh, A.; Desai, A.R.; Dubois, S.G.; Jablonski, A.D.; Kingdon, C.C.; Kruger, E.L.; Townsend, P.A. Remotely estimating photosynthetic capacity, and its response to temperature, in vegetation canopies using imaging spectroscopy. Remote Sens. Environ. 2015, 167, 78–87. [Google Scholar] [CrossRef]
- Schmidtlein, S.; Feilhauer, H.; Bruelheide, H. Mapping plant strategy types using remote sensing. J. Veg. Sci. 2012, 23, 395–405. [Google Scholar] [CrossRef]
- Albert, H.; Violle, C.; Enquist, B.J.; Mcgill, B.J.; Jiang, L.; Hulshof, C.; Jung, V.; Messier, J. The return of the variance: Intraspecific variability in community ecology. Cell 2012, 27, 244–252. [Google Scholar]
- Albert, C.H.; Thuiller, W.; Yoccoz, N.G.; Douzet, R.; Aubert, S.; Lavorel, S. A multi-trait approach reveals the structure and the relative importance of intra- vs. interspecific variability in plant traits. Funct. Ecol. 2010, 24, 1192–1201. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E. Canopy phylogenetic, chemical and spectral assembly in a lowland Amazonian forest. New Phytol. 2011, 189, 999–1012. [Google Scholar] [CrossRef]
- Zagajewski, B.; Tømmervik, H.; Bjerke, J.W.; Raczko, E.; Bochenek, Z.; Klos, A.; Jarocińska, A.; Lavender, S.; Ziólkowski, D. Intraspecific differences in spectral reflectance curves as indicators of reduced vitality in high-arctic plants. Remote Sens. 2017, 9, 1289. [Google Scholar] [CrossRef] [Green Version]
- Siefert, A.; Violle, C.; Chalmandrier, L.; Albert, C.H.; Taudiere, A.; Fajardo, A.; Aarssen, L.W.; Baraloto, C.; Carlucci, M.B.; Cianciaruso, M.V.; et al. A global meta-analysis of the relative extent of intraspecific trait variation in plant communities. Ecol. Lett. 2015, 18, 1406–1419. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Asner, G.P.; Martin, R.E. Airborne spectranomics: Mapping canopy chemical and taxonomic diversity in tropical forests. Front. Ecol. Environ. 2009, 7, 269–276. [Google Scholar] [CrossRef] [Green Version]
- Zhao, Y.; Zeng, Y.; Zheng, Z.; Dong, W.; Zhao, D.; Wu, B.; Zhao, Q. Forest species diversity mapping using airborne LiDAR and hyperspectral data in a subtropical forest in China. Remote Sens. Environ. 2018, 213, 104–114. [Google Scholar] [CrossRef]
- Ustin, S.L.; Asner, G.P.; Gamon, J.A.; Fred Huemmrich, K.; Jacquemoud, S.; Schaepman, M.; Zarco-Tejada, P. Retrieval of quantitative and qualitative information about plant pigment systems from high resolution spectroscopy. Remote Sens. Environ. 2009, 113, S67–S77. [Google Scholar] [CrossRef] [Green Version]
- Tooke, T.R.; Coops, N.C.; Goodwin, N.R.; Voogt, J.A. Extracting urban vegetation characteristics using spectral mixture analysis and decision tree classifications. Remote Sens. Environ. 2009, 113, 398–407. [Google Scholar] [CrossRef]
- Plaza, J.; Hendrix, E.M.T.; García, I.; Martín, G.; Plaza, A. On endmember identification in hyperspectral images without pure pixels: A comparison of algorithms. J. Math. Imaging Vis. 2012, 42, 163–175. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Schweiger, A.K.; Cavender-Bares, J.; Townsend, P.A.; Hobbie, S.E.; Madritch, M.D.; Wang, R.; Tilman, D.; Gamon, J.A. Plant spectral diversity integrates functional and phylogenetic components of biodiversity and predicts ecosystem function. Nat. Ecol. Evol. 2018, 2, 976–982. [Google Scholar] [CrossRef]
- Wold, S.; Sjostrom, M. PLS-regression: A basic tool of chemometrics. Chemom. Intell. Lab. Syst. 2001, 109–130. [Google Scholar] [CrossRef]
- Feilhauer, H.; Asner, G.P.; Martin, R.E. Multi-method ensemble selection of spectral bands related to leaf biochemistry. Remote Sens. Environ. 2015, 164, 57–65. [Google Scholar] [CrossRef]
- Dechant, B.; Cuntz, M.; Vohland, M.; Schulz, E.; Doktor, D. Estimation of photosynthesis traits from leaf reflectance spectra: Correlation to nitrogen content as the dominant mechanism. Remote Sens. Environ. 2017, 196, 279–292. [Google Scholar] [CrossRef]
- Middleton, E.M.; Cheng, Y.B.; Hilker, T.; Black, T.A.; Krishnan, P.; Coops, N.C.; Huemmrich, K.F. Linking foliage spectral responses to canopy-level ecosystem photosynthetic light-use efficiency at a douglas-fir forest in canada. Can. J. Remote Sens. 2009, 35, 166–188. [Google Scholar] [CrossRef]
- Wang, Z.; Townsend, P.A.; Schweiger, A.K.; Couture, J.J.; Singh, A.; Hobbie, S.E.; Cavender-bares, J. Remote Sensing of Environment Mapping foliar functional traits and their uncertainties across three years in a grassland experiment. Remote Sens. Environ. 2019, 221, 405–416. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Anderson, C.B.; Knapp, D.E. Quantifying forest canopy traits: Imaging spectroscopy versus field survey. Remote Sens. Environ. 2015, 158, 15–27. [Google Scholar] [CrossRef]
- Coops, N.C.; Hilker, T.; Wulder, M.A.; St-Onge, B.; Newnham, G.; Siggins, A.; Trofymow, J.A. Estimating canopy structure of Douglas-fir forest stands from discrete-return LiDAR. Trees Struct. Funct. 2007, 21, 295–310. [Google Scholar] [CrossRef] [Green Version]
- Petry, W.K.; Covelo, F.; Abdala-roberts, L.; Galma, A. Interspecific variation in leaf functional and defensive traits in oak species and its underlying climatic drivers. PLoS ONE 2018, 13, e0202548. [Google Scholar]
- Violle, C.; Garnier, E.; Lecoeur, J.; Roumet, C.; Podeur, C.; Navas, A.B.M. Competition, traits and resource depletion in plant communities. Oecologia 2009, 160, 747–755. [Google Scholar] [CrossRef]
- Gamon, J.A.; Surfus, J.S. Assessing leaf pigment content and activity with a reflectometer. New Phytol. 1999, 143, 105–117. [Google Scholar] [CrossRef]
- Fuchs, M.A. Towards a Recovery Strategy for Garry Oak and Associated Ecosystems in Canada: Ecological Assessment and Literature Review; Environment Canada: Victoria, BC, Canada, 2001.
- Bjorkman, A.D.; Vellend, M. Defining historical baselines for conservation: Ecological changes since European settlement on Vancouver Island, Canada. Conserv. Biol. 2010, 24, 1559–1568. [Google Scholar] [CrossRef]
- Barber, C.P.; Cochrane, M.A.; Souza, C.M.; Laurance, W.F. Roads, deforestation, and the mitigating effect of protected areas in the Amazon. Biol. Conserv. 2020, 177, 203–209. [Google Scholar] [CrossRef]
- Forman, T.T.; Alexander, L.E. Roads and their major ecological effects. Annu. Rev. Ecol. Syst. 1998, 29, 207–231. [Google Scholar] [CrossRef] [Green Version]
- Amrheln, C.; Strong, J.E.; Mosher, P.A. Effect of Deicing Salts on Metal and Organic Matter Mobilization in Roadside Soils. Environ. Sci. Technol. 1992, 26, 703–709. [Google Scholar] [CrossRef]
- Banman, I.; Munson, T. Restoring British Columbia’s Garry Oak Ecosystems—Principles and Practices; Victoria, BC, Canada, 2011; ISBN 9780973218145. [Google Scholar]
- Analytical Spectral Devices. FieldSpec ® 3 User Manual; Boulder, CO, USA, 2010. [Google Scholar]
- Wang, Z.; Wang, T.; Darvishzadeh, R.; Skidmore, A.K.; Jones, S.; Suarez, L.; Woodgate, W.; Heiden, U.; Heurich, M.; Hearne, J. Vegetation indices for mapping canopy foliar nitrogen in a mixed temperate forest. Remote Sens. 2016, 8, 491. [Google Scholar] [CrossRef] [Green Version]
- Roth, K.L.; Roberts, D.A.; Dennison, P.E.; Alonzo, M.; Peterson, S.H.; Beland, M. Differentiating plant species within and across diverse ecosystems with imaging spectroscopy. Remote Sens. Environ. 2015, 167, 135–151. [Google Scholar] [CrossRef]
- Mevik, B.-H.; Wehrens, R. The pls Package: Principle Component and Partial Least Squares Regression in R. J. Stat. Softw. 2007, 18, 1–24. [Google Scholar] [CrossRef] [Green Version]
- Curran, P.J. Remote sensing of foliar chemistry. Remote Sens. Environ. 1989, 30, 271–278. [Google Scholar] [CrossRef]
- Wang, Z.; Chlus, A.; Ye, Z.; Geygan, R.; Zheng, T.; Singh, A.; Couture, J.J.; Cavender-Bares, J.; Kruger, E.L.; Townsend, P.A. Foliar functional traits from imaging spectroscopy across NEON domains in the eastern U.S. New Pyhtol. under review.
- Serbin, S.P. Spectroscopic Determination of Leaf Nutritional, Morphological, and Metabolic Traits. 2012. Available online: https://figshare.com/articles/Spectroscopic_determination_of_leaf_nutritional_morphological_and_metabolic_traits/745311 (accessed on 4 February 2019).
- Sibona, E.; Vitali, A.; Meloni, F.; Caffo, L.; Dotta, A.; Lingua, E.; Motta, R.; Garbarino, M. Direct Measurement of Tree Height Provides Different Results on the Assessment of LiDAR accuracy. Forests 2017, 8, 7. [Google Scholar] [CrossRef]
- Curran, P.J.; Dungan, J.L.; Peterson, D.L. Estimating the foliar biochemical concentration of leaves with reflectance spectrometry: Testing the Kokaly and Clark methodologies. Remote Sens. Environ. 2001, 76, 349–359. [Google Scholar] [CrossRef]
- Aubin, I.; Munson, A.D.; Cardou, F.; Burton, P.J.; Isabel, N.; Pedlar, J.H.; Paquette, A.; Taylor, A.R.; Delagrange, S.; Kebli, H.; et al. Traits to stay, traits to move: A review of functional traits to assess sensitivity and adaptive capacity of temperate and boreal trees to climate change. Environ. Rev. 2016, 24, 164–186. [Google Scholar] [CrossRef]
- Asner, G.P.; Martin, R.E.; Carranza-Jiménez, L.; Sinca, F.; Tupayachi, R.; Anderson, C.B.; Martinez, P. Functional and biological diversity of foliar spectra in tree canopies throughout the Andes to Amazon region. New Phytol. 2014, 204, 127–139. [Google Scholar] [CrossRef] [PubMed]


| Leaf Growth | Leaf Structure | Lifeform |
|---|---|---|
| Carbon (%) | Cellulose (%) | DBH (cm) |
| Carotenoid area (mmol/m2) | Fiber (%) | Height (m) |
| Carotenoid mass (ng/mg) | Lignin (%) | Max Crown Width (m) |
| Chlorophyll ab area (mmol/m2) | Leaf Mass Area (LMA) (g m2) | Height to Crown base (m) |
| Chlorophyll ab mass (ng/mg) | Leaf Structure | |
| Nitrogen (%) | Cellulose (%) |
| Group | z | p-value |
|---|---|---|
| Lifeform | 0.01 | 0.497 |
| Leaf growth | −0.045 | 0.512 |
| Leaf Structure | −0.013 | 0.508 |
| Trait | Mean Value | Standard Deviation |
|---|---|---|
| Carbon (%) | 50.082 | 0.219 |
| Carotenoid Area (mmol/m2) | 181.786 | 3.383 |
| Carotenoid Mass (ng/mg) | 1266.166 | 38.087 |
| Cellulose (%) | 15.916 | 0.972 |
| Chlorophyll ab Area (mmol/m2) | 589.166 | 24.965 |
| Chlorophyll ab Mass (ng/mg) | 7529.724 | 251.585 |
| Fiber (%) | 48.806 | 2.032 |
| Lignin (%) | 24.252 | 1.320 |
| Nitrogen (%) | 2.209 | 0.086 |
| LMA (g/m2) | 139.895 | 13.114 |
| PCA 1 | |||||
|---|---|---|---|---|---|
| Lifeform | PC1 (>0.99) | Leaf Growth | PC1 (>0.99) | Leaf Structure | PC1 (0.68) |
| DBH | −0.995 | Chlorophyll ab mass | 0.989 | LMA | −0.995 |
| Height | −0.086 | Carotenoid mass | 0.137 | Lignin | −0.068 |
| Height to crown base | −0.043 | Chlorophyll ab area | 0.053 | Fiber | −0.068 |
| Max crown width | −0.031 | Carotenoid area | 0.005 | Cellulose | −0.027 |
| Nitrogen | <0.0001 | ||||
| Carbon | <−0.0001 |
| PCA 2 | |||||
|---|---|---|---|---|---|
| Lifeform | PC1 (0.98) | Leaf Growth | PC1 (>0.99) | Leaf Structure | PC1 (0.90) |
| DBH | −0.987 | Chlorophyll ab mass | −0.989 | LMA | −0.999 |
| Height | −0.138 | Carotenoid mass | −0.139 | Fiber | 0.013 |
| Height to crown base | −0.069 | Chlorophyll ab area | −0.045 | Lignin | 0.018 |
| Max crown width | −0.033 | Carotenoid area | −0.007 | Cellulose | <−0.001 |
| Nitrogen | <−0.001 | ||||
| Carbon | <−0.001 |
| Chi-Square (χ2) | p-Value | |
|---|---|---|
| Lifeform | 38.24 | <0.001 |
| Leaf Growth | 3.80 | 0.05 |
| Leaf Structure | 6.25 | 0.01 |
| Trait | Functional Trait Category | Agricultural (n = 5) | Meadow (n = 30) |
|---|---|---|---|
| DBH (cm) | Lifeform | 125.14 | 32.12 |
| Chlorophyll ab mass (mmol/m2) | Leaf growth | 9463.48 | 7207.43 |
| LMA (g/m2) | Leaf Structure | 170.85 | 134.74 |
| a) | Distance from CDF (m) | |||
| Trait | First Quartile (25%) | Median Value (50%) | Third Quartile (75%) | p-value |
| DBH (cm) | 30.50 | 27.70 | 22.0 | <0.001 |
| Chlorophyll ab mass (ng/mg) | 6674.25 | 6777.98 | 7035.07 | <0.001 |
| LMA (g/m2) | 135.14 | 131.35 | 130.38 | 0.069 |
| b) | Distance from Road (m) | |||
| Trait | First Quartile (25%) | Median Value (50%) | Third Quartile(75%) | p-value |
| DBH (cm) | 22.00 | 27.70 | 29.00 | 0.004 |
| Chlorophyll ab mass (ng/mg) | 7608.99 | 7208.69 | 7028.97 | 0.002 |
| LMA (g/m2) | 131.22 | 131.35 | 131.48 | 0.243 |
© 2019 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Hacker, P.W.; Coops, N.C.; Townsend, P.A.; Wang, Z. Retrieving Foliar Traits of Quercus garryana var. garryana across a Modified Landscape Using Leaf Spectroscopy and LiDAR. Remote Sens. 2020, 12, 26. https://doi.org/10.3390/rs12010026
Hacker PW, Coops NC, Townsend PA, Wang Z. Retrieving Foliar Traits of Quercus garryana var. garryana across a Modified Landscape Using Leaf Spectroscopy and LiDAR. Remote Sensing. 2020; 12(1):26. https://doi.org/10.3390/rs12010026
Chicago/Turabian StyleHacker, Paul W., Nicholas C. Coops, Philip A. Townsend, and Zhihui Wang. 2020. "Retrieving Foliar Traits of Quercus garryana var. garryana across a Modified Landscape Using Leaf Spectroscopy and LiDAR" Remote Sensing 12, no. 1: 26. https://doi.org/10.3390/rs12010026