An Efficient Processing Approach for Colored Point Cloud-Based High-Throughput Seedling Phenotyping
Abstract
:1. Introduction
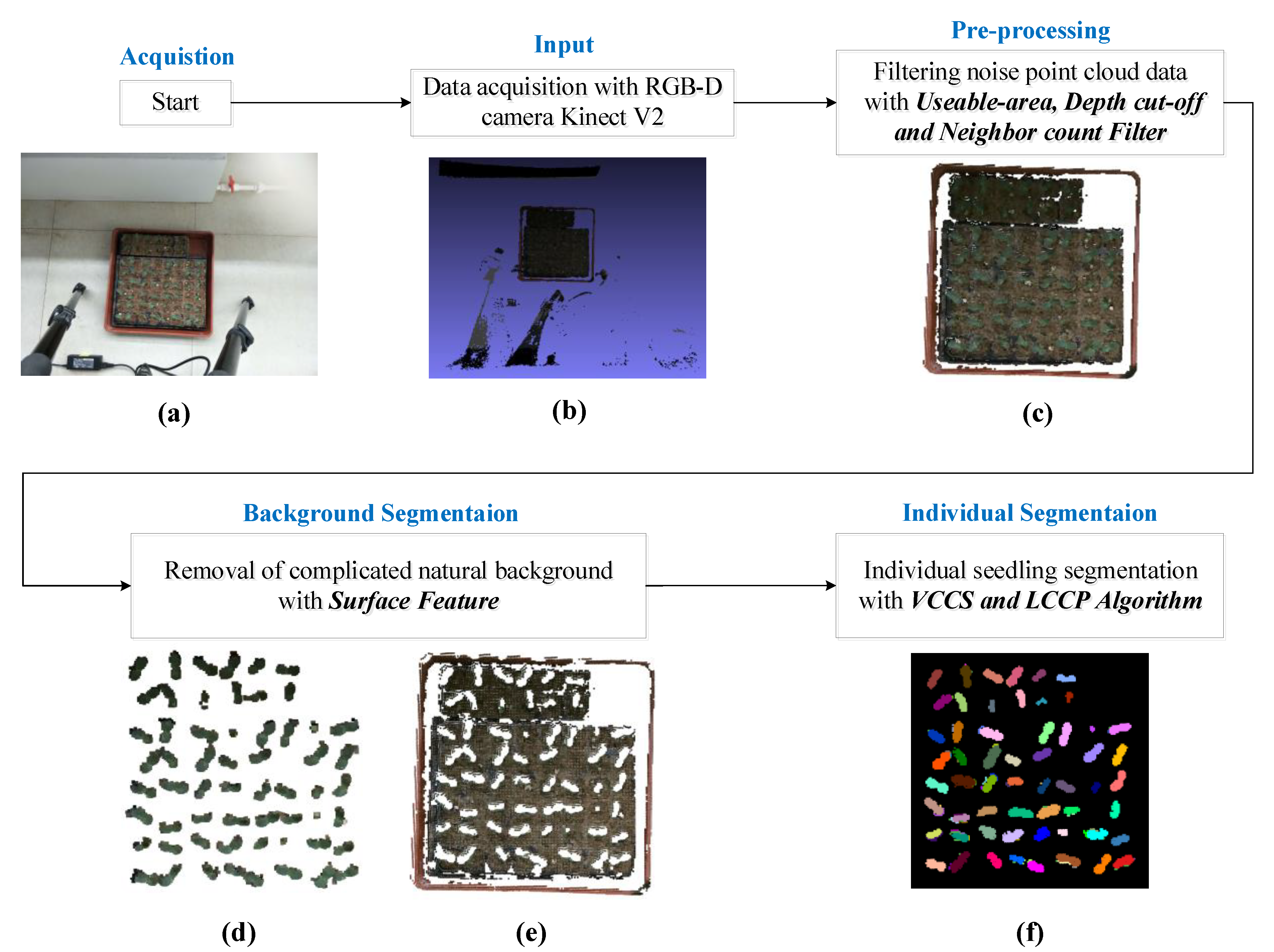
2. Materials and Methods
2.1. Data Acquisition Platform
2.2. Data Pre-Processing
| Algorithm 1. Filtering with usable area, depth cut-off and neighbor count method of original point clouds. |
| Input: original point clouds data (x, y, z, r, g, b in Euclidean space) Output: filtered point clouds data 1. InputCloud ← ocloud % Putting the original point cloud data into the filter container 2. UafRadius ← 180% Setting the radius of considered reliable area as 180 pixels 3. SafFilter (s) ← % Filtering the original point clouds data with Useable-area Outlier 4. InputCloud ← scloud % Putting the point clouds data after useable outlier into the filter container 5. depthMin, depthMax ← 700,950% Setting the outlier threshold as 0.7 m and 0.95 m 6. depthFilter (d) ← % Filtering the point clouds data s with useable Depth Outlier 7. InputCloud ← dcloud %Putting the point clouds data after depth outlier into the filter container 8. MeanK ← 15% Setting the considered adjacent points as 15 9. StddevMulThresh ← 20% Setting the outlier threshold as 20 mm 10. NeighborCountFilter (u) % Filtering the point clouds data u with Neighbor Count Outlier |
2.3. Removal of Natural Background
| Algorithm 2. Removing background with adapted surface feature histogram-based technique of pre-processed point clouds. |
| Input: pre-processed point clouds data (x, y, z, r, g, b in Euclidean space) Output: seedlings point clouds data and background point clouds data 1. InputCloud ← pcloud % Putting the pre-processed point clouds data into the segmentation container 2. initialize and use kd-tree for efficient point access 3. ← 3.5% define radius for calculation of normal 4. ← 11.5% define radius for histogram calculation 5. for all points p in the point clouds do 6. calculate normal by using principal component analysis of points with radius 7. convert RGB into HSI 8. end 9. for all points p in the point clouds do 10. read coordinates of current point p (x, y, z, r, g, b) 11. find neighbor points with radius 12. for all neighbor points do 13. calculate histogram encoding the local geometry 14. end 15. combine z and H, S, I and FPFH as feature histogram and normalize 16. end 17. K-means ← 2% Setting the number of clustering 18. output seedlings point clouds and soil point clouds |
2.4. Segmentation of Individual Seedling
| Algorithm 3. Segmenting individual seedlings with Voxel Cloud Connectivity Segmentation (VCCS) and Locally Convex Connected Patches (LCCP) of seedling point clouds. |
| Input: seedlings point clouds data (x, y, z, r, g, b in Euclidean space) Output: individual seedling point clouds data 1. InputCloud ← scloud %Putting the seedlings point clouds data into the segmentation container 2. initialize and use octree to create voxels 3. ← 0.0038% define radius for voxel 4. ← 0.018% define radius for seed 5. ← 4% define the spatial weight 6. ← 1% define the normal weight 7. use VCCS to create super-voxels 8. ← 10% define radius for histogram calculation 9. use LCCP for convex segmentation 10. output point clouds with each seedling labeled in different color |
3. Results and Discussion
3.1. Removal of Natural Background
3.2. Individual Seedling Segmentation Results
3.3. Height and Leaf Erea Estimation of Each Seedling
3.4. Plant Height Precision Measurement
3.5. Comparison with Similar Research
4. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Furbank, R.T.; Tester, M. Phenomics–technologies to relieve the phenotyping bottleneck. Trends Plant Sci. 2011, 16, 635–644. [Google Scholar] [CrossRef] [PubMed]
- Araus, J.L.; Cairns, J.E. Field high-throughput phenotyping: The new crop breeding frontier. Trends Plant Sci. 2014, 19, 52–61. [Google Scholar] [CrossRef] [PubMed]
- Tong, J.H.; Li, J.B.; Jiang, H.Y. Machine vision techniques for the evaluation of seedling quality based on leaf area. Biosyst. Eng. 2013, 115, 369–379. [Google Scholar] [CrossRef]
- Reynolds, M.; Langridge, P. Physiological breeding. Curr. Opin. Plant Biol. 2016, 31, 162–171. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shakoor, N.; Lee, S.; Mockler, T.C. High throughput phenotyping to accelerate crop breeding and monitoring of diseases in the field. Curr. Opin. Plant Biol. 2017, 38, 184–192. [Google Scholar] [CrossRef]
- Pratap, A.; Gupta, S.; Nair, R.M.; Gupta, S.K.; Basu, P.S. Using plant phenomics to exploit the gains of genomics. Agronomy 2019, 9, 126. [Google Scholar] [CrossRef] [Green Version]
- Pieruschka, R.; Schurr, U. Plant phenotyping: Past, present, and future. Plant Phenomics 2019, 2019, 7507131. [Google Scholar] [CrossRef]
- Granier, C.; Tardieu, F. Multi-scale phenotyping of leaf expansion in response to environmental changes: The whole is more than the sum of parts. Plant Cell Environ. 2009, 32, 1175–1184. [Google Scholar] [CrossRef]
- Schurr, U.; Heckenberger, U.; Herdel, K.; Walter, A.; Feil, R. Leaf development in Ricinus communis during drought stress: Dynamics of growth processes, of cellular structure and of sink-source transition. J. Exp. Bot. 2000, 51, 1515–1529. [Google Scholar] [CrossRef] [Green Version]
- Vos, J.; Evers, J.; Buck-Sorlin, G.H.; Andrieu, B.; Chelle, M.; De Visser, P.H.B. Functional–structural plant modelling: A new versatile tool in crop science. J. Exp. Bot. 2009, 61, 2101–2115. [Google Scholar] [CrossRef]
- Paproki, A.; Sirault, X.; Berry, S. A novel mesh processing based technique for 3D plant analysis. BMC Plant Biol. 2012, 12, 63. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Leister, D.; Varotto, C.; Pesaresi, P.; Salamini, M. Large-scale evaluation of plant growth in Arabidopsis thaliana by non-invasive image analysis. Plant Physiol. Biochem. 1999, 37, 671–678. [Google Scholar] [CrossRef]
- Schmundt, D.; Stitt, M.; Jähne, B.; Schurr, U. Quantitative analysis of the local rates of growth of dicot leaves at a high temporal and spatial resolution, using image sequence analysis. Plant J. 1998, 16, 505–514. [Google Scholar] [CrossRef]
- Arend, D.; Lange, M.; Pape, J.M. Quantitative monitoring of Arabidopsis thaliana growth and development using high-throughput plant phenotyping. Sci. Data 2016, 3, 1–13. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Campbell, Z.C.; Acosta-Gamboa, L.M.; Nepal, N.; Lorence, A. Engineering plants for tomorrow: How high-throughput phenotyping is contributing to the development of better crops. Phytochem. Rev. 2018, 17, 1329–1343. [Google Scholar] [CrossRef]
- Watts-Williams, S.J.; Jewell, N.; Brien, C. Using high-throughput phenotyping to explore growth responses to mycorrhizal fungi and zinc in three plant species. Plant Phenomics 2019, 2019, 5893953. [Google Scholar] [CrossRef]
- Fiorani, F.; Schurr, U. Future scenarios for plant phenotyping. Ann. Rev. Plant Biol. 2013, 64, 267–291. [Google Scholar] [CrossRef] [Green Version]
- Sun, Q.; Sun, L.; Shu, M. Monitoring Maize Lodging Grades via Unmanned Aerial Vehicle Multispectral Image. Plant Phenomics 2019, 2019, 5704154. [Google Scholar] [CrossRef] [Green Version]
- Biskup, B.; Scharr, H.; Schurr, U. A stereo imaging system for measuring structural parameters of plant canopies. Plant Cell Environ. 2007, 30, 1299–1308. [Google Scholar] [CrossRef]
- Li, B.; Xu, X.; Han, J. The estimation of crop emergence in potatoes by UAV RGB imagery. Plant Methods 2019, 15, 15. [Google Scholar] [CrossRef] [Green Version]
- Duan, L.; Yang, W.; Huang, C. A novel machine-vision-based facility for the automatic evaluation of yield-related traits in rice. Plant Methods 2011, 7, 44. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Golzarian, M.R.; Frick, R.A.; Rajendran, K. Accurate inference of shoot biomass from high-throughput images of cereal plants. Plant Methods 2011, 7, 2. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Gehan, M.A.; Fahlgren, N.; Abbasi, A.; Berry, J.C.; Callen, S.T.; Chavez, L.; Doust, A.N.; Feldman, M.J.; Gilbert, K.B.; Hodge, J.G.; et al. PlantCV v2: Image analysis software for high-throughput plant phenotyping. PeerJ 2017, 5, e4088. [Google Scholar] [CrossRef]
- Zhou, J.; Applegate, C.; Alonso, A.D. Leaf-GP: An open and automated software application for measuring growth phenotypes for arabidopsis and wheat. Plant Methods 2017, 13, 117. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Tong, J.; Shi, H.; Wu, C. Skewness correction and quality evaluation of plug seedling images based on Canny operator and Hough transform. Comput. Electron. Agric. 2018, 155, 461–472. [Google Scholar] [CrossRef]
- Mortensen, A.K.; Bender, A.; Whelan, B. Segmentation of lettuce in coloured 3D point clouds for fresh weight estimation. Comput. Electron. Agric. 2018, 154, 373–381. [Google Scholar] [CrossRef]
- Li, L.; Zhang, Q.; Huang, D. A review of imaging techniques for plant phenotyping. Sensors 2014, 14, 20078–20111. [Google Scholar] [CrossRef]
- Vandenberghe, B.; Depuydt, S.; Van Messem, A. How to make sense of 3D representations for plant phenotyping: A compendium of processing and analysis techniques. OSF Prepr. 2018. [Google Scholar] [CrossRef] [Green Version]
- Ivanov, N.; Boissard, P.; Chapron, M. Computer stereo plotting for 3-D reconstruction of a maize canopy. Agric. For. Meteorol. 1995, 75, 85–102. [Google Scholar] [CrossRef]
- Yang, W.; Duan, L.; Chen, G.; Xiong, L.; Liu, Q. Plant phenomics and high-throughput phenotyping: Accelerating rice functional genomics using multidisciplinary technologies. Curr. Opin. Plant Boil. 2013, 16, 180–187. [Google Scholar] [CrossRef]
- Hu, Y.; Wang, L.; Xiang, L. Automatic non-destructive growth measurement of leafy vegetables based on kinect. Sensors 2018, 18, 806. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Jin, S.; Su, Y.; Gao, S. Deep learning: Individual maize segmentation from terrestrial lidar data using faster R-CNN and regional growth algorithms. Front. Plant Sci. 2018, 9, 866. [Google Scholar] [CrossRef] [PubMed]
- Jiang, Y.; Li, C.; Takeda, F. 3D point cloud data to quantitatively characterize size and shape of shrub crops. Hortic. Res. 2019, 6, 1–17. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- An, N.; Welch, S.M.; Markelz, R.C.; Baker, R.L.; Palmer, C.; Ta, J.; Maloof, J.N.; Weinig, C. Quantifying time-series of leaf morphology using 2D and 3D photogrammetry methods for high-throughput plant phenotyping. Comput. Electron. Agric. 2017, 135, 222–232. [Google Scholar] [CrossRef] [Green Version]
- Seitz, S.; Curless, B.; Diebel, J.; Scharstein, D.; Szeliski, R. A Comparison and Evaluation of Multi-View Stereo Reconstruction Algorithms. In Proceedings of the 2006 IEEE Computer Society Conference on Computer Vision and Pattern Recognition—Volume 2 (CVPR 06), Washington, DC, USA, 6 June 2006; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2006; Volume 1, pp. 519–528. [Google Scholar]
- Bamji, C.S.; O’Connor, P.; Elkhatib, T. A 0.13 μm CMOS system-on-chip for a 512× 424 time-of-flight image sensor with multi-frequency photo-demodulation up to 130 MHz and 2 GS/s ADC. IEEE J. Solid State Circuits 2014, 50, 303–319. [Google Scholar] [CrossRef]
- Yamamoto, S.; Hayashi, S.; Saito, S. Proceedings of the Measurement of Growth Information of a Strawberry Plant Using a Natural Interaction Device, Dallas, TX, USA, 29 July–1 August 2012; American Society of Agricultural and Biological Engineers: St Joseph, MI, USA, 2012; Volume 1. [Google Scholar]
- Chéné, Y.; Rousseau, D.; Lucidarme, P. On the use of depth camera for 3D phenotyping of entire plants. Comput. Electron. Agric. 2012, 82, 122–127. [Google Scholar] [CrossRef]
- Ma, X.; Zhu, K.; Guan, H. High-throughput phenotyping analysis of potted soybean plants using colorized depth images based on a proximal platform. Remote Sens. 2019, 11, 1085. [Google Scholar] [CrossRef] [Green Version]
- Paulus, S.; Behmann, J.; Mahlein, A.K. Low-cost 3D systems: Suitable tools for plant phenotyping. Sensors 2014, 14, 3001–3018. [Google Scholar] [CrossRef] [Green Version]
- Fankhauser, P.; Bloesch, M.; Rodríguez, D.; Kaestner, R.; Hutter, M.; Siegwart, R. Kinect v2 for mobile robot navigation: Evaluation and modeling. In Proceedings of the 2015 International Conference on Advanced Robotics (ICAR), Istanbul, Turkey, 31 July 2015; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2015; pp. 388–394. [Google Scholar]
- Corti, A.; Giancola, S.; Mainetti, G. A metrological characterization of the Kinect V2 time-of-flight camera. Robot. Auton. Syst. 2016, 75, 584–594. [Google Scholar] [CrossRef]
- Yang, L.; Zhang, L.; Dong, H. Evaluating and improving the depth accuracy of Kinect for Windows v2. IEEE Sens. J. 2015, 15, 4275–4285. [Google Scholar] [CrossRef]
- Gai, J.; Tang, L.; Steward, B. Plant Localization and Discrimination Using 2D+3D Computer Vision for Robotic Intra-Row Weed Control. In Proceedings of the ASABE Annual International Meeting, Orlando, FL, USA, 17–20 July 2016; American Society of Agricultural and Biological Engineers: St Joseph, MI, USA, 2016; Volume 1. [Google Scholar]
- Vit, A.; Shani, G. Comparing RGB-D sensors for close range outdoor agricultural phenotyping. Sensors 2018, 18, 4413. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Xia, C.; Wang, L.; Chung, B.K. In situ 3D segmentation of individual plant leaves using a RGB-D camera for agricultural automation. Sensors 2015, 15, 20463–20479. [Google Scholar] [CrossRef] [PubMed]
- Paulus, S.; Dupuis, J.; Mahlein, A.K. Surface feature based classification of plant organs from 3D laserscanned point clouds for plant phenotyping. BMC Bioinform. 2013, 14, 238. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Rusu, R.B.; Blodow, N.; Beetz, M. Fast Point Feature Histograms (FPFH) for 3D registration. In Proceedings of the 2009 IEEE International Conference on Robotics and Automation, Kobe, Japan, 12–17 May 2009; Institute of Electrical and Electronics Engineers (IEEE): Piscataway, NJ, USA, 2009; pp. 3212–3217. [Google Scholar]
- Cheng, H.; Jiang, X.; Sun, Y.; Wang, J. Color image segmentation: Advances and prospects. Pattern Recognit. 2001, 34, 2259–2281. [Google Scholar] [CrossRef]
- Christoph Stein, S.; Schoeler, M.; Papon, J. Object partitioning using local convexity. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Seattle, WA, USA, 16–18 June 2014; pp. 304–311. [Google Scholar]
- Papon, J.; Abramov, A.; Schoeler, M. Voxel cloud connectivity segmentation-supervoxels for point clouds. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Portland, OR, USA, 23–28 June 2013; pp. 2027–2034. [Google Scholar]
- Rusu, R.B.; Marton, Z.C.; Blodow, N. Persistent point feature histograms for 3D point clouds. In Proceedings of the 10th International Conference Intelligence Autonomous System (IAS-10), Baden-Baden, Germany, July 2008; pp. 119–128. [Google Scholar]













| Distance | Field of View in mm | Spatial Resolution in mm/pix | ||
|---|---|---|---|---|
| Vertical | Horizontal | Vertical | Horizontal | |
| <500 mm | Not available | |||
| 500 mm | 577 | 701 | 1.36 | 1.37 |
| 750 mm | 866 | 1050 | 2.04 | 2.05 |
| 1000 mm | 1154 | 1400 | 2.72 | 2.73 |
| Seedling Type | Optimal Parameter Pair | Segmentation Accuracy | |
|---|---|---|---|
| Cucumber | 3.5 | 11.5 | 99.97% |
| Tomato | 4.0 | 11.5 | 95.56% |
| Cabbage | 3.5 | 11.0 | 98.30% |
| Lettuce | 3.5 | 11.5 | 96.42% |
| Algorithm | Feature Dimension | Accuracy/% | Time Costs/s |
|---|---|---|---|
| Ours | 37 | 99.97 | 2.761 |
| FPFH [48] | 33 | 96.53 | 2.536 |
| PFH [52] | 125 | 85.28 | 10.238 |
| Paulus et al. [47] | 125 | 98.87 | 10.427 |
| No. | Distance to Kinect v2/mm | Succulents | Chlorophytum | Cyclamen Persicum | |||
|---|---|---|---|---|---|---|---|
| System Measure-Ment/mm | Measure-Ment Error/mm | System Measure-Ment/mm | Measure-Ment Error/mm | System Measure-Ment/mm | Measure-Ment Error/mm | ||
| 1 | 500 | 84 | 3 | 112 | 3 | 99 | 3 |
| 2 | 550 | 85 | 2 | 111 | 4 | 100 | 2 |
| 3 | 600 | 89 | 2 | 114 | 1 | 104 | 2 |
| 4 | 650 | 88 | 1 | 116 | 1 | 106 | 4 |
| 5 | 700 | 86 | 1 | 117 | 2 | 105 | 3 |
| 6 | 750 | 89 | 2 | 116 | 1 | 100 | 2 |
| 7 | 800 | 86 | 1 | 116 | 1 | 99 | 3 |
| 8 | 850 | 86 | 1 | 118 | 3 | 100 | 2 |
| 9 | 900 | 88 | 1 | 118 | 3 | 103 | 1 |
| 10 | 950 | 88 | 1 | 117 | 2 | 106 | 4 |
| 11 | 1000 | 89 | 2 | 116 | 1 | 104 | 2 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Yang, S.; Zheng, L.; Gao, W.; Wang, B.; Hao, X.; Mi, J.; Wang, M. An Efficient Processing Approach for Colored Point Cloud-Based High-Throughput Seedling Phenotyping. Remote Sens. 2020, 12, 1540. https://doi.org/10.3390/rs12101540
Yang S, Zheng L, Gao W, Wang B, Hao X, Mi J, Wang M. An Efficient Processing Approach for Colored Point Cloud-Based High-Throughput Seedling Phenotyping. Remote Sensing. 2020; 12(10):1540. https://doi.org/10.3390/rs12101540
Chicago/Turabian StyleYang, Si, Lihua Zheng, Wanlin Gao, Bingbing Wang, Xia Hao, Jiaqi Mi, and Minjuan Wang. 2020. "An Efficient Processing Approach for Colored Point Cloud-Based High-Throughput Seedling Phenotyping" Remote Sensing 12, no. 10: 1540. https://doi.org/10.3390/rs12101540
APA StyleYang, S., Zheng, L., Gao, W., Wang, B., Hao, X., Mi, J., & Wang, M. (2020). An Efficient Processing Approach for Colored Point Cloud-Based High-Throughput Seedling Phenotyping. Remote Sensing, 12(10), 1540. https://doi.org/10.3390/rs12101540





