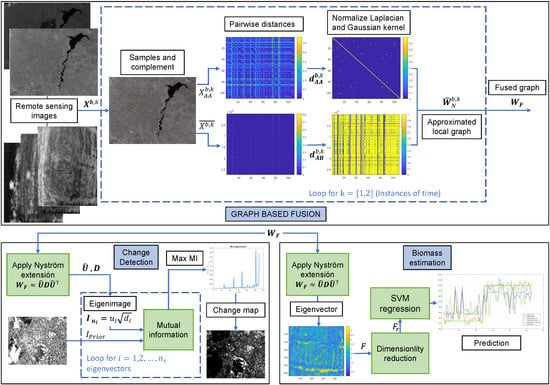
Figure 1.
Graph-based fusion, where k is the time of Events 1 (pre) and 2 (post), b refers to the band, is an image that represents an event, represents the samples from , is the complement, is the pairwise distance between the samples in , is the pairwise distance between and , is the approximated degree matrix, and is the normalized Laplacian calculated by using the Nyström approximation.
Figure 1.
Graph-based fusion, where k is the time of Events 1 (pre) and 2 (post), b refers to the band, is an image that represents an event, represents the samples from , is the complement, is the pairwise distance between the samples in , is the pairwise distance between and , is the approximated degree matrix, and is the normalized Laplacian calculated by using the Nyström approximation.
Figure 2.
Change detection, where is the fused graph, is the approximated eigenvectors, is the eigenvalues, and T in the prior stands for a binarization operator.
Figure 2.
Change detection, where is the fused graph, is the approximated eigenvectors, is the eigenvalues, and T in the prior stands for a binarization operator.
Figure 3.
The proposed methodology based on graph fusion for estimating biomass in rice crops, from q images.
Figure 3.
The proposed methodology based on graph fusion for estimating biomass in rice crops, from q images.
Figure 4.
Datasets used to test the proposed methodology for change detection. From left to right: pre-event, post-event, and reference change map images.
Figure 4.
Datasets used to test the proposed methodology for change detection. From left to right: pre-event, post-event, and reference change map images.
Figure 5.
Images from left to right represent the stages of the crop: vegetative, reproductive, and ripening, respectively for the genotype Tropical Japonica sub-species.
Figure 5.
Images from left to right represent the stages of the crop: vegetative, reproductive, and ripening, respectively for the genotype Tropical Japonica sub-species.
Figure 6.
Change detection maps highlighting the false negatives (FNs), false positives (FPs), and correct changed pixels (Cs). Each row corresponds to a dataset and each column to a method: Kittler–Illingworth (KI), Rayleigh-Rice (rR)-EM, Rayleigh-Rayleigh-Rice (rrR)-EM, unsupervised change detection using the regression homogeneous pixel transformation (U-CD-HPT ), and graph based fusion (GBF)-CD.
Figure 6.
Change detection maps highlighting the false negatives (FNs), false positives (FPs), and correct changed pixels (Cs). Each row corresponds to a dataset and each column to a method: Kittler–Illingworth (KI), Rayleigh-Rice (rR)-EM, Rayleigh-Rayleigh-Rice (rrR)-EM, unsupervised change detection using the regression homogeneous pixel transformation (U-CD-HPT ), and graph based fusion (GBF)-CD.
Figure 7.
Bar charts that evaluate the performance of each method over all metrics and datasets. The count for each method in one of the six possible metrics means that in one dataset, the model outperformed all the competing methods in that metric.
Figure 7.
Bar charts that evaluate the performance of each method over all metrics and datasets. The count for each method in one of the six possible metrics means that in one dataset, the model outperformed all the competing methods in that metric.
Figure 8.
Regression performance by one model for all rice crop growth stages. From left to right, the models are: t-SNE, PCA, and vegetation indices (VIs).
Figure 8.
Regression performance by one model for all rice crop growth stages. From left to right, the models are: t-SNE, PCA, and vegetation indices (VIs).
Table 1.
VIs for biomass estimation.
Table 1.
VIs for biomass estimation.
| Name | Equation |
|---|
| Ratio Vegetation Index (RVI) [40] | |
| Difference Vegetation Index (DVI) [50] | |
| Normalized DVI (NDVI) [40] | |
| Green NDVI (GNDVI) [41] | |
| Corrected Transformed Vegetation Index (CTVI) [50] | |
| Soil-Adjusted Vegetation Index (SAVI) [41] | , with |
| Modified SAVI (MSAVI) [49] | |
Table 2.
Databases used to evaluate the performance of the proposed method.
Table 2.
Databases used to evaluate the performance of the proposed method.
| Place | Event | Pre-Date | Post-Date | Lat | Lon | Size | Band | Sensor |
|---|
| Sardinia Island | Flood | 3 September 1995 | 3 July 1996 | , | , | 479 × 573 | NIR | Landsat-5 TM |
| Omodeo lake | Fire | 25 June 2013 | 10 August 2013 | , | , | 742 × 965 | RED | Landsat-5 TM |
| Alaska | Melt Ice | 24 June 1985 | 13 June 2005 | , | , | 443 × 642 | NIR | Landsat-5 TM |
| Brasil, Madeirinha | Farming building | 15 July 2000 | 16 July 2006 | , | , | 364 × 527 | RED | Landsat-5 TM |
| Colombia, Katios National Park | Fire | 10 March 2019 | 27 April 2019 | , | , | 879 × 1319 | SAR | Sentinel 1 A |
| Colombia, Atlantico | Flood (dam) | 28 April 2010 | 16 March 2011 | , | , | 729 × 1056 | SAR | ALOS/PALSAR |
| San Francisco | Flood | 10 August 2003 | 16 May 2004 | , | , | 275 × 400 | SAR | ERS-2 SAR |
| China, WenChuan | Earthquake | 3 March 2008 | 16 June 2008 | , | , | 301 × 442 | SAR | ESA/ASAR |
| France, Toulouse | Building | 10 February 2009 | 15 July 2013 | , | , | 2604 × 4404 | SAR/NIR | TerraSAR-X Pleiades |
| Canada, Prince George | Fire | 6 July 2017 | 22 August 2017 | , | , | 2479 × 1905 | NIR | Landsat-8 |
| California | Flood | 11 January 2017 | 26 February 2017 | , | , | 3500 × 2000 | NIR/SAR | Landsat-8 Sentinel 1 A |
| U.K., Gloucester-1 | Flood | 5 September 1999 | 17 November 2000 | , | , | 4220 × 2320 | NIR | SPOT |
| Bastrop | Fire | 8 September 2011 | 22 October 2011 | , | , | 1534 × 808 | NIR/NIR | Landsat-5 TM EO-1 ALI |
| U.K., Gloucester-2, UK | Flood | 14 June 2006 | 25 July 2007 | , | , | 4220 × 2320 | NIR/SAR | Quickbird 02 TerraSAR-X |
Table 3.
Parameters used for evaluation of the datasets. The superscripts 1 and 2 stand for pre- and post-event, respectively.
Table 3.
Parameters used for evaluation of the datasets. The superscripts 1 and 2 stand for pre- and post-event, respectively.
| Database | | | |
|---|
| Mulargia | 93 | | |
| Omodeo | 93 | | |
| Alaska | 2 | | |
| Madeirinha | 9 | | |
| Katios National Park | 60 | | |
| Atlantico | 240 | | |
| San Francisco 3 | 4 | | |
| WenChuan | 39 | | |
| Toulouse | 96 | | |
| Prince George | 110 | | |
| California 4 | 270 | | |
| Gloucester-1 | 12 | | |
| Bastrop | 96 | | |
| Gloucester-2 | 76 | | |
Table 4.
Parameters used to evaluate the datasets. The superscripts 1, 2, and 3 stand for bands R, G, and NIR, respectively.
Table 4.
Parameters used to evaluate the datasets. The superscripts 1, 2, and 3 stand for bands R, G, and NIR, respectively.
| Stage | | | |
|---|
| Vegetative | | | |
| Reproductive | | | |
| Ripening | | | |
Table 5.
Performance of the models for the Mulargia dataset. OE, overall error.
Table 5.
Performance of the models for the Mulargia dataset. OE, overall error.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 10.24 | 1.04 | 72.30 | 89.76 | 79.41 | 1.32 | 1.467 |
| rR-EM [17] | 5.72 | 4.01 | 41.73 | 94.28 | 56.05 | 4.06 | 9.881 |
| rrR-EM [18] | 10.14 | 1.06 | 72.04 | 89.86 | 79.29 | 1.33 | 13.895 |
| U-CD-HPT [31] | 9.03 | 2.00 | 58.12 | 90.96 | 69.84 | 2.20 | 107.978 |
| GBF-CD | 12.33 | 0.17 | 93.96 | 87.67 | 90.43 | 0.53 | 19.208 |
Table 6.
Performance of the models for the Omodeo dataset.
Table 6.
Performance of the models for the Omodeo dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 0.00 | 3.42 | 59.04 | 1.00 | 72.62 | 3.26 | 4.850 |
| rR-EM [17] | 0.01 | 3.73 | 56.93 | 1.00 | 70.80 | 3.56 | 14.489 |
| rrR-EM [18] | 0.01 | 2.14 | 69.73 | 1.00 | 81.12 | 2.04 | 9.928 |
| U-CD-HPT [31] | 45.88 | 0.55 | 82.90 | 54.11 | 64.14 | 2.68 | 294.320 |
| GBF-CD | 77.00 | 1.26 | 47.26 | 22.99 | 28.73 | 4.83 | 91.624 |
Table 7.
Performance of the models for the Alaska dataset.
Table 7.
Performance of the models for the Alaska dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 14.13 | 3.57 | 74.23 | 85.86 | 76.98 | 4.70 | 1.424 |
| rR-EM [17] | 8.07 | 10.91 | 50.24 | 91.92 | 59.34 | 10.60 | 7.638 |
| rrR-EM [18] | 12.52 | 4.81 | 68.51 | 87.48 | 73.68 | 5.64 | 8.322 |
| U-CD-HPT [31] | 22.01 | 0.15 | 98.38 | 77.98 | 85.65 | 2.49 | 123.214 |
| GBF-CD | 11.66 | 0.87 | 92.36 | 88.34 | 89.17 | 2.02 | 3.623 |
Table 8.
Performance of the models for the Madeirinha dataset.
Table 8.
Performance of the models for the Madeirinha dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 0.01 | 10.44 | 69.47 | 99.99 | 76.70 | 8.44 | 1.347 |
| rR-EM [17] | 0.01 | 10.19 | 69.98 | 99.99 | 77.18 | 8.23 | 6.171 |
| rrR-EM [18] | 40.31 | 1.32 | 91.45 | 59.69 | 67.27 | 8.81 | 16.320 |
| U-CD-HPT [31] | 61.05 | 0.11 | 98.78 | 38.94 | 50.48 | 11.81 | 77.366 |
| GBF-CD | 24.44 | 1.13 | 94.06 | 75.56 | 80.46 | 5.60 | 4.100 |
Table 9.
Performance of the models for the Katios dataset.
Table 9.
Performance of the models for the Katios dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 67.88 | 5.87 | 39.20 | 32.12 | 28.51 | 12.42 | 1.769 |
| rR-EM [17] | 99.84 | 1.18 | 1.49 | 0.15 | −1.72 | 11.60 | 4.013 |
| rrR-EM [18] | 99.79 | 1.29 | 1.85 | 0.21 | −1.79 | 11.69 | 4.083 |
| U-CD-HPT [31] | 73.00 | 3.58 | 47.03 | 26.99 | 28.82 | 10.90 | 457.230 |
| GBF-CD | 52.05 | 10.63 | 34.74 | 47.95 | 31.96 | 15.00 | 34.481 |
Table 10.
Performance of the models for the Atlantico dataset.
Table 10.
Performance of the models for the Atlantico dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 98.34 | 3.00 | 9.12 | 1.65 | −2.03 | 17.72 | 1.652 |
| rR-EM [17] | 99.69 | 0.29 | 15.70 | 0.30 | 0.01 | 15.63 | 5.099 |
| rrR-EM [18] | 99.93 | 0.08 | 11.62 | 0.06 | −0.04 | 15.49 | – |
| U-CD-HPT [31] | 99.13 | 0.28 | 36.01 | 0.86 | 0.97 | 15.53 | 333.742 |
| GBF-CD | 30.42 | 13.69 | 48.11 | 69.57 | 47.26 | 16.27 | 103.911 |
Table 11.
Performance of the models for the San Francisco dataset.
Table 11.
Performance of the models for the San Francisco dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 1.08 | 63.16 | 18.55 | 98.92 | 12.54 | 55.28 | 1.315 |
| rR-EM [17] | 92.75 | 0.59 | 64.05 | 7.24 | 10.71 | 12.29 | 3.282 |
| rrR-EM [18] | 2.19 | 61.23 | 18.85 | 97.80 | 13.11 | 53.73 | 3.813 |
| U-CD-HPT [31] | 75.81 | 1.52 | 69.62 | 24.19 | 31.43 | 10.92 | 64.899 |
| GBF-CD | 48.82 | 7.64 | 49.34 | 51.17 | 42.85 | 12.87 | 3.213 |
Table 12.
Performance of the models for the WenChuan dataset.
Table 12.
Performance of the models for the WenChuan dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 93.29 | 22.11 | 5.94 | 6.70 | −14.67 | 34.38 | 1.380 |
| rR-EM [17] | 99.79 | 1.07 | 3.72 | 0.20 | −1.41 | 18.10 | 3.318 |
| rrR-EM [18] | 41.61 | 53.95 | 18.40 | 58.39 | 2.38 | 51.83 | 3.678 |
| U-CD-HPT [31] | 99.69 | 2.06 | 3.00 | 0.30 | −2.73 | 18.88 | 65.025 |
| GBF-CD | 35.82 | 22.52 | 37.25 | 64.17 | 32.39 | 24.81 | 6.235 |
Table 13.
Performance of the models for the Toulouse dataset.
Table 13.
Performance of the models for the Toulouse dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 74.42 | 8.33 | 20.97 | 25.57 | 15.66 | 13.59 | 1.380 |
| rR-EM [17] | 74.94 | 8.11 | 21.07 | 25.05 | 15.59 | 13.43 | 3.318 |
| rrR-EM [18] | 52.32 | 22.07 | 15.74 | 47.67 | 13.29 | 24.47 | 3.678 |
| U-CD-HPT [31] | 98.30 | 0.97 | 13.11 | 1.69 | 1.20 | 8.72 | 4449.601 |
| GBF-CD | 54.27 | 17.33 | 18.57 | 45.72 | 17.02 | 20.27 | 839.940 |
Table 14.
Performance of the models for the Prince George dataset.
Table 14.
Performance of the models for the Prince George dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 0.60 | 16.15 | 70.23 | 99.39 | 73.79 | 11.84 | 2.575 |
| rR-EM [17] | 100.00 | 0.00 | – | 0.00 | 0.00 | 27.71 | 4.764 |
| rrR-EM [18] | 54.01 | 1.13 | 93.93 | 45.99 | 53.22 | 15.79 | 723.778 |
| U-CD-HPT [31] | 61.23 | 0.20 | 98.61 | 38.76 | 47.42 | 17.12 | 2075.130 |
| GBF-CD | 54.10 | 0.38 | 97.86 | 45.90 | 54.42 | 15.27 | 240.742 |
Table 15.
Performance of the models for the California dataset.
Table 15.
Performance of the models for the California dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 0.17 | 99.97 | 4.36 | 99.83 | −0.01 | 95.61 | 2.910 |
| rR-EM [17] | 97.74 | 31.71 | 0.32 | 2.26 | −7.66 | 34.60 | 9.989 |
| rrR-EM [18] | 18.01 | 97.85 | 3.69 | 81.98 | −1.43 | 94.36 | 25.521 |
| U-CD-HPT [31] | 58.21 | 2.79 | 40.59 | 41.79 | 38.45 | 5.21 | 2955.937 |
| GBF-CD | 11.93 | 11.79 | 25.44 | 88.06 | 35.07 | 11.80 | 921.624 |
Table 16.
Performance of the models for the Gloucester-1 dataset.
Table 16.
Performance of the models for the Gloucester-1 dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 43.16 | 2.33 | 69.62 | 56.83 | 59.44 | 5.85 | 2.933 |
| rR-EM [17] | 99.99 | 0.04 | 0.03 | 0.00 | −0.07 | 8.64 | 8.202 |
| rrR-EM [18] | 2.35 | 44.06 | 17.27 | 97.65 | 17.24 | 40.47 | 24.540 |
| U-CD-HPT [31] | 44.60 | 2.41 | 68.31 | 55.39 | 57.94 | 6.05 | 3808.564 |
| GBF-CD | 23.80 | 26.57 | 21.26 | 76.19 | 22.86 | 26.33 | 96.464 |
Table 17.
Performance of the models for the Bastrop dataset.
Table 17.
Performance of the models for the Bastrop dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 73.30 | 99.16 | 3.10 | 26.69 | −16.67 | 96.41 | 1.380 |
| rR-EM [17] | 100.00 | 0.00 | – | 0.00 | 0.00 | 10.63 | 3.318 |
| rrR-EM [18] | 100.00 | 0.00 | – | 0.00 | 0.00 | 10.63 | 3.678 |
| U-CD-HPT [31] | 15.50 | 0.39 | 96.17 | 84.49 | 88.84 | 2.00 | 365.296 |
| GBF-CD | 16.83 | 0.23 | 97.71 | 83.16 | 88.75 | 1.99 | 109.347 |
Table 18.
Performance of the models for the Gloucester-2 dataset.
Table 18.
Performance of the models for the Gloucester-2 dataset.
| Model | FN (%) | FP (%) | Recall (%) | Precision (%) | (%) | OE (%) | Time (s) |
|---|
| KI [16] | 90.34 | 4.25 | 13.46 | 9.65 | 6.21 | 9.78 | 1.380 |
| rR-EM [17] | 96.29 | 2.33 | 9.80 | 3.70 | 1.92 | 8.36 | 3.318 |
| rrR-EM [18] | 44.12 | 19.72 | 16.26 | 55.87 | 16.93 | 21.29 | 3.678 |
| U-CD-HPT [31] | 98.36 | 1.57 | 1.63 | 6.63 | 0.08 | 7.78 | 3767.047 |
| GBF-CD | 29.39 | 27.71 | 14.86 | 70.60 | 15.62 | 27.82 | 543.650 |
Table 19.
Performance of each model for biomass estimation. The evaluated metric is the root mean squared error (RMSE).
Table 19.
Performance of each model for biomass estimation. The evaluated metric is the root mean squared error (RMSE).
| | VI | PCA | t-SNE |
|---|
| RMSE | | | |