Feature Dimension Reduction Using Stacked Sparse Auto-Encoders for Crop Classification with Multi-Temporal, Quad-Pol SAR Data
Abstract
:1. Introduction
2. Methodology
2.1. PolSAR Data Structure and Features
2.2. Auto-Encoder
2.3. Stacked Sparse Auto-Encoder
2.4. Architecture of the Proposed Deep CNN Classifier
3. Experiments and Results
3.1. Experimental Sites and PolSAR Data
3.2. Results and Analysis for the Indian Head Site
3.2.1. Polarimetric Feature Extraction
3.2.2. S-SAE Configuration and Optimization
3.2.3. Comparison of Classification Results with the Different Methods
3.2.4. Comparison of the Dimensionality Reduction Features with Different Methods
3.3. Results and Analysis for the Flevoland Site
4. Discussions
4.1. Contribution of Multi-Temporal SAR Data and Decomposed Features
4.2. Network Construction and Parameter Optimization of an S-SAE
4.3. Differences from Existing Works
4.4. Limitations and Future Work
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Boryan, C.; Yang, Z.W.; Mueller, R.; Craig, M. Monitoring US agriculture: The US department of agriculture, national agricultural statistics service, cropland data layer program. Geocarto Int. 2011, 26, 341–358. [Google Scholar] [CrossRef]
- Becker, I.; Vermote, E.; Lindeman, M.; Justice, C. A generalized regression-based model for forecasting winter wheat yields in Kansas and Ukraine using MODIS data. Remote Sens. Environ. 2010, 114, 1312–1323. [Google Scholar] [CrossRef]
- Dong, T.F.; Liu, J.; Qian, B.; Jing, Q.; Croft, H.; Chen, J.; Wang, J.; Huffman, T.; Shang, J.; Chen, P. Deriving maximum light use efficiency from crop growth model and satellite data to improve crop biomass estimation. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 10, 104–117. [Google Scholar] [CrossRef]
- Sabry, R. Terrain and surface modeling using polarimetric SAR data features. IEEE Trans. Geosci. Remote Sens. 2015, 54, 1170–1184. [Google Scholar] [CrossRef]
- Wu, W.; Guo, H.; Li, X. Urban area SAR image man-made target extraction based on the product model and the time–frequency analysis. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 8, 943–952. [Google Scholar] [CrossRef]
- Ren, Y.; Li, X.M.; Gao, G.P.; Busche, T.E. Derivation of sea surface tidal current from spaceborne SAR constellation data. IEEE Trans. Geosci. Remote Sens. 2017, 55, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Jafari, M.; Maghsoudi, Y.; Zoej, M.J.V. A new method for land cover characterization and classification of polarimetric sar data using polarimetric signatures. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 3595–3607. [Google Scholar] [CrossRef]
- Lee, J.S.; Grunes, M.R.; Kwok, R. Classification of multi-look polarimetric SAR imagery based on complex Wishart distribution. Int. J. Remote Sens. 1992, 15, 2299–2311. [Google Scholar] [CrossRef]
- Lardeux, C.; Frison, P.L.; Tison, C.; Souyris, J.C.; Stoll, B.; Fruneau, B.; Rudant, J.P. Support vector machine for multifrequency SAR polarimetric data classification. IEEE Trans. Geosci. Remote Sens. 2009, 47, 4143–4152. [Google Scholar] [CrossRef]
- Maghsoudi, Y.; Collins, M.; Leckie, D.G. Polarimetric classification of boreal forest using nonparametric feature selection and multiple classifiers. Int. J. Appl. Earth Obs. Geoinform. 2012, 19, 139–150. [Google Scholar] [CrossRef]
- Hellmann, M. A new approach for interpretation of SAR-data using polarimetric techniques. Sensing and Managing the Environment. In Proceedings of the IEEE International Geoscience and Remote Sensing, Symposium (IGARSS), Seattle, WA, USA, 6–10 July 1998; pp. 2195–2197. [Google Scholar]
- Cloude, S.R.; Pottier, E. A review of target decomposition theorems in radar polarimetry. IEEE Trans. Geosci. Remote Sens. 1996, 34, 498–518. [Google Scholar] [CrossRef]
- Lee, J.S.; Grunes, M.R.; Ainsworth, T.L.; Pottier, E.; Krogager, E.; Boerner, W.M. Quantitative comparison of classification capability: Fully-polarimetric versus partially polarimetric SAR. In Proceedings of the IEEE International Geoscience & Remote Sensing Symposium(IGARSS), Honolulu, HI, USA, 24–28 July 2000; pp. 1101–1103. [Google Scholar]
- Mohan, S.; Das, A.; Haldar, D.; Maity, S. Monitoring and retrieval of vegetation parameter using multi-frequency polarimetric SAR data. In Proceedings of the International Asia-pacific Conference on Synthetic Aperture Radar (APSAR), Seoul, Korea, 26–30 September 2011; pp. 1–4. [Google Scholar]
- Chen, S.W.; Tao, C.S. PolSAR image classification using polarimetric-feature-driven deep convolutional neural network. IEEE Geosci. Remote Sens. Lett. 2018, 15, 627–631. [Google Scholar] [CrossRef]
- Zhou, Y.; Wang, H.; Xu, F.; Jin, Y.Q. Polarimetric sar image classification using deep convolutional neural networks. IEEE Geosci. Remote Sens. Lett. 2017, 13, 1935–1939. [Google Scholar] [CrossRef]
- Pierce, L.E.; Ulaby, F.T.; Sarabandi, K.; Dobson, M.C. Knowledge-based classification of polarimetric SAR images. IEEE Trans. Geosci. Remote Sens. 1994, 32, 1081–1086. [Google Scholar] [CrossRef]
- Ferrazzoli, P.; Guerriero, L.; Schiavon, G. Experimental and model investigation on radar classification capability. IEEE Trans. Geosci. Remote Sens. 1999, 37, 960–968. [Google Scholar] [CrossRef] [Green Version]
- Skriver, H.; Mattia, F.; Satalino, G.; Balenzano, A.; Pauwels, V.R.N.; Verhoest, N.E.C.; Davidson, M. Crop classification using short-revisit multitemporal SAR data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 4, 423–431. [Google Scholar] [CrossRef]
- Kussul, N.; Lemoine, G.; Gallego, F.J.; Skakun, S.V.; Lavreniuk, M.; Shelestov, A.Y. Parcel-based crop classification in Ukraine using Landsat-8 data and Sentinel-1A data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 2500–2508. [Google Scholar] [CrossRef]
- Veloso, A.; Mermoz, S.; Bouvet, A.; Le Toan, T.; Planells, M.; Dejoux, J.F.; Ceschia, E. Understanding the temporal behavior of crops using Sentinel-1 and Sentinel-2-like data for agricultural applications. Remote Sens. Environ. 2017, 199, 415–426. [Google Scholar] [CrossRef]
- Hoang, H.K.; Bernier, M.; Duchesne, S.; Tran, Y.M. Rice mapping using RADARSAT-2 dual-and quad-pol data in a complex land-use watershed: Cau River Basin (Vietnam). IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2016, 9, 3082–3096. [Google Scholar] [CrossRef] [Green Version]
- White, L.; Millard, K.; Banks, S.; Richardson, M.; Pasher, J.; Duffe, J. Moving to the RADARSAT constellation mission: Comparing synthesized compact polarimetry and dual polarimetry data with fully polarimetric RADARSAT-2 data for image classification of peatlands. Remote Sens. 2017, 9, 573. [Google Scholar] [CrossRef] [Green Version]
- McNairn, H.; Shang, J.; Jiao, X.; Champagne, C. The contribution of ALOS PALSAR multipolarization and polarimetric data to crop classification. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3981–3992. [Google Scholar] [CrossRef] [Green Version]
- Lucas, R.; Rebelo, L.M.; Fatoyinbo, L.; Rosenqvist, A.; Itoh, T.; Shimada, M.; Simard, M.; Souzafilho, P.W.; Thomas, N.; Trettin, C. Contribution of L-band SAR to systematic global mangrove monitoring. Mar. Freshw. Res. 2014, 65, 589–603. [Google Scholar] [CrossRef]
- Li, X.M.; Lehner, S. Algorithm for sea surface wind retrieval from TerraSAR-X and TanDEM-X data. IEEE Trans. Geosci. Remote Sens. 2013, 52, 2928–2939. [Google Scholar] [CrossRef] [Green Version]
- Mattia, F.; Satalino, G.; Balenzano, A.; D’Urso, G.; Capodici, F.; Iacobellis, V.; Milella, P.; Gioia, A.; Rinaldi, M.; Ruggieri, S. Time series of COSMO-SkyMed data for landcover classification and surface parameter retrieval over agricultural sites. In Proceedings of the 2012 IEEE International Geoscience and Remote Sensing Symposium (IGARSS), Munich, Germany, 22–27 July 2012; pp. 6511–6514. [Google Scholar]
- Wei, S.; Zhang, H.; Wang, C.; Wang, Y.; Xu, L. Multi-temporal SAR data large-scale crop mapping based on U-Net model. Remote Sens. 2019, 11, 68. [Google Scholar] [CrossRef] [Green Version]
- Teimouri, N.; Dyrmann, M.; Jørgensen, R.N. A Novel Spatio-Temporal FCN-LSTM Network for Recognizing Various Crop Types Using Multi-Temporal Radar Images. Remote Sens. 2019, 11, 990. [Google Scholar] [CrossRef] [Green Version]
- Zhou, Y.N.; Luo, J.; Feng, L.; Zhou, X. DCN-Based Spatial Features for Improving Parcel-Based Crop Classification Using High-Resolution Optical Images and Multi-Temporal SAR Data. Remote Sens. 2019, 11, 1619. [Google Scholar]
- Zhong, L.; Hu, L.; Zhou, H. Deep learning based multi-temporal crop classification. Remote Sen Environ. 2019, 221, 430–443. [Google Scholar] [CrossRef]
- Yang, H.; Pan, B.; Wu, W.; Tai, J. Field-based rice classification in Wuhua county through integration of multi-temporal Sentinel-1A and Landsat-8 OLI data. Int. J. Appl. Earth Obs. Geoinf. 2018, 69, 226–236. [Google Scholar] [CrossRef]
- Guo, J.; Wei, P.L.; Liu, J.; Jin, B.; Su, B.F.; Zhou, Z.S. Crop classification based on differential characteristics of H/Alpha scattering parameters for multitemporal quad-and dual-polarization SAR images. IEEE Trans. Geosci. Remote Sens. 2018, 56, 6111–6123. [Google Scholar] [CrossRef]
- Ji, S.; Zhang, C.; Xu, A.; Shi, Y.; Duan, Y. 3D convolutional neural networks for crop classification with multi-temporal remote sensing images. Remote Sens. 2018, 10, 75. [Google Scholar] [CrossRef] [Green Version]
- Sonobe, R. Parcel-Based Crop Classification Using Multi-Temporal TerraSAR-X Dual Polarimetric Data. Remote Sens. 2019, 11, 1148. [Google Scholar] [CrossRef] [Green Version]
- Xu, L.; Zhang, H.; Wang, C.; Zhang, B.; Liu, M. Crop Classification Based on Temporal Information Using Sentinel-1 SAR Time-Series Data. Remote Sens. 2019, 11, 53. [Google Scholar] [CrossRef] [Green Version]
- Freeman, A.; Durden, S.L. A three-component scattering model for polarimetric SAR data. IEEE Trans. Geosci. Remote Sens. 1998, 36, 963–973. [Google Scholar] [CrossRef] [Green Version]
- Huynen, J.R. Phenomenological Theory of Radar Targets; Technical University: Delft, The Netherlands, 1978; pp. 653–712. [Google Scholar]
- Yamaguchi, Y.; Moriyama, T.; Ishido, M.; Yamada, H. Four-component scattering model for polarimetric SAR image decomposition. IEEE Trans. Geosci. Remote Sens. 2005, 104, 1699–1706. [Google Scholar] [CrossRef]
- Cloude, S.R.; Pottoer, E. An entropy based classification scheme for land applications of polarimetric SAR. IEEE Trans. Geosci. Remote Sens. 1997, 35, 68–78. [Google Scholar] [CrossRef]
- Jolliffe, I.T. Principal Component Analysis; Springer: Heidelberg/Berlin, Germany, 2002; pp. 1094–1096. [Google Scholar]
- Roweis, S.T.; Saul, L.K. Nonlinear dimensionality reduction by locally linear embedding. Science 2000, 290, 2323–2326. [Google Scholar] [CrossRef] [Green Version]
- Lee, J.-M.; Yoo, C.; Choi, S.W.; Vanrolleghem, P.A.; Lee, I.-B. Nonlinear process monitoring using kernel principal component analysis. Chem. Eng. Sci. 2004, 59, 223–234. [Google Scholar] [CrossRef]
- Hong, C.; Yeung, D.Y. Robust locally linear embedding. Pattern Recognit. 2006, 39, 1053–1065. [Google Scholar]
- Mou, L.; Ghamisi, P.; Zhu, X.X. Deep recurrent neural networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2017, 55, 3639–3655. [Google Scholar] [CrossRef] [Green Version]
- Shrivastava, A.; Gupta, A.; Girshick, R. Training region-based object detectors with online hard example mining. In Proceedings of the IEEE Conference on Computer Vision & Pattern Recognition(CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 761–769. [Google Scholar]
- Hinton, G.E.; Salakhutdinov, R.R. Reducing the dimensionality of data with neural networks. Science 2006, 313, 504–507. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Chen, Y.; Lin, Z.; Xing, Z.; Gang, W.; Gu, Y. Deep learning-based classification of hyperspectral data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2017, 7, 2094–2107. [Google Scholar] [CrossRef]
- Paul, S.; Kumar, D.N. Spectral-spatial classification of hyperspectral data with mutual information based segmented stacked autoencoder approach. ISPRS J. Photogramm. Remote Sens. 2018, 138, 265–280. [Google Scholar] [CrossRef]
- Lee, J.S.; Grunes, M.R.; Ainsworth, T.L.; Du, L.J.; Schuler, D.L.; Cloude, S.R. Unsupervised classification using polarimetric decomposition and the complex Wishart classifier. IEEE Trans. Geosci. Remote Sens. 2002, 37, 2249–2258. [Google Scholar]
- Bai, Y.; Peng, D.; Yang, X.; Chen, L.; Yang, W. Supervised feature selection for polarimetric SAR classification. In Proceedings of the 2014 12th International Conference on Signal Processing (ICSP), Hangzhou, China, 19–23 October 2014; pp. 1006–1010. [Google Scholar]
- Dong, G.; Liao, G.; Liu, H.; Kuang, G. A Review of the Autoencoder and Its Variants: A Comparative Perspective from Target Recognition in Synthetic-Aperture Radar Images. IEEE Geosci. Remote Sens. Mag. 2018, 6, 44–68. [Google Scholar] [CrossRef]
- Ng, A. Sparse Autoencoder; CS294A Lecture notes; Stanford: San Francisco, CA, USA, 2011; Volume 72, pp. 1–19. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the IEEE Conference on Computer Vision & Pattern Recognition(CVPR), Boston, MA, USA, 8–12 June 2015; pp. 1–9. [Google Scholar]
- Caves, R. Final Report: Technical Assistance for the Implementation of the AgriSAR 2009 Campaign; Tech. Rep. 22689/09; ESA: Paris, France, 2009. [Google Scholar]
- Caves, R.; Davidson, G.; Padda, J.; Ma, A. Data Analysis-Crop Classification; Tech. Rep. 22689/09/NL/FF/ef; ESA: Paris, France, 2011. [Google Scholar]
- Caves, R.; Davidson, G.; Padda, J.; Ma, A. Data Analysis-Multi-Temporal Filtering; Tech. Rep. 22689/09/NL/FF/ef; ESA: Paris, France, 2011. [Google Scholar]
- Tao, C.; Chen, S.; Li, Y.; Xiao, S. PolSAR land cover classification based on roll-invariant and selected hidden polarimetric features in the rotation domain. Remote Sens. 2017, 9, 660. [Google Scholar]
















| Experimental Sites | Acquisition Dates | Category Code | Crop Type | Number of Pixels | Proportion |
|---|---|---|---|---|---|
| Indian Head(Canada) | 21 April 15 May 8 June 2 July 26 July 19 August 12 September | H1 | Lentil | 215,659 | 10.62% |
| H2 | Durum Wheat | 98,927 | 4.87% | ||
| H3 | Spring Wheat | 571,205 | 28.13% | ||
| H4 | Field Pea | 252,277 | 12.43% | ||
| H5 | Oat | 70,541 | 3.47% | ||
| H6 | Canola | 452,068 | 22.27% | ||
| H7 | Grass | 23,452 | 1.16% | ||
| H8 | Mixed Pasture | 14,608 | 0.72% | ||
| H9 | Mixed Hay | 27,135 | 1.34% | ||
| H10 | Barley | 106,022 | 5.22% | ||
| H11 | Summer fallow | 22,067 | 1.09% | ||
| H12 | Flax | 127,757 | 6.29% | ||
| H13 | Canary seed | 45,915 | 2.26% | ||
| H14 | Chemical fallow | 2682 | 0.13% | ||
| Flevoland(Netherlands) | 21 April 15 May 8 June 2 July 26 July 19 August 12 September | F1 | Carrots | 440 | 0.1% |
| F2 | Flower bulbs | 11,499 | 2.58% | ||
| F3 | Fruit | 10,198 | 2.29% | ||
| F4 | Grass | 33,787 | 7.58% | ||
| F5 | Lucerne | 2255 | 0.51% | ||
| F6 | Maize | 18,253 | 4.09% | ||
| F7 | Misc | 31,573 | 7.08% | ||
| F8 | Onions | 41,001 | 9.19% | ||
| F9 | Peas | 7105 | 1.59% | ||
| F10 | Potato | 100,040 | 22.43% | ||
| F11 | Spring barley | 6340 | 1.42% | ||
| F12 | Spring wheat | 17,991 | 4.03% | ||
| F13 | Sugarbeet | 58,403 | 13.09% | ||
| F14 | Winter wheat | 107,142 | 24.02% |
| Feature Extraction Methods | Features | Dimension |
|---|---|---|
| Features based on measured data | Polarization intensities(, , ) | 3 |
| Amplitude of HH-VV correlation() | 1 | |
| Phase difference of HH-VV() | 1 | |
| Co-polarized ratio() | 1 | |
| Cross-polarized ratio() | 1 | |
| Co-polarization ratio() | 1 | |
| Degrees of polarization(,) | 2 | |
| Incoherent decomposition | Freeman decomposition | 5 |
| Yamaguchi decomposition | 7 | |
| Cloude decomposition | 3 | |
| Huynen decomposition | 9 | |
| Other Decomposition | Null angle parameters | 2 |
| Sum | 36 | |
| Algorithm | Architecture | Method | Classification Accuracy (%) | |||||
|---|---|---|---|---|---|---|---|---|
| 0.3% PRA Ratio | 1% PRA Ratio | 5% PRA Ratio | ||||||
| OA | Kappa | OA | Kappa | OA | Kappa | |||
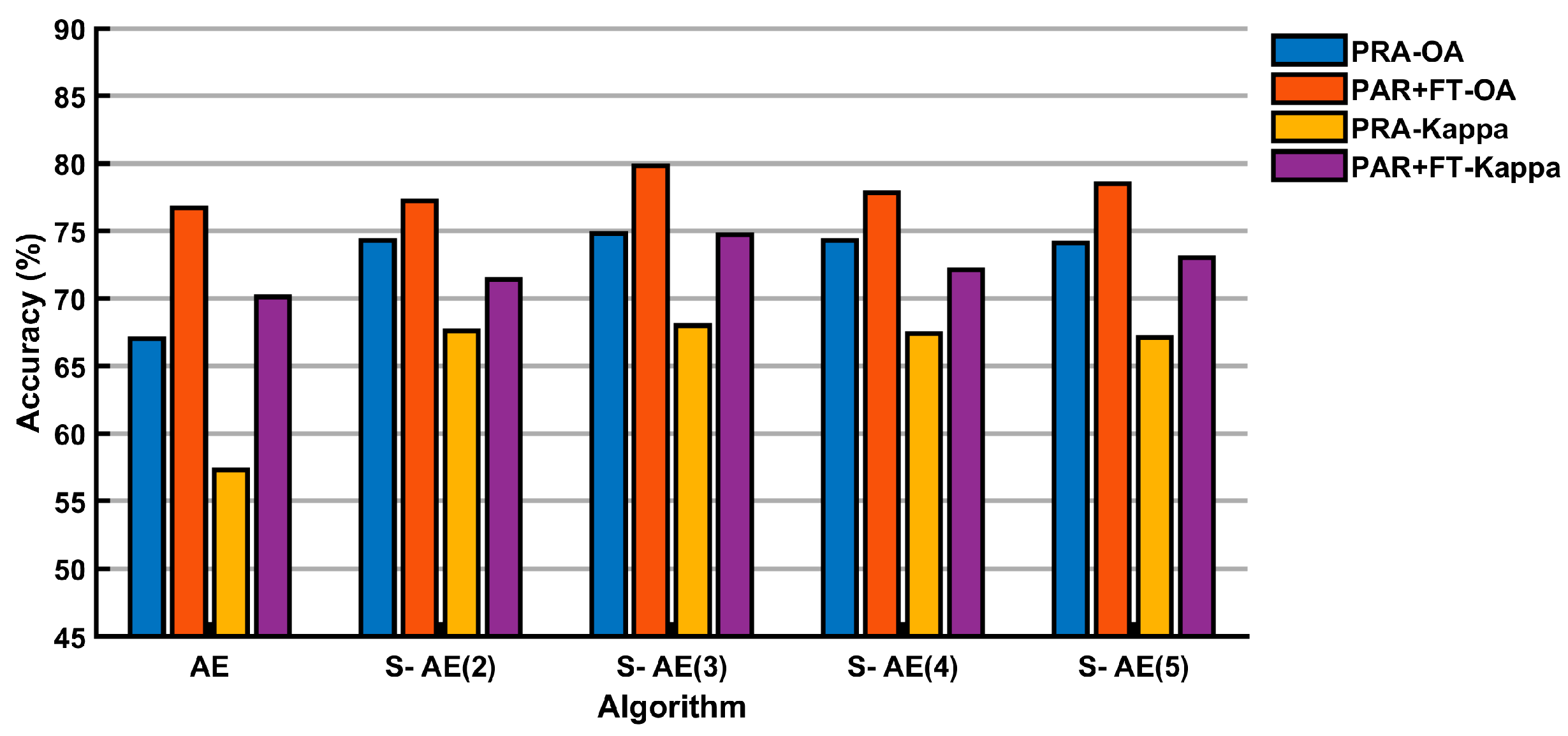
| AE | 252 - 9 | PRA | 63.61 | 52.30 | 67.01 | 57.26 | 70.35 | 62.01 |
| PRA+FT | 73.10 | 65.75 | 76.74 | 70.51 | 78.91 | 73.55 | ||
| S-AE2 | 252 - 100 - 9 | PRA | 73.09 | 65.83 | 74.34 | 67.57 | 76.23 | 70.12 |
| PRA+FT | 76.42 | 70.31 | 77.23 | 71.35 | 77.74 | 72.05 | ||
| S-AE3 | 252 - 100 - 50 - 9 | PRA | 73.08 | 65.79 | 74.77 | 68.01 | 76.56 | 70.48 |
| PRA+FT | 76.45 | 70.27 | 79.75 | 74.67 | 79.31 | 74.07 | ||
| S-AE4 | 252 - 100 - 50 -110 - 9 | PRA | 72.91 | 65.56 | 74.30 | 67.38 | 77.00 | 71.03 |
| PRA+FT | 74.16 | 67.23 | 77.82 | 72.13 | 77.51 | 71.73 | ||
| S-AE5 | 252 - 100 - 50 -110 - 30 - 9 | PRA | 72.37 | 64.83 | 74.12 | 67.14 | 76.47 | 70.73 |
| PRA+FT | 75.91 | 69.58 | 78.46 | 73.00 | 78.14 | 72.56 | ||
| Parameters | |||
|---|---|---|---|
| 0.0024 | 0.02 | 0.001 | |
| 0.6 | 0.2 | 0.4 | |
| 0.5 | 0.55 | 0.25 |
| Methods | OA (%) | Kappa (%) | Classification Accuracy of the 14 Crops (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H13 | H14 | |||
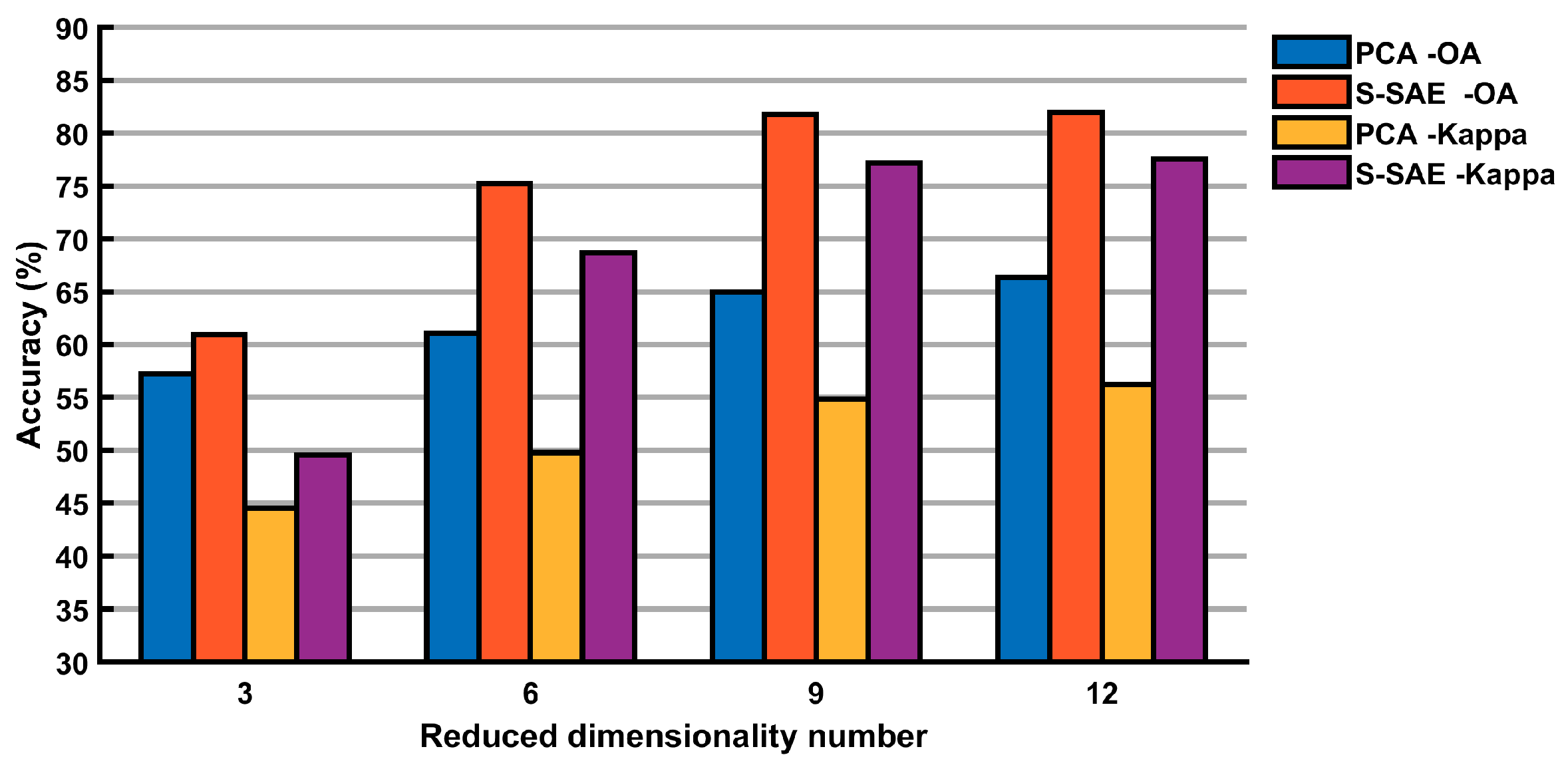
| PCA | 65.00 | 54.88 | 88 | 1 | 96 | 48 | 1 | 90 | 44 | 2 | 33 | 1 | 8 | 18 | 1 | 0 |
| LLE | 65.19 | 55.23 | 90 | 1 | 96 | 48 | 1 | 89 | 48 | 3 | 36 | 1 | 19 | 21 | 1 | 0 |
| AE(1) | 67.21 | 57.55 | 83 | 1 | 97 | 83 | 3 | 90 | 4 | 6 | 37 | 3 | 25 | 23 | 1 | 0 |
| AE(2) | 76.74 | 70.51 | 88 | 2 | 98 | 92 | 22 | 94 | 35 | 7 | 37 | 29 | 35 | 56 | 29 | 2 |
| S-AE(3) | 79.75 | 74.67 | 90 | 5 | 98 | 93 | 26 | 95 | 62 | 2 | 57 | 40 | 43 | 65 | 34 | 0 |
| S-AE(4) | 80.47 | 75.62 | 92 | 5 | 98 | 94 | 24 | 94 | 68 | 3 | 59 | 43 | 49 | 65 | 44 | 0 |
| S-SAE | 81.77 | 77.20 | 92 | 7 | 98 | 94 | 31 | 96 | 72 | 7 | 48 | 41 | 58 | 69 | 53 | 25 |
| Methods | OA (%) | Kappa (%) | Classification Accuracy of the 14 Crops (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| H1 | H2 | H3 | H4 | H5 | H6 | H7 | H8 | H9 | H10 | H11 | H12 | H13 | H14 | |||
| PCA | 87.59 | 84.87 | 96 | 55 | 96 | 92 | 56 | 98 | 78 | 50 | 79 | 61 | 76 | 80 | 49 | 23 |
| LLE | 88.30 | 85.80 | 97 | 51 | 94 | 94 | 62 | 99 | 76 | 62 | 79 | 63 | 86 | 84 | 60 | 61 |
| AE(1) | 87.86 | 85.29 | 91 | 44 | 92 | 97 | 63 | 99 | 88 | 59 | 71 | 70 | 89 | 79 | 75 | 31 |
| AE(2) | 93.03 | 91.58 | 96 | 70 | 96 | 99 | 79 | 99 | 88 | 55 | 83 | 80 | 89 | 87 | 86 | 92 |
| S-AE(3) | 94.74 | 93.65 | 98 | 69 | 97 | 99 | 81 | 100 | 85 | 58 | 88 | 84 | 95 | 94 | 93 | 61 |
| S-AE(4) | 94.95 | 93.92 | 98 | 71 | 97 | 99 | 81 | 100 | 83 | 59 | 82 | 92 | 91 | 94 | 98 | 79 |
| S-SAE | 95.44 | 94.51 | 99 | 78 | 97 | 99 | 80 | 100 | 89 | 57 | 86 | 88 | 97 | 94 | 94 | 75 |
| Methods | 1% | 5% | 10% | |||
|---|---|---|---|---|---|---|
| OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | |
| Complex Wishart | 60.86 | 55.89 | 61.05 | 56.08 | 61.26 | 56.28 |
| LLE + SVM | 65.19 | 55.23 | 69.93 | 61.60 | 72.75 | 65.35 |
| S-SAE + SVM | 81.77 | 77.20 | 82..35 | 78.01 | 84.06 | 80.17 |
| Chen + SVM | 66.99 | 57.45 | 71.61 | 63.77 | 72.13 | 64.47 |
| Chen + CNN | 91.74 | 90.04 | 96.35 | 95.55 | 97.93 | 97.51 |
| LSTM | 69.67 | 61.31 | 80.74 | 76.07 | 82.83 | 78.76 |
| LLE + CNN | 88.30 | 85.80 | 96..66 | 96.00 | 99.23 | 98.95 |
| S-SAE + CNN | 95.44 | 94.51 | 99.08 | 98.89 | 99.61 | 99.53 |
| Parameters | |||
|---|---|---|---|
| 0.001 | 0.02 | 0.001 | |
| 0.6 | 0.2 | 0.4 | |
| 0.25 | 0.55 | 0.25 |
| Methods | OA (%) | Kappa (%) | Classification Accuracy of the 14 Crops (%) | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| F1 | F2 | F3 | F4 | F5 | F6 | F7 | F8 | F9 | F10 | F11 | F12 | F13 | F14 | |||
| PCA | 87.30 | 84.94 | 0 | 94 | 91 | 89 | 64 | 83 | 70 | 87 | 59 | 95 | 57 | 32 | 90 | 96 |
| LLE | 87.90 | 85.69 | 5 | 95 | 79 | 92 | 62 | 86 | 67 | 89 | 80 | 94 | 56 | 32 | 94 | 97 |
| AE(1) | 86.62 | 84.16 | 12 | 93 | 92 | 82 | 45 | 84 | 72 | 84 | 79 | 96 | 46 | 18 | 95 | 94 |
| AE(2) | 88.09 | 85.89 | 13 | 94 | 88 | 90 | 50 | 87 | 70 | 88 | 68 | 95 | 49 | 34 | 93 | 97 |
| S-AE(3) | 88.98 | 86.95 | 0 | 98 | 86 | 86 | 82 | 74 | 74 | 86 | 81 | 96 | 62 | 39 | 97 | 98 |
| S-AE(4) | 89.93 | 87.98 | 3 | 97 | 91 | 87 | 36 | 87 | 80 | 88 | 63 | 97 | 74 | 40 | 94 | 98 |
| SAE | 91.09 | 89.47 | 34 | 96 | 92 | 91 | 55 | 91 | 79 | 90 | 80 | 98 | 64 | 44 | 95 | 97 |
| S-SAE | 91.63 | 90.11 | 35 | 97 | 92 | 93 | 67 | 90 | 77 | 87 | 85 | 98 | 86 | 41 | 97 | 98 |
| Methods | 1% | 5% | 10% | |||
|---|---|---|---|---|---|---|
| OA (%) | Kappa (%) | OA (%) | Kappa (%) | OA (%) | Kappa (%) | |
| Complex Wishart | 77.50 | 73.92 | 76.74 | 73.08 | 76.11 | 72.34 |
| LLE + SVM | 80.22 | 76.07 | 82.63 | 77.59 | 84.15 | 79.24 |
| S-SAE + SVM | 82.66 | 79.22 | 84.81 | 81.83 | 86.06 | 83.36 |
| Chen + SVM | 75.42 | 72.33 | 78.37 | 75.01 | 80.24 | 78.55 |
| Chen + CNN | 81.19 | 77.62 | 93.57 | 92.40 | 96.41 | 95.76 |
| LSTM | 73.93 | 68.51 | 76.52 | 71.72 | 79.77 | 75.71 |
| LLE + CNN | 87.90 | 85.69 | 94.99 | 94.09 | 97.28 | 96.30 |
| S-SAE + CNN | 91.63 | 90.11 | 95.90 | 95.17 | 97.57 | 97.14 |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Guo, J.; Li, H.; Ning, J.; Han, W.; Zhang, W.; Zhou, Z.-S. Feature Dimension Reduction Using Stacked Sparse Auto-Encoders for Crop Classification with Multi-Temporal, Quad-Pol SAR Data. Remote Sens. 2020, 12, 321. https://doi.org/10.3390/rs12020321
Guo J, Li H, Ning J, Han W, Zhang W, Zhou Z-S. Feature Dimension Reduction Using Stacked Sparse Auto-Encoders for Crop Classification with Multi-Temporal, Quad-Pol SAR Data. Remote Sensing. 2020; 12(2):321. https://doi.org/10.3390/rs12020321
Chicago/Turabian StyleGuo, Jiao, Henghui Li, Jifeng Ning, Wenting Han, Weitao Zhang, and Zheng-Shu Zhou. 2020. "Feature Dimension Reduction Using Stacked Sparse Auto-Encoders for Crop Classification with Multi-Temporal, Quad-Pol SAR Data" Remote Sensing 12, no. 2: 321. https://doi.org/10.3390/rs12020321
APA StyleGuo, J., Li, H., Ning, J., Han, W., Zhang, W., & Zhou, Z.-S. (2020). Feature Dimension Reduction Using Stacked Sparse Auto-Encoders for Crop Classification with Multi-Temporal, Quad-Pol SAR Data. Remote Sensing, 12(2), 321. https://doi.org/10.3390/rs12020321






