1. Introduction
Conventionally, forest inventory has been implemented by field survey which is time-consuming and labor-intensive [
1,
2]. Moreover, because forest attribute information is affected by topography and the environment, the parameters that can be measured manually are very limited [
3]. Over the poast three decades, airborne light detection and ranging (LiDAR) technology has been increasingly used to estimate forest structural attributes [
4,
5,
6]. Airborne LiDAR data provide detailed three-dimensional (3D) information of vertical forest structures to estimate forestry parameters at both individual and stand tree levels [
7]. With the technology development, compared to airborne LiDAR, unmanned aerial vehicles (UAV LiDAR) have gained a reputation in forest inventory surveying because of their increased data resolution, lower costs, flight flexibility, and simple operation [
6,
8,
9,
10].
Forest inventory requires accurate individual tree structures to directly obtain forest structural attributes (i.e., crown width and tree height) or estimate height-related attributes by establishing regression models (such as diameter at breast height (DBH), volume, and aboveground biomass (AGB)) [
11,
12,
13,
14]. Thus, the segmentation of individual trees becomes the key step to highly accurate forestry parameter estimation. However, the existing problems of individual tree segmentation using LiDAR data are mainly caused by: (1) adjacent, overlapped tree crowns, and various tree sizes, which result in serious omission errors in multi-layered forest stands [
15]; (2) irregular tree shapes (such as a branch of a tree extending outward widely and resembling a small tree), which lead to commission errors [
7]; (3) suppressed or small trees located below tall trees, which are difficult detected because LiDAR systems are less likely to penetrate the dense forest canopy and have less information of suppressed trees.
To solve the aforementioned issues, many methods have been developed. Some studies make use of height variation of canopy height model (CHM) to find treetops by local maxima (LM) filtering [
16] or moving variable window [
17,
18]. Then, tree crowns are mainly delineated by image-processing algorithms designed for edge detection and feature extraction, such as region growing [
19], watershed analysis [
17,
20], spatial wavelet analysis [
21], and template matching [
22,
23]. The CHM-based tree detection algorithms are fast and efficient but easy to produce omission and commission errors [
24]. The tree detection accuracy is sensitive to the resolution of CHM and the noise caused by irregular tree shapes and overlapped trees. In addition, because of a variety of canopy sizes, it is difficult to select the optimal resolution and smoothing factor suitable for the whole scene [
7,
25,
26]. To improve the detection rate, the vertical and horizontal structures of point clouds are analyzed. Liu et al. [
7] integrated a multiscale CHM-based algorithm and a point-based vertical profile analysis algorithm to improve individual tree segmentation accuracy. Paris et al. [
24] improved the segmentation accuracies of dominant trees by combining CHM data and horizontal profiles of point clouds and extracted subdominant trees by analyzing vertical profiles. By using a marker-controlled watershed segmentation method, Harikumar et al. [
27] first segmented a point cloud into a set of segments, each of which was projected onto a novel 3D space to separate subdominant from dominant trees.
Because of the increased spatial resolution of UAV LiDAR data, some studies, to improve segmentation accuracy, segmented individual trees directly from point clouds. The existing point-based clustering methods (such as region growing [
17], k-means clustering [
26], normalized cut [
28], and mean shift [
29,
30]) have been widely used for individual tree segmentation in terms of 3D data characteristics, geometric structures, and height variation of trees [
31,
32,
33]. Compared with other clustering methods, mean shift does not require seed points or number of clusters before clustering. It proves to be robust to segment various kinds of trees [
34,
35]. Mean shift aims to move each data point to the densest area within a certain neighborhood by iteratively performing shift operations based on a kernel density function. The kernel function determines the weights of neighboring points for re-estimating the local maximum. Eventually, the points will converge to the local maxima of density [
29]. Ferraz et al. [
30] developed a fixed bandwidth mean shift method, in which a horizontal and a vertical kernel functions were designed to shift data points to a denser and higher region until it converged to the treetop. Ferraz et al. [
36] further improved it by establishing a self-calibrated bandwidth model, which relates tree heights to crown widths and depths by extracting easily recognized individual trees. The method required complicated pre-processing and the established allometric model might not be suitable for all tree species [
29]. Hu et al. [
29] proposed an adaptive mean shift-based clustering approach, in which the key parameter—kernel size—was automatically changed to reduce over- and under-segmentations according to the estimated crown diameters of distinct trees, and nearly 30% more suppressed trees are identified compared to the fixed bandwidth-based methods. However, point-based methods are generally highly computational when having been directly applied to the whole scene [
7,
33]. Hamraz et al. [
35], to improve computational efficiency, generated vertical profiles of point clouds in different directions within a local region, then detected crown boundary for each profile. This method greatly improved computational efficiency and required no prior knowledge of the shapes and sizes of tree crowns [
35].
In summary, in mean shift methods, the kernel bandwidth is a major factor, which has a strong impact on the segmentation accuracy and clustering speed. The fixed kernel bandwidth was widely used in segmentation, but it cannot adapt to heterogeneous crown sizes, and thereby might result in omission errors or commission errors. For example, if a small kernel bandwidth is applied to large objects, commission errors will occur (that is, a single tree might be segmented into several clusters); whereas if a large kernel bandwidth is applied to small objects, omission errors will greatly appear (that is, more than two trees are clustered into one segment). Our previous research in [
37] used fixed bandwidth mean shift method to segment individual trees and reduced omission errors by normalized Cut algorithm (NCut). The method improved tree segmentation accuracy and also increased computational complexity.
Therefore, in this paper, to reduce omission and commission errors, we propose a self-adaptive mean shift method, where an adaptive kernel bandwidth is used according to local tree structure information for individual tree segmentation. Moreover, because mean shift methods are generally highly computational when directly applied to LiDAR data, a 3D space division strategy is used to improve tree segmentation efficiency. In addition, point density is another important factor affecting individual tree detection, we also evaluate the effects of point density on individual tree detection. The remainder of the paper is organized as follows:
Section 2 details the proposed self-adaptive mean shift segmentation method.
Section 3 describes the study area and the test data including UAV LiDAR data and field survey data, and then analyzes the segmentation results. Concluding remarks are given in
Section 4.
2. Methodology
In this paper, the proposed method segments individual trees by the following steps. Ground points are separated from non-ground points, and then non-ground points are normalized for the data pre-processing. Then, the optimal kernel bandwidth for mean shift is automatically estimated without any prior knowledge of crown size to extract individual tree points. The proposed self-adaptive mean shift method is iteratively implemented within a given area until all trees are segmented.
In the pre-processing process, because ground points affect segmentation results, we remove them by a cloth simulation (CSF) algorithm [
38]. Afterward, to reduce the influence of topography on the following tree segmentation, we normalize non-ground points according to the produced digital terrain model (DTM) interpolated by the filtered ground points. The details can be seen in [
37].
2.1. Self-Adaptive Kernel Bandwidth Determination
The kernel bandwidth is a critical parameter of the mean shift method. In this study, it represents the region where the local maximum exists, thereby we adaptively determine it according to the related tree crown size. The kernel bandwidth is determined by the following steps.
3D space division, which locates the global maximum point in a point cloud as the center point to divide the point cloud into a set of angular sectors at multiple directions.
Crown surface generation, which generates a vertical profile for each sector and extracts LiDAR canopy surface points to simulate a crown surface.
Kernel bandwidth determination, which, from the generated crown surface, a crown boundary is delineated to automatically determine the kernel bandwidth according to the between-tree gap and height variation.
The rest of this section details the delineation of the kernel bandwidth.
2.1.1. 3D Space Division
We first locate the highest point in a point cloud as the center point,
Xc = (
xc,
yc,
zc), which is defined as the apex of the highest tree. With the center point,
Xc, we divide the 3D space into
n angular sectors within a given search radius
R (
R must be larger than the largest trees in the scene to be processed) (see
Figure 1). For each angular sector
θj Є [2π
j/
n, 2π(
j + 1)/
n] (
j = 0,1…
n-1), we search for the points
Xi = (
xi, yi, zi) (
i = 1, 2….
n, n is the number of the points) belonging to. The set
Xi belonging to angular sector
θj is defined as:
2.1.2. Crown Surface Simulation
For each angular sector (see
Figure 2a), we generate a vertical profile of LiDAR points and extract potential canopy surface points. We first calculate the distance
Di (
i = 1, 2….
n),
between
Xc and
Xi. Then, a vertical profile is generated in terms of coordinates
z and
d (see
Figure 2b). In this study, only LiDAR canopy surface points are kept to simulate the crown surface. As seen in
Figure 2c, in doz plane, points are divided into a set of grids with a given grid size
ɛ, each of which the maximum height is kept to simulate the crown surface.
2.1.3. Kernel Bandwidth Determination
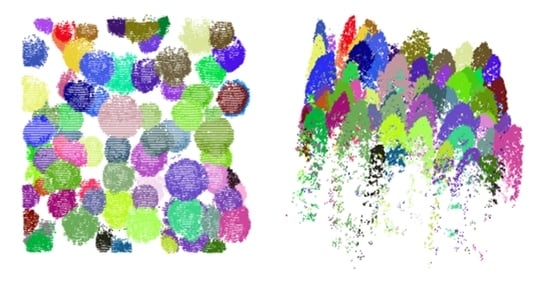
After the crown surface simulation, the crown boundary can be delineated according to between-tree gaps and height variations. In the canopy surface points, the local minimum points always exist between adjacent tree crowns. Starting from Xc, only the first local minimum point is remained for further analysis. The distance between center point Xc and the local minimum point is identified as a crown radius. To smooth extended branches and reduce false local minimum points, a Gaussian filtering is first applied to the simulated crown surface points. After estimating all crown radii at multiple directions, the average crown radius is calculated as the kernel bandwidth.
The determination of kernel bandwidth is completely based on the coordinate information of LiDAR points without any prior knowledge. Moreover, the division of the 3D space decreases the complexity of 3D forest structure and improves the computational efficiency.
2.2. Mean Shift-Based Individual Tree Segmentation
With the determined kernel bandwidth, the individual tree points are extracted by mean shift, which gains its reputations for great performance in individual tree segmentation. In this study, a horizontal and a vertical kernel functions (g
s and
gr) are designed to shift data points
Xi to treetop
Xc. The offset vector is defined by:
where the superscripts
s and
r refer to the horizontal and vertical directions, respectively.
hs and
hr represent the horizontal and vertical bandwidths. g
s is the horizontal kernel function that follows Gaussian function:
gr is specially-designed for assigning a larger weight to the highest voxel:
where mask (
xcr,xir) represents a mask of foreground object; dist(
xcr,xir) is the distance between
Xi and the boundary of the mask. They are defined by,
More details can be found in literature [
31].
After detecting and extracting the tallest tree points, the method will automatically search the tallest point in the remaining points. The method was iteratively implemented within a given area until all individual tree points were segmented, and eliminate clusters containing less than 50 points to reduce interference from branches.