An Optimized Object-Based Random Forest Algorithm for Marsh Vegetation Mapping Using High-Spatial-Resolution GF-1 and ZY-3 Data
Abstract
:1. Introduction
2. Study Area and Data Source
2.1. Study Area
2.2. Data Source
2.2.1. Remotely Sensed and Ancillary Data
2.2.2. Field and Validation Data
2.3. Data Preparation
2.3.1. Data Preprocessing
2.3.2. Calculation of Spectral Indices and Textural Information
3. Method
3.1. Multi-“Scales” Segmentation
3.2. Object-Based RF Model Sevelopment and Classification
3.2.1. Parameter Optimization
3.2.2. Variable Selection Algorithms
- (1)
- RFE Algorithm
- Train the RF model on the training set using all features.
- Calculate model performance.
- Rank feature importance.
- for each subset size Si, i = 1 … S do
- Keep the Si most important features.
- Preprocess the data.
- Train the model on the training set using Si predictors.
- Calculate model performance.
- Recalculate the rankings for each predictor.
- end
- Calculate the performance profile over the Si.
- Determine the appropriate number of predictors.
- Use the model corresponding to the optimal Si.
- (2)
- Boruta Algorithm
- Extend the information system by adding copies of all features (at least five shadow features).
- Remove their relevance to the response by adding features.
- Run RF classification on the expanded feature set and calculate z-scores.
- Find the maximum z-score among shadow features (MZSF) and then assign a hit for each feature that scored better than MZSF.
- For each feature with undetermined importance, perform a two-sided test of equality with the MZSF.
- Features that are significantly less important than MZSF are called “not important”; permanently remove them from the feature set.
- Features that are significantly more important than MZSF are called “important.”
- Remove all shadow attributes.
- Repeat the procedure until you have specified importance for all attributes.
- (3)
- VSURF Algorithm
- Preliminary elimination and ranking
- Sort features by feature importance in descending order (99 RF runs).
- Eliminate features of lower importance (let m denote the number of remaining features).
- Variable selection
- For interpretation: construct a nested set of RF models involving the k first features, for k = 1 to m and select the features involved in the model that cause the smallest out-of-bag error. This leads to the consideration of m’ features.
- For prediction: starting with the ordered features reserved for interpretation, construct an incremental sequence of RF models by invoking and testing the features in a stepwise way. Select the features of the last model.
3.2.3. Accuracy Assessment
4. Results
4.1. Parameter Optimization
4.2. Variable Selection
4.2.1. RFE-based Variable Selection Result
4.2.2. Boruta-based Variable Selection Result
4.2.3. VSURF-based Variable Selection Result
4.3. Visual Comparison and Accuracy Assessment of Classification Results
5. Discussion
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Appendix A

References
- Henderson, F.M.; Lewis, A.J. Radar detection of wetland ecosystems: A review. Int. J. Remote Sens. 2008, 29, 5809–5835. [Google Scholar] [CrossRef]
- Betbeder, J.; Rapinel, S.; Corpetti, T.; Pottier, E.; Corgne, S.; Hubert-Moy, L. Multitemporal classification of TerraSAR-X data for wetland vegetation mapping. J. Appl. Remote Sens. 2014, 8, 083648. [Google Scholar] [CrossRef]
- Zhou, D.; Gong, H.; Wang, Y.; Khan, S.; Zhao, K. Driving forces for the marsh wetland degradation in the Honghe National Nature Reserve in Sanjiang Plain, Northeast China. Environ. Model. Assess. 2009, 14, 101–111. [Google Scholar] [CrossRef]
- Adam, E.; Mutanga, O.; Rugege, D. Multispectral and hyperspectral remote sensing for identification and mapping of marsh wetland vegetation: A review. Wetl. Ecol. Manag. 2010, 18, 281–296. [Google Scholar] [CrossRef]
- Millard, K.; Richardson, M. Wetland mapping with LiDAR derivatives, SAR polarimetric decompositions, and LiDAR–SAR fusion using a random forest classifier. Can. J. Remote Sens. 2013, 39, 290–307. [Google Scholar] [CrossRef]
- Zedler, J.B.; Kercher, S. Wetland resources: Status, trends, ecosystem services, and restorability. Annu. Rev. Environ. Resour. 2005, 30, 39–74. [Google Scholar] [CrossRef] [Green Version]
- Kokaly, R.F.; Despain, D.G.; Clark, R.N.; Livo, K.E. Mapping vegetation in Yellowstone National Park using spectral feature analysis of AVIRIS data. Remote Sens. Environ. 2003, 84, 437–456. [Google Scholar] [CrossRef] [Green Version]
- Ramsey, E.; Rangoonwala, A.; Middleton, B.; Lu, Z. Satellite optical and radar data used to track wetland forest impact and short-term recovery from Hurricane Katrina. Wetlands 2009, 29, 66–79. [Google Scholar] [CrossRef] [Green Version]
- Jenkins, R.B.; Frazier, P.S. High-resolution remote sensing of upland swamp boundaries and vegetation for baseline mapping and monitoring. Wetlands 2010, 30, 531–540. [Google Scholar] [CrossRef]
- Frohn, R.C.; Autrey, B.C.; Lane, C.R.; Reif, M. Segmentation and object-oriented classification of wetlands in a karst Florida landscape using multi-season Landsat-7 ETM+ imagery. Int. J. Remote Sens. 2011, 32, 1471–1489. [Google Scholar] [CrossRef]
- Tuxen, K.; Schile, L.; Stralberg, D.; Siegel, S.; Parker, T.; Vasey, M.; Callaway, J.; Kelly, M. Mapping changes in tidal wetland vegetation composition and pattern across a salinity gradient using high spatial resolution imagery. Wetl. Ecol. Manag. 2011, 19, 141–157. [Google Scholar] [CrossRef]
- Zhang, Y.; Lu, D.; Yang, B.; Sun, C.; Sun, M. Coastal wetland vegetation classification with a Landsat Thematic Mapper image. Int. J. Remote Sens. 2011, 32, 545–561. [Google Scholar] [CrossRef]
- Abeysinghe, T.; Simic Milas, A.; Arend, K.; Hohman, B.; Reil, P.; Gregory, A.; Vázquez-Ortega, A. Mapping invasive phragmites australis in the Old Woman Creek Estuary using UAV remote sensing and machine learning classifiers. Remote Sens. 2019, 11, 1380. [Google Scholar] [CrossRef] [Green Version]
- Wietecha, M.; Jełowicki, Ł.; Mitelsztedt, K.; Miścicki, S.; Stereńczak, K. The capability of species-related forest stand characteristics determination with the use of hyperspectral data. Remote Sens. Environ. 2019, 231, 111232. [Google Scholar] [CrossRef]
- Sang, X.; Guo, Q.; Wu, X.; Fu, Y.; Xie, T.; He, C.; Zang, J. Intensity and stationarity analysis of land use change based on CART algorithm. Sci. Rep. 2019, 9, 1–12. [Google Scholar] [CrossRef] [Green Version]
- Li, W.; El-Askary, H.; Qurban, M.A.; Li, J.; ManiKandan, K.P.; Piechota, T. Using multi-indices approach to quantify mangrove changes over the Western Arabian Gulf along Saudi Arabia coast. Ecol. Indic. 2019, 102, 734–745. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Mohammadimanesh, F.; Homayouni, S.; Gill, E. The first wetland inventory map of newfoundland at a spatial resolution of 10 m using sentinel-1 and sentinel-2 data on the google earth engine cloud computing platform. Remote Sens. 2019, 11, 43. [Google Scholar] [CrossRef] [Green Version]
- Amani, M.; Salehi, B.; Mahdavi, S.; Brisco, B. Spectral analysis of wetlands using multi-source optical satellite imagery. ISPRS J. Photogramm. 2018, 144, 119–136. [Google Scholar] [CrossRef]
- Fu, B.; Wang, Y.; Campbell, A.; Li, Y.; Zhang, B.; Yin, S.; Jin, X. Comparison of object-based and pixel-based Random Forest algorithm for marsh wetland vegetation mapping using high spatial resolution GF-1 and SAR data. Ecol. Indic. 2017, 73, 105–117. [Google Scholar] [CrossRef]
- Dronova, I.; Gong, P.; Wang, L. Object-based analysis and change detection of major wetland cover types and their classification uncertainty during the low water period at Poyang Lake, China. Remote Sens. Environ. 2011, 115, 3220–3236. [Google Scholar] [CrossRef]
- Boyden, J.; Joyce, K.E.; Boggs, G.; Wurm, P. Object-based mapping of native vegetation and para grass (Urochloa mutica) on a monsoonal wetland of Kakadu NP using a Landsat 5 TM Dry-season time series. J. Spat. Sci. 2013, 58, 53–77. [Google Scholar] [CrossRef]
- Dronova, I. Object-based image analysis in wetland research: A review. Remote Sens. 2015, 7, 6380–6413. [Google Scholar] [CrossRef] [Green Version]
- Dronova, I.; Gong, P.; Wang, L.; Zhong, L. Mapping dynamic cover types in a large seasonally flooded wetland using extended principal component analysis and object-based classification. Remote Sens. Environ. 2015, 158, 193–206. [Google Scholar] [CrossRef]
- Mui, A.; He, Y.; Weng, Q. An object-based approach to delineate wetlands across landscapes of varied disturbance with high spatial resolution satellite imagery. ISPRS J. Photogramm. 2015, 109, 30–46. [Google Scholar] [CrossRef] [Green Version]
- Dronova, I.; Gong, P.; Clinton, N.E.; Wang, L.; Fu, W.; Qi, S.; Liu, Y. Landscape analysis of wetland plant functional types: The effects of image segmentation scale, vegetation classes and classification methods. Remote Sen. Environ. 2012, 127, 357–369. [Google Scholar] [CrossRef]
- Chen, Y.; Niu, Z.; Johnston, C.A.; Hu, S. A Unifying Approach to Classifying Wetlands in the Ontonagon River Basin, Michigan, Using Multi-temporal Landsat-8 OLI Imagery. Can. J. Remote Sens. 2018, 44, 373–389. [Google Scholar] [CrossRef]
- Ludwig, C.; Walli, A.; Schleicher, C.; Weichselbaum, J.; Riffler, M. A highly automated algorithm for wetland detection using multi-temporal optical satellite data. Remote Sens. Environ. 2019, 224, 333–351. [Google Scholar] [CrossRef]
- Mahdianpari, M.; Salehi, B.; Mohammadimanesh, F.; Motagh, M. Random forest wetland classification using ALOS-2 L-band, RADARSAT-2 C-band, and TerraSAR-X imagery. ISPRS J. Photogramm. 2017, 130, 13–31. [Google Scholar] [CrossRef]
- Merchant, M.A.; Warren, R.K.; Edwards, R.; Kenyon, J.K. An object-based assessment of multi-wavelength SAR, optical imagery and topographical datasets for operational wetland mapping in Boreal Yukon, Canada. Can. J. Remote Sens. 2019, 45, 308–332. [Google Scholar] [CrossRef]
- Mohammadimanesh, F.; Salehi, B.; Mahdianpari, M.; Motagh, M.; Brisco, B. An efficient feature optimization for wetland mapping by synergistic use of SAR intensity, interferometry, and polarimetry data. Int. J. Appl. Earth Obs. 2018, 73, 450–462. [Google Scholar] [CrossRef]
- Ghosh, A.; Joshi, P.K. A comparison of selected classification algorithms for mapping bamboo patches in lower Gangetic plains using very high resolution WorldView 2 imagery. Int. J. Appl. Earth Obs. 2014, 26, 298–311. [Google Scholar] [CrossRef]
- Räsänen, A.; Kuitunen, M.; Tomppo, E.; Lensu, A. Coupling high-resolution satellite imagery with ALS-based canopy height model and digital elevation model in object-based boreal forest habitat type classification. ISPRS J. Photogramm. 2014, 94, 169–182. [Google Scholar] [CrossRef] [Green Version]
- Speiser, J.L.; Miller, M.E.; Tooze, J.; Ip, E. A comparison of Random Forest variable selection methods for classification prediction modeling. Expert Syst. Appl. 2019, 134, 93–101. [Google Scholar] [CrossRef]
- Chunling, L.; Zhaoguang, B. Characteristics and typical applications of GF-1 satellite. In Proceedings of the 2015 IEEE International Geoscience and Remote Sensing Symposium, Milan, Italy, 26–31 July 2015; pp. 1246–1249. [Google Scholar]
- Cao, H.; Gao, W.; Zhang, X.; Liu, X.; Fan, B.; Li, S. Overview of ZY-3 satellite research and application. In Proceedings of the 63rd IAC (International Astronautical Congress), Naples, Italy, 1–5 October 2012. [Google Scholar]
- Johnston, K.; Ver Hoef, J.M.; Krivoruchko, K.; Lucas, N. Using ArcGIS Geostatistical Analyst; Esri: Redlands, CA, USA, 2001. [Google Scholar]
- Exelis, V.I.S. ENVI 5.3; Exelis VIS: Boulder, CO, USA, 2015. [Google Scholar]
- Kaufman, Y.J.; Wald, A.E.; Remer, L.A.; Gao, B.; Li, R.; Flynn, L. The modis 2.1-μm channel-correlation with visible reflectance for use in remote sensing of aerosol. IEEE Trans. Geosci. Remote 1997, 35, 1286–1298. [Google Scholar] [CrossRef]
- Laben, C.A.; Brower, B.V. Process for enhancing the spatial resolution of multispectral imagery using pan-sharpening. U.S. Patent 6,011,875, 4 January 2000. [Google Scholar]
- Cho, M.A.; Malahlela, O.; Ramoelo, A. Assessing the utility WorldView-2 imagery for tree species mapping in South African subtropical humid forest and the conservation implications: Dukuduku forest patch as case study. Int. J. Appl. Earth Obs. 2015, 38, 349–357. [Google Scholar] [CrossRef]
- Xu, K.; Tian, Q.; Yang, Y.; Yue, J.; Tang, S. How up-scaling of remote-sensing images affects land-cover classification by comparison with multiscale satellite images. Int. J. Remote Sens. 2019, 40, 2784–2810. [Google Scholar] [CrossRef]
- Rampi, L.P.; Knight, J.F.; Pelletier, K.C. Wetland mapping in the upper midwest United States. Photogramm. Eng. Rem. Sens. 2014, 80, 439–448. [Google Scholar] [CrossRef]
- Maxwell, A.E.; Warner, T.A.; Strager, M.P. Predicting palustrine wetland probability using random forest machine learning and digital elevation data-derived terrain variables. Photogramm. Eng. Rem. Sens. 2016, 82, 437–447. [Google Scholar] [CrossRef]
- Shawky, M.; Moussa, A.; Hassan, Q.K.; El-Sheimy, N. Pixel-based geometric assessment of channel networks/orders derived from global spaceborne digital elevation models. Remote Sens. 2019, 11, 235. [Google Scholar] [CrossRef] [Green Version]
- Haralick, R.M.; Shanmugam, K.; Dinstein, I.H. Textural features for image classification. IEEE Trans. Syst. Man Cybern. 1973, 6, 610–621. [Google Scholar] [CrossRef] [Green Version]
- Szantoi, Z.; Escobedo, F.; Abd-Elrahman, A.; Smith, S.; Pearlstine, L. Analyzing fine-scale wetland composition using high resolution imagery and texture features. Int. J. Appl. Earth Obs. 2013, 23, 204–212. [Google Scholar] [CrossRef]
- Hidayat, S.; Matsuoka, M.; Baja, S.; Rampisela, D. Object-based image analysis for sago palm classification: The most important features from high-resolution satellite imagery. Remote Sens. 2018, 10, 1319. [Google Scholar] [CrossRef] [Green Version]
- Tian, S.; Zhang, X.; Tian, J.; Sun, Q. Random forest classification of wetland landcovers from multi-sensor data in the arid region of Xinjiang, China. Remote Sens. 2016, 8, 954. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Li, G.; Moran, E.; Dutra, L.; Batistella, M. The roles of textural images in improving land-cover classification in the Brazilian Amazon. Int. J. Remote Sens. 2014, 35, 8188–8207. [Google Scholar] [CrossRef] [Green Version]
- Szantoi, Z.; Escobedo, F.; Abd-Elrahman, A.; Pearlstine, L.; Dewitt, B.; Smith, S. Classifying spatially heterogeneous wetland communities using machine learning algorithms and spectral and textural features. Environ. Monit. Assess. 2015, 187, 262. [Google Scholar] [CrossRef]
- eCognition Developer, T. 9.0 User Guide; Trimble Germany GmbH: Munich, Germany, 2014. [Google Scholar]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring Vegetation Systems in the Great Plains with ERTS. In Third Earth Resources Technology Satellite-1 Symposium; NASA: Washington, DC, USA, 1973; pp. 309–317. [Google Scholar]
- Major, D.J.; Baret, F.; Guyot, G. A ratio vegetation index adjusted for soil brightness. Int. J. Remote Sens. 1990, 11, 727–740. [Google Scholar] [CrossRef]
- Gitelson, A.; Spivak, L.; Zakarin, E.; Kogan, F.; Lebed, L. Estimation of seasonal dynamics of pasture and crop productivity in Kazakhstan using NOAA/AVHRR data. In Proceedings of the IGARSS’96. 1996 International Geoscience and Remote Sensing Symposium, Lincoln, NE, USA, 31–31 May 1996. [Google Scholar]
- Mallick, K.; Bhattacharya, B.K.; Patel, N.K. Estimating volumetric surface moisture content for cropped soils using a soil wetness index based on surface temperature and NDVI. Agr. Forest Meteorol. 2009, 149, 1327–1342. [Google Scholar] [CrossRef]
- Sörensen, R.; Zinko, U.; Seibert, J. On the calculation of the topographic wetness index: Evaluation of different methods based on field observations. Hydrol. Earth Syst. Sci. 2006, 10, 101–112. [Google Scholar] [CrossRef] [Green Version]
- Inglada, J. Automatic recognition of man-made objects in high resolution optical remote sensing images by SVM classification of geometric image features. ISPRS J. Photogramm. 2007, 62, 236–248. [Google Scholar] [CrossRef]
- Moffett, K.B.; Gorelick, S.M. Distinguishing wetland vegetation and channel features with object-based image segmentation. Int. J. Remote Sens. 2013, 34, 1332–1354. [Google Scholar] [CrossRef]
- Drǎguţ, L.; Tiede, D.; Levick, S.R. ESP: A tool to estimate scale parameter for multiresolution image segmentation of remotely sensed data. Int. J. Geogr. Inf. Sci. 2010, 24, 859–871. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Phiri, D.; Morgenroth, J.; Xu, C. Four decades of land cover and forest connectivity study in Zambia—An object-based image analysis approach. Int. J. Appl. Earth. Obs. 2019, 79, 97–109. [Google Scholar] [CrossRef]
- Genuer, R.; Poggi, J.M.; Tuleau-Malot, C. Variable selection using random forests. Pattern Recogn. Lett. 2010, 31, 2225–2236. [Google Scholar] [CrossRef] [Green Version]
- Silveira, E.M.; Silva, S.H.G.; Acerbi-Junior, F.W.; Carvalho, M.C.; Carvalho, L.M.T.; Scolforo, J.R.S.; Wulder, M.A. Object-based random forest modelling of aboveground forest biomass outperforms a pixel-based approach in a heterogeneous and mountain tropical environment. Int. J. Appl. Earth Obs. 2019, 78, 175–188. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. The randomforest package. R News 2002, 2, 18–22. [Google Scholar]
- Team, R.C. R: A language and environment for statistical computing. Available online: http://http://cran.fhcrc.org/web/packages/dplR/vignettes/intro-dplR.pdf (accessed on 15 April 2020).
- Kuhn, M. Variable Selection Using the Caret Package. Available online: http://cran.r-project.org/web/packages/caret/vignettes/caretSelection.pdf (accessed on 15 April 2020).
- Kursa, M.B.; Rudnicki, W.R. Feature selection with the Boruta package. J. Stat. Softw. 2010, 36, 1–13. [Google Scholar] [CrossRef] [Green Version]
- Genuer, R.; Poggi, J.M.; Tuleaumalot, C. VSURF: An R package for variable selection using Random Forests. R. J. 2015, 7, 19–33. [Google Scholar] [CrossRef] [Green Version]
- Olofsson, P.; Foody, G.M.; Herold, M.; Stehman, S.V.; Woodcock, C.E.; Wulder, M.A. Good practices for estimating area and assessing accuracy of land change. Remote Sens. Environ. 2014, 148, 42–57. [Google Scholar] [CrossRef]
- Phiri, D.; Morgenroth, J.; Xu, C.; Hermosilla, T. Effects of pre-processing methods on Landsat OLI-8 land cover classification using OBIA and random forests classifier. Int. J. Appl. Earth. Obs. 2018, 73, 170–178. [Google Scholar] [CrossRef]
- Foody, G.M. Thematic map comparison: Evaluating the Statistical significance of differences in classification accuracy. Photogramm. Eng. Rem. Sens. 2004, 70, 627–634. [Google Scholar] [CrossRef]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Thanh Noi, P.; Kappas, M. Comparison of random forest, k-nearest neighbor, and support vector machine classifiers for land cover classification using Sentinel-2 imagery. Sensors 2018, 18, 18. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Stumpf, A.; Kerle, N. Object-oriented mapping of landslides using Random Forests. Remote Sens. Environ. 2011, 115, 2564–2577. [Google Scholar] [CrossRef]
- Nguyen, U.; Glenn, E.P.; Dang, T.D.; Pham, L.T. Mapping vegetation types in semi-arid riparian regions using random forest and object-based image approach: A case study of the Colorado River Ecosystem, Grand Canyon, Arizona. Ecol. Inform. 2019, 50, 43–50. [Google Scholar] [CrossRef]
- Zhang, Y.; Zhang, H.; Lin, H. Improving the impervious surface estimation with combined use of optical and SAR remote sensing images. Remote Sens. Environ. 2014, 141, 155–167. [Google Scholar] [CrossRef]
- Ming, D.; Zhou, T.; Wang, M.; Tan, T. Land cover classification using random forest with genetic algorithm-based parameter optimization. J. Appl. Remote Sens. 2016, 10, 035021. [Google Scholar] [CrossRef]
- Zhang, H.; Li, Q.; Liu, J.; Shang, J.; Du, X.; McNairn, H.; Champagne, C.; Dong, T.; Liu, M. Image classification using rapideye data: Integration of spectral and textual features in a random forest classifier. IEEE J. STARS. 2017, 10, 5334–5349. [Google Scholar] [CrossRef]
- Georganos, S.; Grippa, T.; Vanhuysse, S.; Lennert, M.; Shimoni, M.; Kalogirou, S.; Wolff, E. Less is more: Optimizing classification performance through feature selection in a very-high-resolution remote sensing object-based urban application. GISci. Remote Sens. 2018, 55, 221–242. [Google Scholar] [CrossRef]
- Lagrange, A.; Fauvel, M.; Grizonnet, M. Large-scale feature selection with Gaussian mixture models for the classification of high dimensional remote sensing images. IEEE Trans. Comput. Imaging 2017, 3, 230–242. [Google Scholar] [CrossRef] [Green Version]
- Millard, K.; Richardson, M. On the importance of training data sample selection in random forest image classification: A case study in peatland ecosystem mapping. Remote Sens. 2015, 7, 8489–8515. [Google Scholar] [CrossRef] [Green Version]
- Shih, H.C.; Stow, D.A.; Tsai, Y.H. Guidance on and comparison of machine learning classifiers for Landsat-based land cover and land use mapping. Int. J. Remote Sens. 2019, 40, 1248–1274. [Google Scholar] [CrossRef]
- Lim, J.; Kim, K.M.; Jin, R. Tree species classification using Hyperion and Sentinel-2 Data with machine learning in South Korea and China. ISPRS. Int. J. Geo-Inf. 2019, 8, 150. [Google Scholar] [CrossRef] [Green Version]
- Gumbricht, T. Detecting trends in wetland extent from MODIS derived soil moisture estimates. Remote Sens. 2018, 10, 611. [Google Scholar] [CrossRef] [Green Version]
- Berhane, T.; Lane, C.; Wu, Q.; Autrey, B.; Anenkhonov, O.; Chepinoga, V.; Liu, H. Decision-tree, rule-based, and random forest classification of high-resolution multispectral imagery for wetland mapping and inventory. Remote Sens. 2018, 10, 580. [Google Scholar] [CrossRef] [Green Version]
- McCarthy, M.J.; Radabaugh, K.R.; Moyer, R.P.; Muller-Karger, F.E. Enabling efficient, large-scale high-spatial resolution wetland mapping using satellites. Remote Sens. Environ. 2018, 208, 189–201. [Google Scholar] [CrossRef]










| Classification Type | Vegetation Associations | Class Codes |
|---|---|---|
| Forest | Quercus mongolica Fisch. ex Ledeb., Populus davidiana Dode, Betula platyphylla Sukaczev | A |
| Cropland | Zea mays L., Sorghum abyssinicum (Fresen.) Kuntze | B |
| Deep-water herbaceous vegetation | Carex pseudocuraica F.Schmidt, Carex lasiocarpa Ehrh. | C |
| Shallow-water herbaceous vegetation | Calamagrostis angustifolia Kom., Carex tato Chang | D |
| Shrub | Salix brachypoda (Trautv. & C. A. Mey.) Kom., Spiraea salicifolia L. | E |
| Open water | None | F |
| Paddy field | Oryza sativa L. | G |
| Sensor | Panchromatic (nm) | Blue (nm) | Green (nm) | Red (nm) | Near IR (nm) | Spatial Resolution | Radiometric Resolution | Acquisition Time |
|---|---|---|---|---|---|---|---|---|
| GF-1 | 450–900 | 450–520 | 520–590 | 630–690 | 770–890 | 2 m (Pan), 8 m (MS) | 10 bit | 2016.09.21 |
| ZY-3 | - | 450–520 | 520–590 | 630–690 | 770–890 | 5.8 m (MS) | 10 bit | 2016.09.23 |
| Sample Types | A | B | C | D | E | F | G | Total | |
|---|---|---|---|---|---|---|---|---|---|
| GF-1 | Training | 72 | 38 | 39 | 65 | 62 | 77 | 49 | 402 |
| Testing | 32 | 46 | 21 | 109 | 86 | 69 | 49 | 412 | |
| ZY-3 | Training | 70 | 37 | 46 | 76 | 76 | 61 | 49 | 415 |
| Testing | 122 | 47 | 48 | 91 | 118 | 30 | 26 | 482 |
| Additional Data | Description | Reference |
|---|---|---|
| NDVI | [52] | |
| RVI | [53] | |
| GNDVI | [54] | |
| SWI | [55] | |
| Slope | 12.5 m ALOS DEM with a vertical resolution of 4–5 m | [56] |
| TWI | ||
| Texture measurements | Mean, variance, homogeneity, contrast, dissimilarity, entropy, second moment, standard deviation, and correlation for 4 spectral bands of GF-1 and ZY-3 data | [57] |
| Geometry measurements | Area, roundness, main direction, rectangular fit, asymmetry, border index, compactness, max difference, and shape index of GF-1 and ZY-3 data | [58] |
| Multiresolution Segmentation | Sensor | Large Scale | Small Scale | |
|---|---|---|---|---|
| GF-1 | 150 | 50 | ||
| ZY-3 | 150 | 30 | ||
| Scenario | Sensor | Number of Variables | Candidate Image Layers | |
| 1 | GF-1 and ZY-3 | 24 | Four spectral bands, NDVI, RVI, GNDVI, SWI | |
| 2 | GF-1 and ZY-3 | 26 | Four spectral bands, NDVI, RVI, GNDVI, SWI, slope, TWI | |
| 3 | GF-1 and ZY-3 | 35 | Four spectral bands, NDVI, RVI, GNDVI, SWI, slope, TWI; nine geometric data layers | |
| 4 | GF-1 and ZY-3 | 131 | Four spectral bands, NDVI, RVI, GNDVI, SWI, slope, TWI; nine geometric data layers; 96 textural data layers | |
| Scenario | Sensor | mtry | ntrees | Overall Accuracy(%) | Kappa(%) |
|---|---|---|---|---|---|
| 1 | GF-1 | 6 | 1450 | 81.87 | 78.36 |
| ZY-3 | 4 | 1400 | 70.26 | 64.51 | |
| 2 | GF-1 | 6 | 1550 | 83.47 | 80.27 |
| ZY-3 | 5 | 1250 | 73.61 | 68.43 | |
| 3 | GF-1 | 5 | 1400 | 84.00 | 80.90 |
| ZY-3 | 7 | 1550 | 74.72 | 69.77 | |
| 4 | GF-1 | 10 | 1500 | 83.73 | 80.60 |
| ZY-3 | 13 | 1350 | 73.98 | 68.85 |
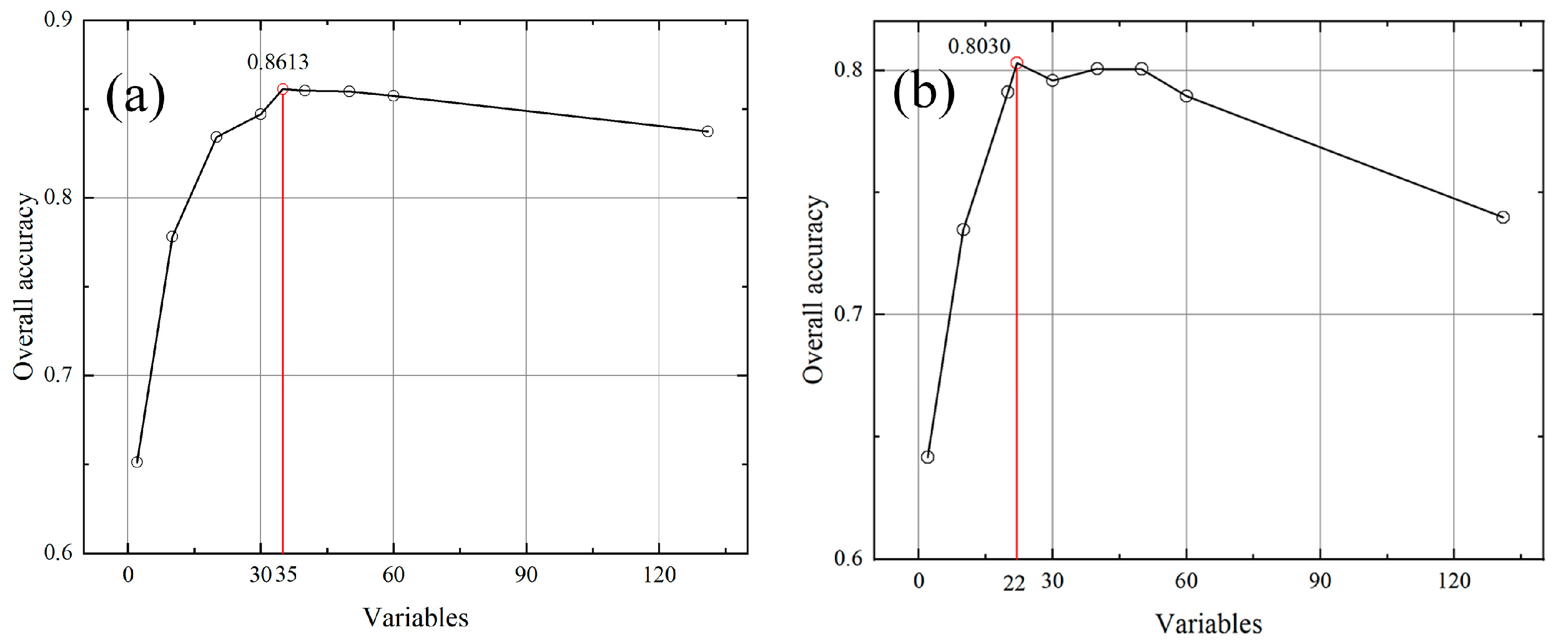
| Sensor | Order of Variables | OA (%) | SD(OA) (%) | Kappa (%) | SD(Kappa) (%) |
|---|---|---|---|---|---|
| GF-1 | 2 | 65.11 | 8.77 | 58.65 | 7.80 |
| 10 | 77.81 | 4.40 | 73.70 | 5.25 | |
| 20 | 83.42 | 3.15 | 80.32 | 3.39 | |
| 30 | 84.70 | 3.22 | 81.86 | 3.41 | |
| 35 | 86.13 | 3.43 | 83.68 | 3.49 | |
| 40 | 86.03 | 3.83 | 83.41 | 3.76 | |
| 50 | 85.99 | 3.00 | 83.37 | 3.99 | |
| 60 | 85.74 | 3.38 | 83.08 | 3.41 | |
| 131 | 83.73 | 3.04 | 83.54 | 3.02 | |
| ZY-3 | 2 | 64.15 | 9.04 | 59.13 | 8.64 |
| 10 | 73.47 | 5.14 | 68.39 | 5.79 | |
| 20 | 79.11 | 4.97 | 75.04 | 4.17 | |
| 22 | 80.30 | 4.72 | 76.44 | 4.06 | |
| 30 | 79.59 | 4.86 | 75.98 | 4.25 | |
| 40 | 80.07 | 4.45 | 76.88 | 4.33 | |
| 50 | 80.06 | 4.45 | 76.74 | 4.36 | |
| 60 | 78.95 | 4.41 | 74.83 | 4.25 | |
| 131 | 73.98 | 4.02 | 68.85 | 4.27 |
| Sensor | Order of Variables | OA (%) | SD (OA) (%) | Kappa (%) | SD (Kappa) (%) |
|---|---|---|---|---|---|
| GF-1 | 2 | 65.11 | 6.48 | 59.38 | 7.79 |
| 10 | 77.64 | 4.75 | 72.84 | 4.24 | |
| 20 | 83.50 | 3.16 | 80.50 | 3.70 | |
| 30 | 84.50 | 3.68 | 79.73 | 4.33 | |
| 40 | 84.51 | 3.92 | 80.88 | 4.65 | |
| 50 | 84.93 | 3.94 | 81.47 | 4.67 | |
| 60 | 84.58 | 4.07 | 81.58 | 4.79 | |
| 76 | 85.07 | 3.58 | 81.89 | 3.22 | |
| 80 | 84.84 | 3.17 | 80.78 | 3.76 | |
| 131 | 83.73 | 3.32 | 79.28 | 3.95 | |
| ZY-3 | 2 | 60.57 | 8.38 | 56.54 | 8.14 |
| 10 | 68.69 | 7.25 | 64.47 | 7.22 | |
| 20 | 73.73 | 5.84 | 69.52 | 6.06 | |
| 30 | 75.40 | 4.68 | 71.44 | 5.23 | |
| 40 | 76.44 | 4.43 | 72.05 | 4.98 | |
| 50 | 76.30 | 4.35 | 72.11 | 4.57 | |
| 62 | 76.58 | 4.31 | 72.14 | 4.48 | |
| 70 | 76.45 | 4.22 | 73.25 | 4.39 | |
| 80 | 76.04 | 4.15 | 72.94 | 4.37 | |
| 131 | 73.98 | 4.02 | 68.85 | 4.27 |
| Sensor | Order of Variables | OA (%) | SD (OA) (%) | Kappa (%) | SD (Kappa) (%) |
|---|---|---|---|---|---|
| GF-1 | 2 | 64.63 | 6.41 | 58.00 | 6.29 |
| 10 | 78.54 | 4.57 | 73.90 | 5.37 | |
| 20 | 83.51 | 5.17 | 79.54 | 5.58 | |
| 30 | 85.03 | 4.93 | 82.47 | 4.07 | |
| 40 | 85.21 | 3.13 | 83.10 | 3.11 | |
| 43 | 85.60 | 3.63 | 83.31 | 3.54 | |
| 50 | 85.41 | 3.99 | 83.06 | 3.96 | |
| 60 | 84.80 | 3.51 | 82.37 | 3.58 | |
| 70 | 84.25 | 3.48 | 82.68 | 3.52 | |
| 131 | 83.73 | 3.03 | 81.49 | 3.01 | |
| ZY-3 | 2 | 61.54 | 7.75 | 53.96 | 8.37 |
| 10 | 69.25 | 6.51 | 64.81 | 7.54 | |
| 20 | 74.32 | 5.48 | 69.87 | 6.34 | |
| 30 | 76.87 | 4.97 | 72.32 | 5.11 | |
| 33 | 77.70 | 4.68 | 73.25 | 4.69 | |
| 40 | 77.21 | 4.52 | 73.11 | 4.36 | |
| 45 | 76.89 | 4.28 | 72.84 | 4.23 | |
| 50 | 76.32 | 4.31 | 71.97 | 4.15 | |
| 60 | 76.11 | 4.08 | 71.54 | 4.18 | |
| 131 | 73.98 | 4.02 | 68.85 | 4.27 |
| Sensor | Scenario | Estimate (%) | Standard Error (%) | 95% Confidence Intervals (%) | ||
|---|---|---|---|---|---|---|
| GF-1 | 1 | Overall | 81.87 | 3.97 | 77.59 | 85.63 |
| Kappa | 78.36 | 3.60 | 74.90 | 82.78 | ||
| 3 | Overall | 84.00 | 3.32 | 79.89 | 87.56 | |
| Kappa | 80.90 | 3.83 | 76.54 | 84.59 | ||
| 4(RFE) | Overall | 86.13 | 3.43 | 79.60 | 87.32 | |
| Kappa | 83.68 | 3.49 | 76.67 | 83.57 | ||
| 4(Boruta) | Overall | 85.07 | 3.58 | 79.60 | 87.32 | |
| Kappa | 81.89 | 3.22 | 76.67 | 83.57 | ||
| 4(VSURF) | Overall | 85.60 | 3.63 | 79.60 | 87.32 | |
| Kappa | 83.31 | 3.54 | 76.67 | 83.57 | ||
| ZY-3 | 1 | Overall | 70.26 | 4.96 | 67.54 | 73.18 |
| Kappa | 64.51 | 4.24 | 61.78 | 66.79 | ||
| 3 | Overall | 74.42 | 4.65 | 71.62 | 77.14 | |
| Kappa | 69.77 | 4.37 | 66.80 | 72.55 | ||
| 4(RFE) | Overall | 80.30 | 4.72 | 77.43 | 83.25 | |
| Kappa | 76.44 | 4.06 | 74.37 | 78.89 | ||
| 4(Boruta) | Overall | 76.58 | 4.85 | 73.06 | 89.51 | |
| Kappa | 71.95 | 4.21 | 68.67 | 75.57 | ||
| 4(VSURF) | Overall | 77.70 | 4.77 | 74.24 | 80.53 | |
| Kappa | 73.24 | 4.16 | 70.12 | 76.57 | ||
| A | B | C | D | E | F | G | T | U | CI | |||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Scenario 1 GF-1 | A | 82 | 0 | 0 | 0 | 3 | 0 | 0 | 85 | 96.5 | 93.2 | 98.8 |
| B | 0 | 34 | 0 | 0 | 0 | 0 | 0 | 34 | 100.0 | 100.0 | 100.0 | |
| C | 0 | 0 | 30 | 3 | 4 | 3 | 1 | 41 | 73.2 | 69.9 | 76.8 | |
| D | 0 | 0 | 2 | 41 | 0 | 0 | 22 | 65 | 63.1 | 60.4 | 68.0 | |
| E | 1 | 8 | 3 | 2 | 69 | 0 | 0 | 83 | 83.1 | 80.6 | 86.3 | |
| F | 0 | 0 | 2 | 0 | 1 | 35 | 0 | 38 | 92.1 | 88.3 | 95.8 | |
| G | 0 | 0 | 0 | 13 | 0 | 0 | 16 | 29 | 55.1 | 53.0 | 58.8 | |
| T | 83 | 42 | 37 | 59 | 77 | 38 | 39 | |||||
| P | 98.8 | 81.0 | 81.1 | 69.5 | 89.6 | 92.1 | 41.0 | |||||
| CI | 95.1 | 77.8 | 77.9 | 66.4 | 84.8 | 87.7 | 38.5 | |||||
| 100.0 | 84.1 | 85.1 | 72.5 | 93.6 | 96.9 | 44.2 | ||||||
| Scenario 3 GF-1 | A | 82 | 0 | 0 | 0 | 4 | 0 | 0 | 86 | 95.3 | 93.8 | 98.6 |
| B | 0 | 37 | 0 | 3 | 1 | 1 | 0 | 42 | 88.1 | 85.8 | 92.6 | |
| C | 0 | 1 | 34 | 0 | 0 | 5 | 2 | 42 | 81.0 | 77.8 | 84.5 | |
| D | 0 | 0 | 1 | 45 | 1 | 1 | 22 | 70 | 64.3 | 60.7 | 67.4 | |
| E | 1 | 4 | 1 | 2 | 71 | 0 | 0 | 79 | 89.9 | 86.9 | 93.3 | |
| F | 0 | 0 | 1 | 0 | 0 | 31 | 0 | 32 | 96.9 | 93.2 | 98.9 | |
| G | 0 | 0 | 0 | 9 | 0 | 0 | 15 | 24 | 62.5 | 59.7 | 65.4 | |
| T | 83 | 42 | 37 | 59 | 77 | 38 | 39 | |||||
| P | 98.8 | 88.1 | 91.9 | 71.2 | 93.5 | 78.9 | 41.0 | |||||
| CI | 94.0 | 85.1 | 87.8 | 67.3 | 88.9 | 75.4 | 38.4 | |||||
| 100.0 | 93.2 | 94.9 | 74.6 | 97.4 | 81.8 | 44.6 | ||||||
| Scenario 4 (RFE) GF-1 | A | 83 | 0 | 0 | 0 | 1 | 0 | 0 | 84 | 98.8 | 96.0 | 100.0 |
| B | 0 | 36 | 1 | 1 | 0 | 0 | 0 | 38 | 94.7 | 91.6 | 97.1 | |
| C | 0 | 0 | 31 | 1 | 4 | 1 | 1 | 38 | 81.6 | 77.7 | 84.0 | |
| D | 0 | 2 | 0 | 51 | 3 | 0 | 22 | 78 | 65.4 | 62.7 | 68.4 | |
| E | 0 | 4 | 0 | 1 | 69 | 0 | 0 | 74 | 93.2 | 90.1 | 96.7 | |
| F | 0 | 0 | 5 | 0 | 0 | 37 | 0 | 42 | 88.1 | 84.6 | 91.2 | |
| G | 0 | 0 | 0 | 5 | 0 | 0 | 16 | 21 | 76.2 | 73.2 | 80.0 | |
| T | 83 | 42 | 37 | 59 | 77 | 38 | 39 | |||||
| P | 100.0 | 88.1 | 83.8 | 86.4 | 89.6 | 97.4 | 41.0 | |||||
| CI | 100.0 | 88.4 | 80.4 | 83.3 | 86.9 | 94.7 | 38.1 | |||||
| 100.0 | 97.1 | 87.0 | 89.1 | 92.6 | 99.9 | 44.4 | ||||||
| Scenario 1 ZY-3 | A | 55 | 0 | 0 | 0 | 11 | 1 | 0 | 67 | 82.1 | 78.5 | 85.3 |
| B | 0 | 19 | 0 | 2 | 4 | 0 | 0 | 25 | 76.0 | 72.6 | 79.5 | |
| C | 0 | 0 | 20 | 6 | 2 | 5 | 2 | 35 | 57.1 | 54.3 | 60.4 | |
| D | 0 | 0 | 3 | 22 | 1 | 0 | 8 | 34 | 64.7 | 61.7 | 67.9 | |
| E | 3 | 5 | 2 | 2 | 38 | 6 | 1 | 57 | 66.7 | 62.8 | 70.1 | |
| F | 0 | 0 | 3 | 0 | 1 | 15 | 0 | 19 | 78.9 | 75.3 | 82.2 | |
| G | 0 | 1 | 0 | 10 | 1 | 0 | 20 | 32 | 62.5 | 59.3 | 65.8 | |
| T | 58 | 25 | 28 | 42 | 58 | 27 | 31 | |||||
| P | 94.8 | 76.0 | 71.4 | 52.4 | 65.5 | 55.6 | 64.5 | |||||
| CI | 91.5 | 72.6 | 68.1 | 48.6 | 62.4 | 52.3 | 61.7 | |||||
| 97.4 | 79.7 | 74.9 | 56.3 | 68.9 | 58.4 | 67.8 | ||||||
| Scenario 3ZY-3 | A | 56 | 0 | 0 | 0 | 3 | 1 | 0 | 60 | 93.3 | 91.0 | 96.2 |
| B | 0 | 19 | 0 | 0 | 1 | 0 | 0 | 20 | 95.0 | 91.8 | 98.3 | |
| C | 0 | 0 | 22 | 4 | 2 | 5 | 0 | 33 | 66.7 | 63.5 | 70.0 | |
| D | 0 | 0 | 0 | 24 | 2 | 0 | 20 | 46 | 52.2 | 49.1 | 55.4 | |
| E | 2 | 6 | 2 | 1 | 50 | 1 | 1 | 63 | 79.4 | 76.3 | 82.8 | |
| F | 0 | 0 | 4 | 0 | 0 | 20 | 0 | 24 | 83.3 | 80.5 | 86.4 | |
| G | 0 | 0 | 0 | 13 | 0 | 0 | 10 | 23 | 43.5 | 40.2 | 47.1 | |
| T | 58 | 25 | 28 | 42 | 58 | 27 | 31 | |||||
| P | 96.6 | 76.0 | 78.6 | 57.1 | 86.2 | 74.1 | 32.3 | |||||
| CI | 93.2 | 72.3 | 75.6 | 54.0 | 82.9 | 71.5 | 29.1 | |||||
| 99.4 | 78.9 | 81.2 | 60.8 | 89.6 | 77.7 | 35.4 | ||||||
| Scenario 4 (RFE) ZY-3 | A | 58 | 1 | 0 | 0 | 4 | 1 | 0 | 64 | 90.6 | 87.4 | 93.7 |
| B | 0 | 19 | 0 | 2 | 0 | 0 | 0 | 21 | 90.5 | 87.5 | 93.2 | |
| C | 0 | 0 | 20 | 3 | 1 | 5 | 1 | 30 | 66.7 | 63.2 | 70.3 | |
| D | 0 | 2 | 0 | 30 | 1 | 0 | 12 | 45 | 66.7 | 62.4 | 70.6 | |
| E | 0 | 3 | 1 | 2 | 52 | 1 | 1 | 60 | 86.7 | 83.3 | 90.0 | |
| F | 0 | 0 | 6 | 1 | 0 | 20 | 0 | 27 | 74.1 | 71.5 | 77.7 | |
| G | 0 | 0 | 1 | 4 | 0 | 0 | 17 | 22 | 77.3 | 74.1 | 80.4 | |
| T | 58 | 25 | 28 | 42 | 58 | 27 | 31 | |||||
| P | 100.0 | 76.0 | 71.4 | 71.4 | 89.7 | 74.1 | 54.8 | |||||
| CI | 100.0 | 73.1 | 68.1 | 68.5 | 86.8 | 70.9 | 51.5 | |||||
| 100.0 | 79.4 | 74.3 | 71.6 | 92.4 | 77.0 | 57.9 | ||||||
| Sensor | Comparisons | Scenario 1 | Scenario 3 | Scenario 4 (RFE) | Scenario 4 (Boruta) | Scenario 4 (VSURF) |
|---|---|---|---|---|---|---|
| GF-1 | Scenario 1 | – | 2.00 | 2.67 | 1.42 | 3.15 |
| Scenario 3 | – | 2.91 | 1.14 | 1.80 | ||
| Scenario 4 (RFE) | – | 2.23 | 0.25 | |||
| Scenario 4 (Boruta) | – | 0.06 | ||||
| Scenario 4 (VSURF) | – | |||||
| ZY-3 | Scenario 1 | – | 3.52 | 8.25 | 4.58 | 5.27 |
| Scenario 3 | – | 5.53 | 3.41 | 4.19 | ||
| Scenario 4 (RFE) | – | 3.32 | 1.87 | |||
| Scenario 4 (Boruta) | – | 1.69 | ||||
| Scenario 4 (VSURF) | – |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lou, P.; Fu, B.; He, H.; Li, Y.; Tang, T.; Lin, X.; Fan, D.; Gao, E. An Optimized Object-Based Random Forest Algorithm for Marsh Vegetation Mapping Using High-Spatial-Resolution GF-1 and ZY-3 Data. Remote Sens. 2020, 12, 1270. https://doi.org/10.3390/rs12081270
Lou P, Fu B, He H, Li Y, Tang T, Lin X, Fan D, Gao E. An Optimized Object-Based Random Forest Algorithm for Marsh Vegetation Mapping Using High-Spatial-Resolution GF-1 and ZY-3 Data. Remote Sensing. 2020; 12(8):1270. https://doi.org/10.3390/rs12081270
Chicago/Turabian StyleLou, Peiqing, Bolin Fu, Hongchang He, Ying Li, Tingyuan Tang, Xingchen Lin, Donglin Fan, and Ertao Gao. 2020. "An Optimized Object-Based Random Forest Algorithm for Marsh Vegetation Mapping Using High-Spatial-Resolution GF-1 and ZY-3 Data" Remote Sensing 12, no. 8: 1270. https://doi.org/10.3390/rs12081270






