Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Sites
2.2. Data Collection
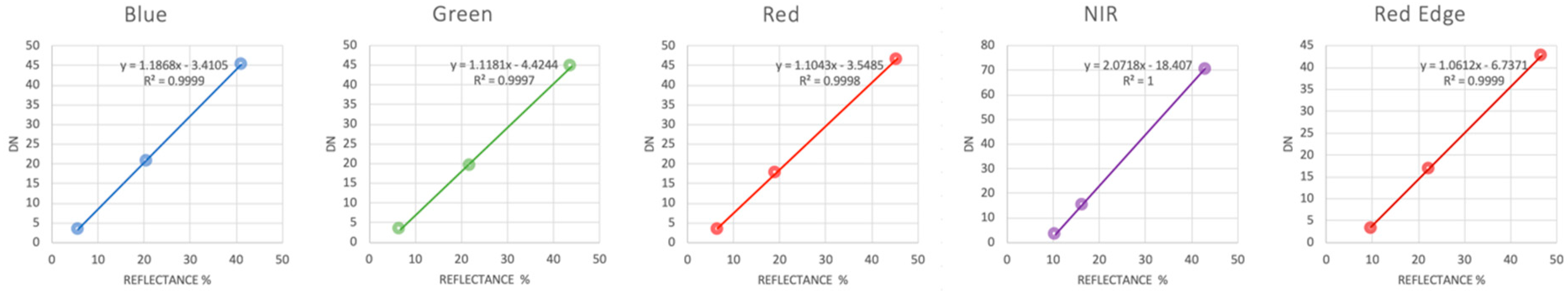
2.3. Data Processing
2.4. Classifications
2.4.1. Conventional Classification
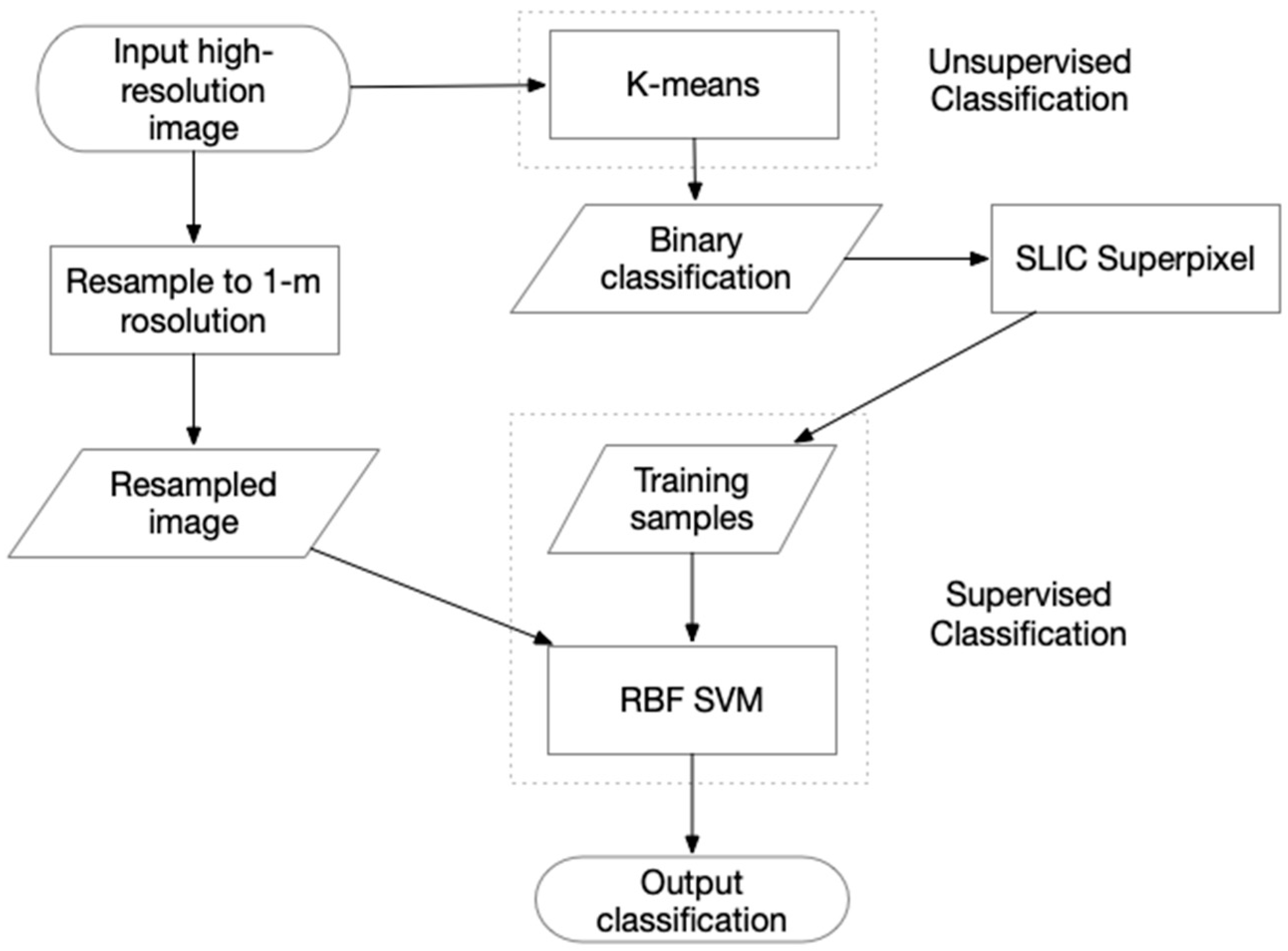
2.4.2. An Improved Semi-Supervised Classifier Based on k-means and SVM
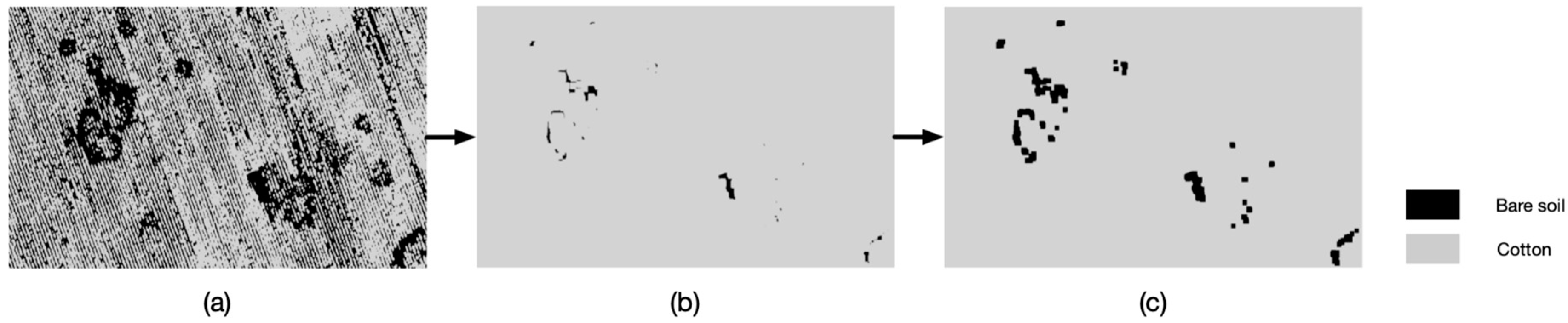
2.4.3. An Improved Classification Based on k-means Segmentation
2.5. Accuracy Assessment
- (a)
- A region with more than 10 adjacent cotton plants infected with CRR was marked as a CRR-infested region.
- (b)
- In a CRR-infested area, a region with more than 10 adjacent healthy cotton plants was regarded as a non-infested area.
3. Results
3.1. The Newly Proposed Classification Methods
3.2. Comparison Between Newly Proposed and Conventional Classification Methods
4. Discussion
5. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Pammel, L.H. Root rot of cotton or “cotton blight”. Texas Agric. Exp. Stn. Ann. Bull. 1888, 4, 50–65. [Google Scholar]
- Smith, H.E.; Elliott, F.C.; Bird, L.S. Root rot losses of cotton can be reduced. Misc. Publ. Tex. Agric. Exp. Stn. No. 361 1962. [Google Scholar]
- Yang, C.; Odvody, G.N.; Fernandez, C.J.; Landivar, J.A.; Minzenmayer, R.R.; Nichols, R.L.; Thomasson, J.A. Monitoring cotton root rot progression within a growing season using airborne multispectral imagery. J. Cotton Sci. 2014, 93, 85–93. [Google Scholar]
- Smith, R. South Texas Cotton Root Rot Draws Study. Available online: https://www.farmprogress.com/south-texas-cotton-root-rot-draws-study (accessed on 23 September 2019).
- Yang, C.; Odvody, G.N.; Fernandez, C.J.; Landivar, J.A.; Minzenmayer, R.R.; Nichols, R.L. Evaluating unsupervised and supervised image classification methods for mapping cotton root rot. Precis. Agric. 2015, 16, 201–215. [Google Scholar] [CrossRef]
- Isakeit, T.; Minzenmayer, R.R.; Drake, D.R.; Morgan, G.D.; Mott, D.A.; Fromme, D.D.; Multer, W.L.; Jungman, M.; Abrameit, A. Fungicide management of cotton root rot (Phymatotrichopsis omnivora): 2011 results. In Proceedings of the Beltwide Cotton Conference, San Antonio, TX, USA, 3–6 January 2012; pp. 235–238. [Google Scholar]
- Isakeit, T.; Minzenmayer, R.; Abrameit, A.; Moore, G.; Scasta, J.D. Control of phymatotrichopsis root rot of cotton with flutriafol. In Proceedings of the Beltwide Cotton Conference, San Antonio, TX, USA, 9 January 2013; pp. 200–203. [Google Scholar]
- Isakeit, T. Management of cotton root rot. In Proceedings of the Beltwide Cotton Conference, San Antonio, TX, USA, 3–5 January 2018; p. 43. [Google Scholar]
- Yang, C.; Everitt, J.H.; Fernandez, C.J. Comparison of airborne multispectral and hyperspectral imagery for mapping cotton root rot. Biosyst. Eng. 2010, 107, 131–139. [Google Scholar] [CrossRef]
- Chenhai, Y.; Minzenmayer, R.R.; Extension, T.A.; Nichols, R.L.; Incorporated, C.; Isakeit, T.; Thomasson, J.A.; Fernandez, C.J.; Landivar, J.A. Monitoring cotton root rot infection in fungicide-treated cotton fields using airborne imagery. In Proceedings of the Beltwide Cotton Conferences, New Orleans, LA, USA, 6–8 January 2013. [Google Scholar]
- Taubenhaus, J.J.; Ezekiel, W.N.; Neblette, C.B. Airplane photography in the study of cotton root rot. Phytopathology 1929, 19, 1025–1029. [Google Scholar]
- Nixon, P.R.; Lyda, S.D.; Heilman, M.D.; Bowen, R.L. Incidence and control of cotton root rot observed with color infrared photography. MP Tex. Agric. Exp. Stn. 1975. [Google Scholar]
- Nixon, P.R.; Escobar, D.E.; Bowen, R.L. A multispectral false-color video imaging system for remote sensing applications. In Proceedings of the 11th Biennial Workshop on Color Aerial Photography and Videography in the Plant Sciences and Related Fields, Weslaco, TX, USA, 1 september 1987; pp. 295–305. [Google Scholar]
- Yang, C.; Fernandez, C.J.; Everitt, J.H. Mapping phymatotrichum root rot of cotton using airborne three-band digital imagery. Trans. ASAE 2005, 48, 1619–1626. [Google Scholar] [CrossRef]
- Song, X.; Yang, C.; Wu, M.; Zhao, C.; Yang, G.; Hoffmann, W.C.; Huang, W. Evaluation of Sentinel-2A satellite imagery for mapping cotton root rot. Remote Sens. 2017, 9, 906. [Google Scholar] [CrossRef] [Green Version]
- Yang, C.; Greenberg, S.M.; Everitt, J.H.; Fernandez, C.J. Assessing cotton defoliation, regrowth control and root rot infection using remote sensing technology. Int. J. Agric. Biol. Eng. 2011, 4, 1–11. [Google Scholar]
- Huang, Y.; Thomson, S.J.; Brand, H.J.; Reddy, K.N. Development and evaluation of low-altitude remote sensing systems for crop production management. Int. J. Agric. Biol. Eng. 2016, 9, 1–11. [Google Scholar]
- Easterday, K.; Kislik, C.; Dawson, T.E.; Hogan, S.; Kelly, M. Remotely sensed water limitation in vegetation: Insights from an experiment with unmanned aerial vehicles (UAVs). Remote Sens. 2019, 11, 1853. [Google Scholar] [CrossRef] [Green Version]
- Zhou, X.; Zheng, H.B.; Xu, X.Q.; He, J.Y.; Ge, X.K.; Yao, X.; Cheng, T.; Zhu, Y.; Cao, W.X.; Tian, Y.C. Predicting grain yield in rice using multi-temporal vegetation indices from UAV-based multispectral and digital imagery. ISPRS J. Photogramm. Remote Sens. 2017, 130, 246–255. [Google Scholar] [CrossRef]
- Albetis, J.; Duthoit, S.; Guttler, F.; Jacquin, A.; Goulard, M.; Poilvé, H.; Féret, J.B.; Dedieu, G. Detection of Flavescence dorée grapevine disease using unmanned aerial vehicle (UAV) multispectral imagery. Remote Sens. 2017, 9, 308. [Google Scholar] [CrossRef] [Green Version]
- Romero, M.; Luo, Y.; Su, B.; Fuentes, S. Vineyard water status estimation using multispectral imagery from an UAV platform and machine learning algorithms for irrigation scheduling management. Comput. Electron. Agric. 2018, 147, 109–117. [Google Scholar] [CrossRef]
- Mattupalli, C.; Moffet, C.A.; Shah, K.N.; Young, C.A. Supervised classification of RGB Aerial imagery to evaluate the impact of a root rot disease. Remote Sens. 2018, 10, 917. [Google Scholar] [CrossRef] [Green Version]
- Duan, B.; Fang, S.; Zhu, R.; Wu, X.; Wang, S.; Gong, Y.; Peng, Y. Remote estimation of rice yield with unmanned aerial vehicle (uav) data and spectral mixture analysis. Front. Plant Sci. 2019, 10, 204. [Google Scholar] [CrossRef] [Green Version]
- Herrmann, I.; Bdolach, E.; Montekyo, Y.; Rachmilevitch, S.; Townsend, P.A.; Karnieli, A. Assessment of maize yield and phenology by drone-mounted superspectral camera. Precis. Agric. 2020, 21, 51–76. [Google Scholar] [CrossRef]
- Cai, Y.; Guan, K.; Nafziger, E.; Chowdhary, G.; Peng, B.; Jin, Z.; Wang, S.; Wang, S. Detecting In-season crop nitrogen stress of corn for field trials using UAV-and cubesat-based multispectral sensing. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2019, 12, 5153–5166. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, H.; Niu, Y.; Han, W. Mapping maizewater stress based on UAV multispectral remote sensing. Remote Sens. 2019, 11, 605. [Google Scholar] [CrossRef] [Green Version]
- Su, J.; Liu, C.; Coombes, M.; Hu, X.; Wang, C.; Xu, X.; Li, Q.; Guo, L.; Chen, W. Wheat yellow rust monitoring by learning from multispectral UAV aerial imagery. Comput. Electron. Agric. 2018, 155, 157–166. [Google Scholar] [CrossRef]
- Matese, A.; Toscano, P.; Di Gennaro, S.F.; Genesio, L.; Vaccari, F.P.; Primicerio, J.; Belli, C.; Zaldei, A.; Bianconi, R.; Gioli, B. Intercomparison of UAV, aircraft and satellite remote sensing platforms for precision viticulture. Remote Sens. 2015, 7, 2971–2990. [Google Scholar] [CrossRef] [Green Version]
- Ball, G.H.; Hall, D.J. ISODATA, a Novel Method of Data Analysis and Pattern Classification; Stanford Research Institute: Menlo Park, CA, USA, 1965. [Google Scholar]
- Hartigan, J.A.; Wong, M.A. Algorithm AS 136: A K-means clustering algorithm. J. R. Stat. Soc. Ser. C Appl. Stat. 1979, 28, 100–108. [Google Scholar] [CrossRef]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Aha, D.W.; Kibler, D.; Albert, M.K. Instance-based learning algorithms. Mach. Learn. 1991, 6, 37–66. [Google Scholar] [CrossRef] [Green Version]
- Burges, C.J.C. A tutorial on support vector machines for pattern recognition. Data Min. Knowl. Discov. 1998, 2, 121–167. [Google Scholar] [CrossRef]
- Chang, C.C.; Lin, C.J. LIBSVM: A Library for support vector machines. ACM Trans. Intell. Syst. Technol. 2011, 2, 27. [Google Scholar] [CrossRef]
- Dempster, A.P.; Laird, N.M.; Rubin, D.B. Maximum likelihood from incomplete data via the EM algorithm. J. R. Stat. Soc. Ser. B 1977, 39, 1–22. [Google Scholar]
- MacQueen, J. Some methods for classification and analysis of multivariate observations. Proc. Fifth Berkeley Symp. Math. Stat. Probab. 1967, 1, 281–297. [Google Scholar]
- De Maesschalck, R.; Jouan-Rimbaud, D.; Massart, D.L. The Mahalanobis distance. Chemom. Intell. Lab. Syst. 2000, 50, 1–18. [Google Scholar] [CrossRef]
- Pal, M.; Mather, P.M. Support vector machines for classification in remote sensing. Int. J. Remote Sens. 2005, 26, 1007–1011. [Google Scholar] [CrossRef]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Gong, P.; Howarth, P.J. Land-use classification of SPOT HRV data using a cover-frequency method. Int. J. Remote Sens. 1992, 13, 1459–1471. [Google Scholar] [CrossRef]
- Lu, D.; Weng, Q. A survey of image classification methods and techniques for improving classification performance. Int. J. Remote Sens. 2007, 28, 823–870. [Google Scholar] [CrossRef]
- Fauvel, M.; Benediktsson, J.A.; Chanussot, J.; Sveinsson, J.R. Spectral and spatial classification of hyperspectral data using SVMs and morphological profiles. IEEE Trans. Geosci. Remote Sens. 2008, 46, 3804–3814. [Google Scholar] [CrossRef] [Green Version]
- Huang, X.; Weng, C.; Lu, Q.; Feng, T.; Zhang, L. Automatic labelling and selection of training samples for high-resolution remote sensing image classification over urban areas. Remote Sens. 2015, 7, 16024–16044. [Google Scholar] [CrossRef] [Green Version]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Huang, C.; Davis, L.S.; Townshend, J.R.G. An assessment of support vector machines for land cover classification. Int. J. Remote Sens. 2002, 23, 725–749. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The measurement of observer agreement for categorical data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [Green Version]
- Rafieyan, V. Effect of cultural distance on translation of culture-bound texts. Int. J. Educ. Lit. Stud. 2016, 4, 67–73. [Google Scholar]













| Overall accuracy | 90.69% | |||||
| Kappa coefficient | 0.7277 | |||||
| Class types determined from the reference source (Ground-truth) | Commission | Omission | ||||
| Class types determined from classified map | Infested plants | Healthy plants | Totals | |||
| Infested plants | 2157162 | 684748 | 2841910 | 24.09% | 18.24% | |
| Healthy plants | 481191 | 9205114 | 9686305 | 4.97% | 6.92% | |
| Totals | 2638353 | 9889862 | 12528215 | |||
| Overall accuracy | 92.63% | |||||
| Kappa coefficient | 0.7786 | |||||
| Class types determined from reference source (Ground-truth) | Commission | Omission | ||||
| Class types determined from classified map | Infested plants | Healthy plants | Totals | |||
| Infested plants | 2182161 | 467623 | 2649784 | 17.65% | 17.29% | |
| Healthy plants | 456192 | 9422239 | 9878431 | 4.62% | 4.73% | |
| Totals | 2638353 | 9889862 | 12528215 | |||
| Overall Accuracy | Kappa Coefficient | ||||||||||
| CH | WP | SH | Mean | Std. Dev. | CH | WP | SH | Mean | Std. Dev. | ||
| U | 2-class k-means | 78.60% | 78.76% | 81.44% | 79.60%a | 1.60% | 0.5106 | 0.4527 | 0.6162 | 0.5265A | 0.0829 |
| C-U | 3 to 2-class k-means | 88.89% | 87.26% | 79.45% | 85.20%ab | 5.05% | 0.6868 | 0.5751 | 0.5392 | 0.6004A | 0.0770 |
| 5 to 2-class k-means | 88.67% | 88.81% | 83.49% | 86.99%ab | 3.03% | 0.6085 | 0.5293 | 0.6452 | 0.5943A | 0.0592 | |
| 10 to 2-class k-means | 90.97% | 88.01% | 81.14% | 86.71%ab | 5.04% | 0.6986 | 0.5885 | 0.5911 | 0.6261A | 0.0628 | |
| S | SVM | 92.02% | 78.66% | 87.48% | 86.05%ab | 6.79% | 0.7587 | 0.4481 | 0.7345 | 0.6471A | 0.1728 |
| Minimum distance | 88.12% | 86.14% | 82.79% | 85.68%ab | 2.69% | 0.6753 | 0.5604 | 0.6346 | 0.6234A | 0.0583 | |
| Maximum likelihood | 91.71% | 77.92% | 87.65% | 85.76%ab | 7.09% | 0.7498 | 0.4419 | 0.7422 | 0.6446A | 0.1756 | |
| Mahalanobis distance | 89.60% | 87.13% | 86.27% | 87.67%ab | 1.73% | 0.7076 | 0.5764 | 0.7144 | 0.6661A | 0.0778 | |
| PA | KMSVM | 90.69% | 84.47% | 88.15% | 87.77%ab | 3.13% | 0.7277 | 0.6048 | 0.7494 | 0.6940A | 0.0780 |
| KMSEG | 92.62% | 85.80% | 87.06% | 88.49%b | 3.63% | 0.7786 | 0.6428 | 0.7379 | 0.7198A | 0.0697 | |
| Error of Commission (CRR class) | Error of Omission (CRR class) | ||||||||||
| CH | WP | SH | Mean | Std. Dev. | CH | WP | SH | Mean | Std. Dev. | ||
| U | 2-class k-means | 50.43% | 56.88% | 27.16% | 44.82%a | 15.63% | 7.84% | 14.10% | 18.00% | 13.31%A | 5.13% |
| C-U | 3 to 2-class k-means | 30.04% | 40.17% | 17.43% | 29.21%ab | 11.39% | 17.22% | 28.28% | 41.43% | 28.98%BCD | 12.12% |
| 5 to 2-class k-means | 14.26% | 24.23% | 19.36% | 19.28%ab | 4.99% | 44.58% | 51.63% | 25.30% | 40.50%D | 13.63% | |
| 10 to 2-class k-means | 10.85% | 37.51% | 21.20% | 23.19%ab | 13.44% | 34.95% | 29.83% | 30.73% | 31.84%CD | 2.73% | |
| S | SVM | 18.50% | 57.07% | 16.18% | 30.58%ab | 22.97% | 19.66% | 15.04% | 16.69% | 17.13%AB | 2.34% |
| Minimum distance | 32.90% | 43.70% | 22.15% | 32.92%ab | 10.78% | 14.48% | 24.71% | 23.19% | 20.79%ABC | 5.52% | |
| Maximum likelihood | 19.38% | 57.88% | 18.53% | 31.93%ab | 22.48% | 20.17% | 13.23% | 12.39% | 15.26%AB | 4.27% | |
| Mahalanobis distance | 28.74% | 40.79% | 20.73% | 30.09%ab | 10.10% | 15.18% | 26.86% | 13.25% | 18.43%ABC | 7.36% | |
| PA | KMSVM | 24.09% | 32.34% | 9.26% | 21.90%ab | 11.70% | 18.24% | 25.17% | 9.97% | 17.79%ABC | 7.61% |
| KMSEG | 17.64% | 7.46% | 23.30% | 16.13%b | 8.03% | 17.29% | 11.95% | 5.09% | 11.44%A | 6.12% | |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wang, T.; Thomasson, J.A.; Yang, C.; Isakeit, T.; Nichols, R.L. Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing. Remote Sens. 2020, 12, 1310. https://doi.org/10.3390/rs12081310
Wang T, Thomasson JA, Yang C, Isakeit T, Nichols RL. Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing. Remote Sensing. 2020; 12(8):1310. https://doi.org/10.3390/rs12081310
Chicago/Turabian StyleWang, Tianyi, J. Alex Thomasson, Chenghai Yang, Thomas Isakeit, and Robert L. Nichols. 2020. "Automatic Classification of Cotton Root Rot Disease Based on UAV Remote Sensing" Remote Sensing 12, no. 8: 1310. https://doi.org/10.3390/rs12081310





