Can Agricultural Management Induced Changes in Soil Organic Carbon Be Detected Using Mid-Infrared Spectroscopy?
Abstract
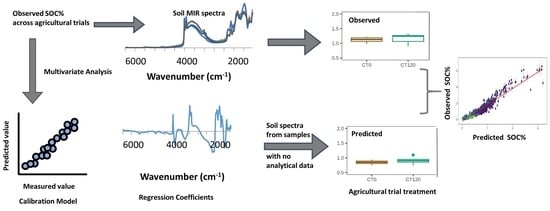
:1. Introduction
2. Materials and Methods
2.1. Site Descriptions
2.2. Laboratory Analyses
2.3. Spectral Modeling
2.4. Statistical Analyses
3. Results
3.1. Predictive Model Performance
3.2. Comparing Predictions between Instruments at Woodwell and the KSSL
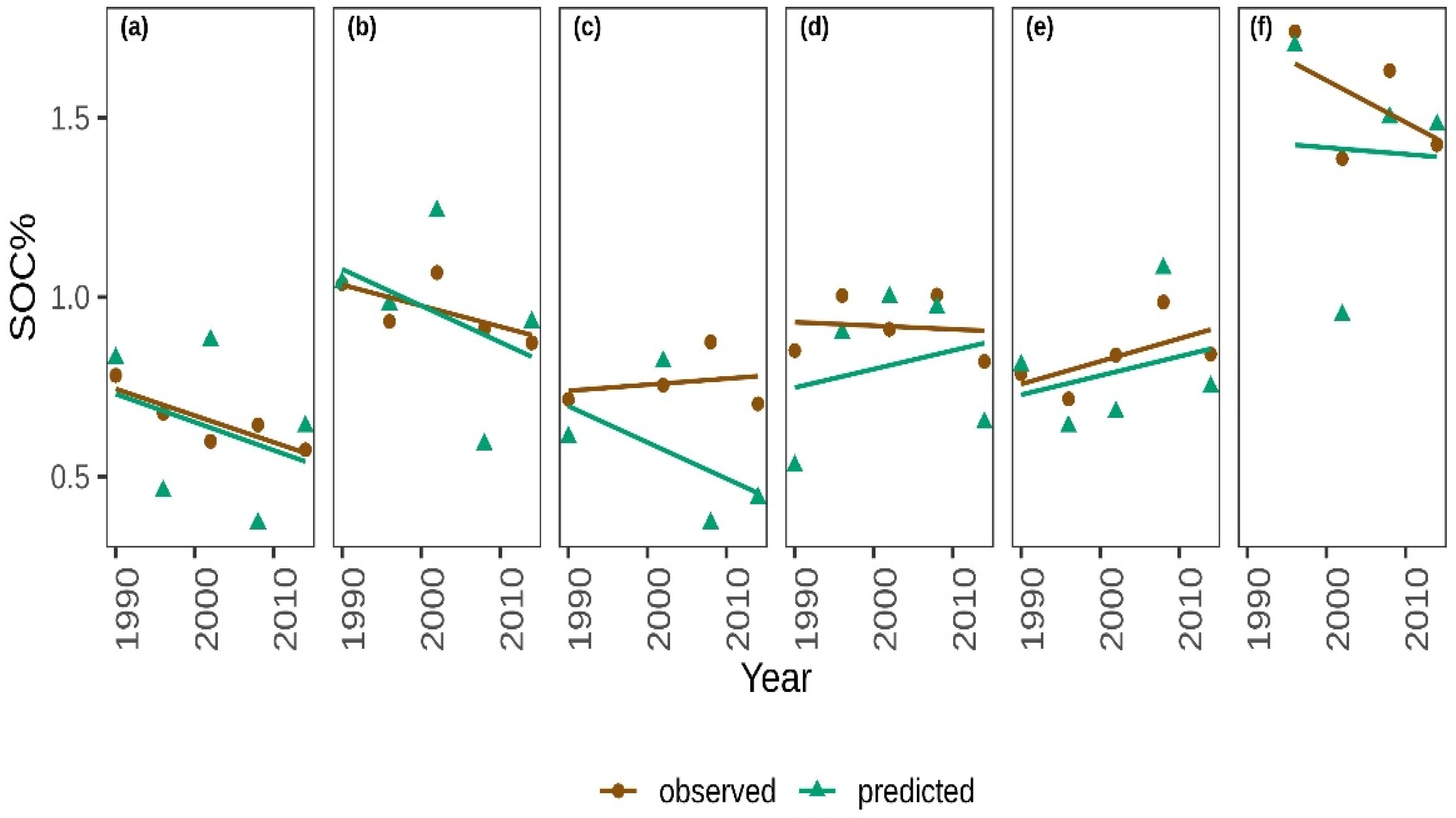
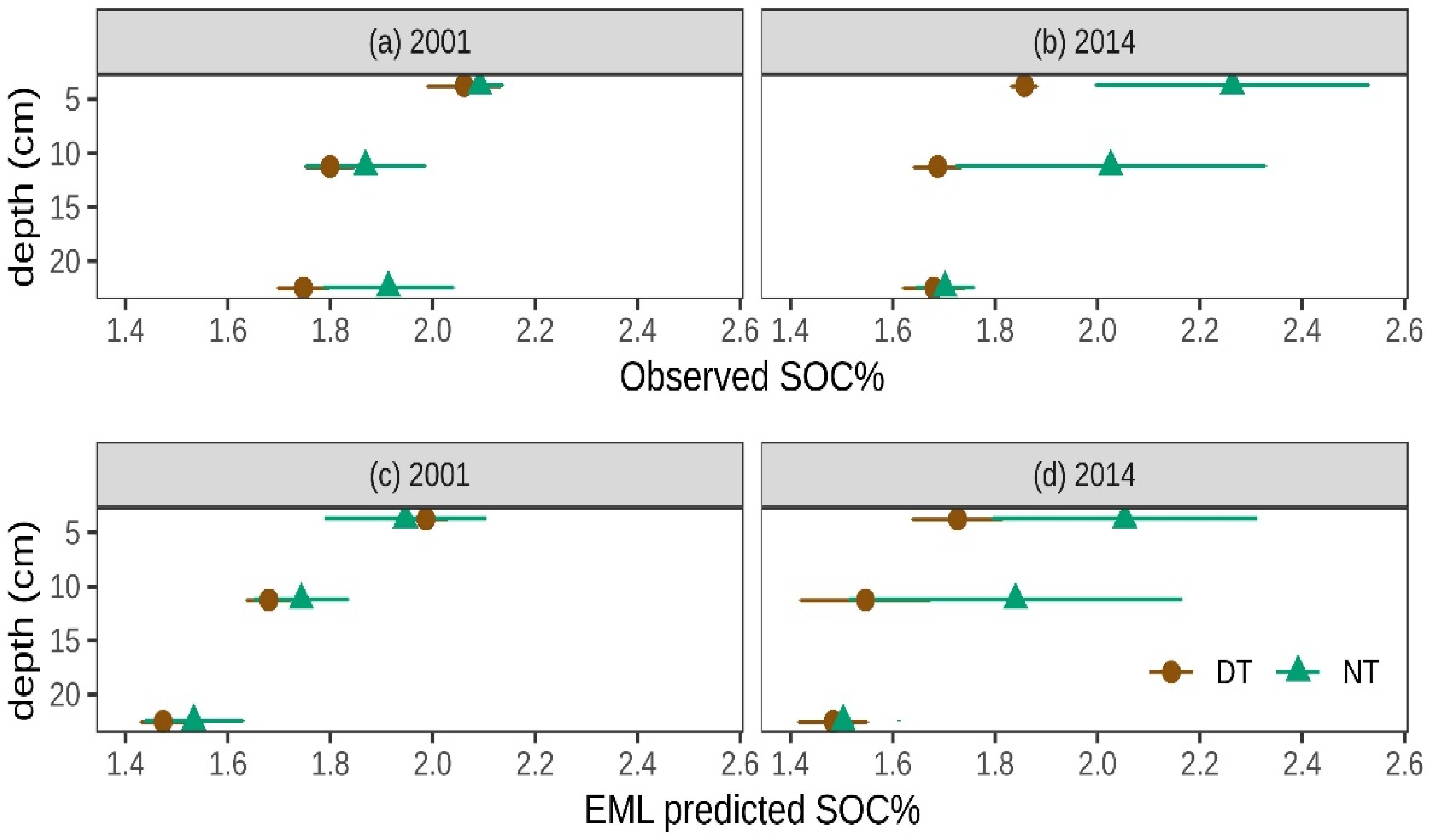
3.3. Detecting Changes in SOC% Across LTR Trials
4. Discussion
4.1. How Well Can Spectroscopy Predict Soil Organic Carbon?
4.2. Can Spectroscopy Detect Changes in Soil Organic Carbon Due to Management?
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Bossio, D.A.; Cook-Patton, S.C.; Ellis, P.W.; Fargione, J.; Sanderman, J.; Smith, P.; Wood, S.; Zomer, R.J.; von Unger, M.; Emmer, I.M.; et al. The Role of Soil Carbon in Natural Climate Solutions. Nat. Sustain. 2020, 3, 391–398. [Google Scholar] [CrossRef]
- CFI 2018-Carbon Credits (Carbon Farming Initiative—Measurement of Soil Carbon Sequestration in Agricultural Systems) Methodology Determination 2018. Available online: www.legislation.gov.au/Details/F2018L00089 (accessed on 20 March 2021).
- Smith, P.; Soussana, J.F.; Angers, D.; Schipper, L.; Chenu, C.; Rasse, D.P.; Batjes, N.H.; van Egmond, F.; McNeill, S.; Kuhnert, M.; et al. How to Measure, Report and Verify Soil Carbon Change to Realize the Potential of Soil Carbon Sequestration for Atmospheric Greenhouse Gas Removal. Glob. Chang. Biol. 2020, 26, 219–241. [Google Scholar] [CrossRef] [Green Version]
- Bellon-Maurel, V.; McBratney, A. Near-Infrared (NIR) and Mid-Infrared (MIR) Spectroscopic Techniques for Assessing the Amount of Carbon Stock in Soils-Critical Review and Research Perspectives. Soil Biol. Biochem. 2011, 43, 1398–1410. [Google Scholar] [CrossRef]
- Nocita, M.; Stevens, A.; van Wesemael, B.; Aitkenhead, M.; Bachmann, M.; Barthès, B.; Dor, E.B.; Brown, D.J.; Clairotte, M.; Csorba, A.; et al. Soil Spectroscopy: An Alternative to Wet Chemistry for Soil Monitoring. Adv. Agron. 2015, 132, 139–159. [Google Scholar] [CrossRef]
- Gholizadeh, A.; Luboš, B.; Saberioon, M.; Vašát, R. Visible, near-Infrared, and Mid-Infrared Spectroscopy Applications for Soil Assessment with Emphasis on Soil Organic Matter Content and Quality: State-of-the-Art and Key Issues. Appl. Spectrosc. 2013, 67, 1349–1362. [Google Scholar] [CrossRef] [PubMed]
- Viscarra Rossel, R.A.; Walvoort, D.J.J.; McBratney, A.B.; Janik, L.J.; Skjemstad, J.O. Visible, near Infrared, Mid Infrared or Combined Diffuse Reflectance Spectroscopy for Simultaneous Assessment of Various Soil Properties. Geoderma 2006, 131, 59–75. [Google Scholar] [CrossRef]
- Shepherd, K.D.; Walsh, M.G. Development of Reflectance Spectral Libraries for Characterization of Soil Properties. Soil Sci. Soc. Am. J. 2002, 66, 988–998. [Google Scholar] [CrossRef]
- Soriano-Disla, J.M.; Janik, L.J.; Viscarra Rossel, R.A.; MacDonald, L.M.; McLaughlin, M.J. The Performance of Visible, near-, and Mid-Infrared Reflectance Spectroscopy for Prediction of Soil Physical, Chemical, and Biological Properties. Appl. Spectrosc. Rev. 2014, 49, 139–186. [Google Scholar] [CrossRef]
- Clairotte, M.; Grinand, C.; Kouakoua, E.; Thébault, A.; Saby, N.P.A.; Bernoux, M.; Barthès, B.G. National Calibration of Soil Organic Carbon Concentration Using Diffuse Infrared Reflectance Spectroscopy. Geoderma 2016, 276, 41–52. [Google Scholar] [CrossRef]
- Ng, W.; Minasny, B.; Montazerolghaem, M.; Padarian, J.; Ferguson, R.; Bailey, S.; McBratney, A.B. Convolutional Neural Network for Simultaneous Prediction of Several Soil Properties Using Visible/near-Infrared, Mid-Infrared, and Their Combined Spectra. Geoderma 2019, 352, 251–267. [Google Scholar] [CrossRef]
- Reeves, J.B. Near- versus Mid-Infrared Diffuse Reflectance Spectroscopy for Soil Analysis Emphasizing Carbon and Laboratory versus on-Site Analysis: Where Are We and What Needs to Be Done? Geoderma 2010, 158, 3–14. [Google Scholar] [CrossRef]
- Griffiths, P.R.; de Haseth, J.A. Fourier Transform Infrared Spectrometry, 2nd ed.; John Wiley & Sons: Hoboken, NJ, USA, 2006. [Google Scholar]
- Baldock, J.A.; Hawke, B.; Sanderman, J.; MacDonald, L.M. Predicting Contents of Carbon and Its Component Fractions in Australian Soils from Diffuse Reflectance Mid-Infrared Spectra. Soil Res. 2013, 51, 577–595. [Google Scholar] [CrossRef] [Green Version]
- Dangal, S.; Sanderman, J.; Wills, S.; Ramirez-Lopez, L. Accurate and Precise Prediction of Soil Properties from a Large Mid-Infrared Spectral Library. Soil Syst. 2019, 3, 11. [Google Scholar] [CrossRef] [Green Version]
- Wijewardane, N.K.; Ge, Y.; Wills, S.; Libohova, Z. Predicting Physical and Chemical Properties of US Soils with a Mid-Infrared Reflectance Spectral Library. Soil Sci. Soc. Am. J. 2018, 82, 722–732. [Google Scholar] [CrossRef] [Green Version]
- Dangal, S.R.S.; Sanderman, J. Is Standardization Necessary for Sharing of a Large Mid-Infrared Soil Spectral Library? Sensors 2020, 20, 6729. [Google Scholar] [CrossRef] [PubMed]
- Pittaki-Chrysodonta, Z.; Hartemink, A.E.; Sanderman, J.; Ge, Y.; Huang, J. Evaluating Three Calibration Transfer Methods for Predictions of Soil Properties Using Mid-infrared Spectroscopy. Soil Sci. Soc. Am. J. 2021, 85, 501–519. [Google Scholar] [CrossRef]
- Feudale, R.N.; Woody, N.A.; Tan, H.; Myles, A.J.; Brown, S.D.; Ferré, J. Transfer of Multivariate Calibration Models: A Review. Chemom. Intell. Lab. Syst. 2002, 64, 181–192. [Google Scholar] [CrossRef]
- Workman, J.J. A Review of Calibration Transfer Practices and Instrument Differences in Spectroscopy. Appl. Spectrosc. 2018, 72, 340–365. [Google Scholar] [CrossRef] [PubMed]
- Seybold, C.A.; Ferguson, R.; Wysocki, D.; Bailey, S.; Anderson, J.; Nester, B.; Schoeneberger, P.; Wills, S.; Libohova, Z.; Hoover, D.; et al. Application of Mid-Infrared Spectroscopy in Soil Survey. Soil Sci. Soc. Am. J. 2019, 83, 1746–1749. [Google Scholar] [CrossRef]
- CAR 2020 Climate Action Reserve 2001–2020. Available online: www.climateactionreserve.org/how/projects/ (accessed on 20 March 2021).
- Sanderman, J.; Savage, K.; Dangal, S.R.S. Mid-Infrared Spectroscopy for Prediction of Soil Health Indicators in the United States. Soil Sci. Soc. Am. J. 2020, 84, 251–261. [Google Scholar] [CrossRef] [Green Version]
- Robertson, G.P.; Hamilton, S.K. Long-Term Ecological Research at the Kellogg Biological Station LTER Site: Conceptual and Experimental Framework. In The Ecology of Agricultural Landscapes: Long-Term Research on the Path to Sustainability; Hamilton, S.K., Doll, J.E., Robertson, G.P., Eds.; Oxford University Press: New York, NY, USA, 2015; pp. 1–32. [Google Scholar]
- Jin, V.L.; Schmer, M.R.; Wienhold, B.J.; Stewart, C.E.; Varvel, G.E.; Sindelar, A.J.; Follett, R.F.; Mitchell, R.B.; Vogel, K.P. Twelve Years of Stover Removal Increases Soil Erosion Potential without Impacting Yield. Soil Sci. Soc. Am. J. 2015, 79, 1169–1178. [Google Scholar] [CrossRef]
- Follett, R.F.; Vogel, K.P.; Varvel, G.E.; Mitchell, R.B.; Kimble, J. Soil Carbon Sequestration by Switchgrass and No-Till Maize Grown for Bioenergy. Bioenergy Res. 2012, 5, 866–875. [Google Scholar] [CrossRef] [Green Version]
- Sindelar, A.J.; Schmer, M.R.; Jin, V.L.; Wienhold, B.J.; Varvel, G.E. Long-Term Corn and Soybean Response to Crop Rotation and Tillage. Agron. J. 2015, 107, 2241–2252. [Google Scholar] [CrossRef] [Green Version]
- Hepperly, P.; Seidel, R.; Pimentel, D.; Hanson, J.; Douds, D. Organic farming enhances soil carbon and its benefits. In Soil Carbon Management; Kimble, J.M., Rice, C.W., Reed, D., Mooney, S., Follett, R.F., Lal, R., Eds.; CRC Press: Boca Raton, FL, USA, 2007; pp. 130–150. [Google Scholar]
- Sanderson, M.A.; Johnson, H.; Liebig, M.A.; Hendrickson, J.R.; Duke, S.E. Kentucky. Bluegrass Invasion Alters Soil Carbon and Vegetation Structure on Northern Mixed-Grass Prairie of the United States. Invasive Plant Sci. Manag. 2017, 10, 9–16. [Google Scholar] [CrossRef]
- Liebig, M.A.; Gross, J.R.; Kronberg, S.L.; Phillips, R.L. Grazing Management Contributions to Net Global Warming Potential: A Long-Term Evaluation in the Northern Great Plains. J. Environ. Qual. 2010, 39, 799–809. [Google Scholar] [CrossRef] [PubMed]
- Gollany, H.T.; DelGrosso, S.J.; Dell, C.J.; Adler, P.R.; Polumsky, R.W. Assessing the Effectiveness of Agricultural Conservation Practices in Maintaining Soil Organic Carbon under Contrasting Agroecosystems and Changing Climate. Soil Sci. Soc. Am. J. 2021, 1–18. [Google Scholar] [CrossRef]
- Cavigelli, M.A.; Teasdale, J.R.; Conklin, A.E. Long-Term Agronomic Performance of Organic and Conventional Field Crops in the Mid-Atlantic Region. Agron. J. 2008, 100, 785–794. [Google Scholar] [CrossRef] [Green Version]
- White, K.E.; Cavigelli, M.A.; Conklin, A.E.; Rasmann, C. Economic Performance of Long-Term Organic and Conventional Crop Rotations in the Mid-Atlantic. Agron. J. 2019, 111, 1358–1370. [Google Scholar] [CrossRef]
- Roudier, P. Clhs: A R Package for Conditioned Latin Hypercube Sampling. 2011. Available online: https://cran.r-project.org/web/packages/clhs/clhs.pdf (accessed on 20 March 2021).
- R. Core Team (2020). R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna Austria, 2020; Available online: https://www.r-project.org/ (accessed on 20 March 2021).
- Ramirez-Lopez, L.; Behrens, T.; Schmidt, K.; Stevens, A.; Demattê, J.A.M.; Scholten, T. The Spectrum-Based Learner: A New Local Approach for Modeling Soil Vis-NIR Spectra of Complex Datasets. Geoderma 2013, 195–196. [Google Scholar] [CrossRef]
- Ramirez-Lopez, L.; Stevens, A. Resemble: Regression and Similarity Evaluation for Memory-Based Learning in Spectral Chemometrics, R Package Version. 2019. Available online: https://cran.r-project.org/web/packages/resemble/resemble.pdf(accessed on 20 March 2021).
- Microsoft Corporation; Weston, S. DoSNOW: Foreach Parallel Adaptor for the “Snow”. Available online: https://cran.r-project.org/web/packages/doSNOW/doSNOW.pdf (accessed on 20 March 2021).
- Lin, L.I.-K. A Concordance Correlation Coefficient to Evaluate Reproducibility. Biometrics 1989, 45, 255–268. [Google Scholar] [CrossRef]
- De Mendiburu, F.; Yaseen, M. Statistical Procedures for Agricultural Research; John Wiley & Sons: Hoboken, NJ, USA, 1984. [Google Scholar]
- Ben-Shachar, M.; Lüdecke, D.; Makowski, D. Effectsize: Estimation of Effect Size Indices and Standardized Parameters. J. Open Source Softw. 2020, 5, 1–7. [Google Scholar] [CrossRef]
- Wickham, H. Ggplot2: Elegant Graphics for Data Analysis; Springer: New York, NY, USA, 2016; ISBN 978-3-319-24277-4. [Google Scholar]
- Ahlmann-Eltze, C. Ggsignif: Significance Brackets for “Ggplot2”. R Package Version 0.6.0. 2019. Available online: https://cran.r-project.org/web/packages/ggsignif/ggsignif.pdf (accessed on 20 March 2021).
- Brown, D.J. Using a Global VNIR Soil-Spectral Library for Local Soil Characterization and Landscape Modeling in a 2nd-Order Uganda Watershed. Geoderma 2007, 140, 444–453. [Google Scholar] [CrossRef]
- Seidel, M.; Hutengs, C.; Ludwig, B.; Thiele-Bruhn, S.; Vohland, M. Strategies for the Efficient Estimation of Soil Organic Carbon at the Field Scale with Vis-NIR Spectroscopy: Spectral Libraries and Spiking vs. Local Calibrations. Geoderma 2019, 354, 113856. [Google Scholar] [CrossRef]
- Lobsey, C.R.; Viscarra Rossel, R.A.; Roudier, P.; Hedley, C.B. Rs-Local Data-Mines Information from Spectral Libraries to Improve Local Calibrations. Eur. J. Soil Sci. 2017, 68, 840–852. [Google Scholar] [CrossRef] [Green Version]
- Ge, Y.; Morgan, C.L.S.; Grunwald, S.; Brown, D.J.; Sarkhot, D.V. Comparison of Soil Reflectance Spectra and Calibration Models Obtained Using Multiple Spectrometers. Geoderma 2011, 161, 202–211. [Google Scholar] [CrossRef]
- Viscarra Rossel, R.A.; Jeon, Y.S.; Odeh, I.O.A.; McBratney, A.B. Using a Legacy Soil Sample to Develop a Mid-IR Spectral Library. Soil Res. 2008, 46, 1–16. [Google Scholar] [CrossRef]
- Walkley, A.; Black, I.A. An Examination of the Degtjareff Method for Determining Soil Organic Matter, and a Proposed Modification of the Chromic Acid Titration Method. Soil Sci. 1934, 37, 29–38. [Google Scholar] [CrossRef]
- Pribyl, D.W. A Critical Review of the Conventional SOC to SOM Conversion Factor. Geoderma 2010, 156, 75–83. [Google Scholar] [CrossRef]
- Cambardella, C.A.; Moorman, T.B.; Novak, J.M.; Parkin, T.B.; Karlen, D.L.; Turco, R.F.; Konopka, A.E. Field-Scale Variability of Soil Properties in Central Iowa Soils. Soil Sci. Soc. Am. J. 1994, 58, 1501–1511. [Google Scholar] [CrossRef]
- Zhang, Y.; Hartemink, A.E. Quantifying Short-Range Variation of Soil Texture and Total Carbon of a 330-Ha Farm. Catena 2021, 201, 105200. [Google Scholar] [CrossRef]
- Post, W.M.; Kwon, K.C. Soil Carbon Sequestration and Land-Use Change: Processes and Potential. Glob. Chang. Biol. 2000, 6, 317–327. [Google Scholar] [CrossRef] [Green Version]
- Sanderman, J.; Baldock, J.A. Accounting for Soil Carbon Sequestration in National Inventories: A Soil Scientist’s Perspective. Environ. Res. Lett. 2010, 5, 1–7. [Google Scholar] [CrossRef]
- Atkinson, A.C.; Bailey, R.A. One Hundred Years of the Design of Experiments on and off the Pages of Biometrika. Biometrika 2001, 88, 53–97. [Google Scholar] [CrossRef]
- Van Es, H.M.; Gomes, C.P.; Sellmann, M.; van Es, C.L. Spatially-Balanced Complete Block Designs for Field Experiments. Geoderma 2007, 140, 346–352. [Google Scholar] [CrossRef]
- Jin, V.L.; Wienhold, B.J.; Mikha, M.M.; Schmer, M.R. Cropping System Partially Offsets Tillage-Related Degradation of Soil Organic Carbon and Aggregate Properties in a 30-Yr Rainfed Agroecosystem. Soil Tillage Res. 2021, 209, 104968. [Google Scholar] [CrossRef]





| LTR Trial | ID | Treatment | a Crop | Years | Depths (cm) | n | b ANOVA Model |
|---|---|---|---|---|---|---|---|
| KBS | T1 T2 T3 T4 T6 T8 | Conventional No-till Reduced input Biologically based Perennial rotation Never tilled | CSW CSW CSW CSW A G | 1989-2017 | (0–25) | 28 | SOC~T |
| Lincoln-REAP | NT-None DT-All | No till, no stover removal Disk till, full stover removal | CC CC | 2001, 2011, 2014 | 0–7.5 7.5–15 15–30 (0–150) | 24 24 24 | SOC~Y*T*D |
| Lincoln-TCSE | Disk No-till Plow | Disk till No till Moldboard plow | CC CS | 1999, 2004, 2011 | 0–15 15–30 (0–150) | 54 54 | SOC~Y*T*D |
| Rodale | Conventional_Till Conventional_No till Conventional_Reduced Till Organic Legume_Till Organic Legume_ReducedTill Organic Manure_Till Organic Manure_ReducedTill | CS CS CR CR CR CR | 1981, 1987, 1995, 2002, 2008, 2012, 2015, 2018 | (0–20) | 32 | SOC~Y*T | |
| Mandan | HGP MGP | High grazing pressure Moderate grazing pressure | G | 1959, 1991, 2003, 2014 | 0–7.5 7.5–15 (0–60) | 24 24 | SOC~Y*T*D |
| Pendleton | CT0 CT120 NTA0 NTA120 NTB0 NTB120 | Moldboard Till, 0N+ Moldboard Till, 120N No-till 1982, 0N No-Till 1982, 120N No-till 1997, 0N No-Till 1997, 120N | WF WP | 2005, 2009, 2014 | 0–10 10–20 20–30 30–60 (0–60) | 72 | SOC~Y*T |
| Beltsville | nt ct 2Yor 3Yor 6Yor | No-till (3-year rotation) Chisel till (3-year rotation) 2-year organic rotation 3-year organic rotation 6-year organic rotation | CSW CSW CS CSW CSWAAA | 1996, 2006, 2011, 2016 | 5–10 (0–50) | 56 | SOC~Y*T |
| Trial | Slope | Intercept | Bias | R2 | RMSE a (%) | CCC b | RPD c | MAE d (%) | n e |
|---|---|---|---|---|---|---|---|---|---|
| All trials | 0.90 | 0.35 | 0.23 | 0.91 | 0.24 | 0.92 | 3.40 | 0.28 | 1377 |
| Beltsville | 1.25 | 0.10 | 0.26 | 0.87 | 0.23 | 0.81 | 2.76 | 0.28 | 390 |
| KBS | 0.73 | 0.31 | 0.08 | 0.70 | 0.16 | 0.80 | 1.82 | 0.15 | 28 |
| Mandan | 0.83 | 0.41 | 0.09 | 0.94 | 0.33 | 0.96 | 4.01 | 0.33 | 199 |
| Pendleton | 1.15 | 0.11 | 0.25 | 0.84 | 0.12 | 0.63 | 2.50 | 0.25 | 264 |
| Lincoln-REAP | 0.80 | 0.53 | 0.23 | 0.89 | 0.10 | 0.73 | 2.89 | 0.23 | 179 |
| Rodale | 0.99 | 0.41 | 0.38 | 0.81 | 0.15 | 0.54 | 2.37 | 0.39 | 78 |
| Lincoln-TCSE | 0.80 | 0.48 | 0.26 | 0.92 | 0.14 | 0.84 | 3.55 | 0.28 | 239 |
| Slope | Intercept | Bias | R2 | RMSE a (%) | CCC b | RPD c | MAE d (%) | n e | |
|---|---|---|---|---|---|---|---|---|---|
| Woodwell | 0.95 | 0.32 | 0.25 | 0.91 | 0.24 | 0.90 | 3.28 | 0.28 | 240 |
| KSSL | 0.98 | 0.05 | 0.02 | 0.93 | 0.21 | 0.96 | 3.75 | 0.15 | 240 |
| Trial | Model | Observed SOC% p-Value | m.ES a | EML SOC% p-Value | e.ES b |
|---|---|---|---|---|---|
| KBS | Treatment | 2.53 × 10−11 *** | 0.49 | 0.0005 *** | 0.55 |
| n | 28 | 28 | |||
| Lincoln-REAP | Year | 0.571 | 0 | 0.684 | 0.01 |
| Treatment | 0.035 * | 0.08 | 0.169 | 0.17 | |
| Depth | 0.009 ** | 0.41 | 0.002 *** | 0.32 | |
| Year × Treatment | 0.290 | 0.05 | 0.287 | 0.05 | |
| Year × Depth | 0.674 | 0 | 0.941 | 0.03 | |
| Treatment × Depth | 0.774 | 0.02 | 0.790 | 0.02 | |
| Year × Treatment × Depth | 0.362 | 0.04 | 0.617 | 0.08 | |
| n | 72 | 72 | |||
| Lincoln-TCSE | Year | 0.563 | 0 | 0.252 | 0.01 |
| Treatment | 0.000 *** | 0.17 | 0.003 ** | 0.11 | |
| Depth | 1.84 × 10−11 *** | 0.38 | 4.01 × 10−7 *** | 0.24 | |
| Year × Treatment | 0.944 | 0 | 0.960 | 0 | |
| Year × Depth | 0.838 | 0 | 0.925 | 0 | |
| Treatment × Depth | 0.853 | 0 | 0.936 | 0 | |
| Year × Treatment × Depth | 0.925 | 0 | 0.916 | 0 | |
| n | 54 | 54 | |||
| Rodale | Treatment | 0.019 * | 0.51 | 0.001 ** | 0.47 |
| n | 32 | 32 | |||
| Mandan | Year | 4.94 × 10−5 *** | 0.31 | 0.0001 *** | 0.34 |
| Treatment | 0.031 * | 0.08 | 0.068 + | 0.11 | |
| Depth | 2.50 × 10−15 *** | 0.79 | 4.07 × 10−15 *** | 0.79 | |
| Year × Treatment | 0.881 | 0.01 | 0.491 | 0 | |
| Year × Depth | 6.72 × 10−5 *** | 0.20 | 0.003 ** | 0.33 | |
| Treatment × Depth | 0.929 | 0 | 0.652 | 0 | |
| Year × Treatment × Depth | 0.424 | 0 | 0.831 | 0.02 | |
| n | 48 | 48 | |||
| Pendleton | Year | 0.001 ** | 0.10 | 0.042 * | 0.21 |
| Treatment | 5.14 × 10−7 *** | 0.41 | 8.38 × 10−6 *** | 0.47 | |
| Year × Treatment | 0.147 | 0.02 | 0.962 | 0.15 | |
| n | 72 | 72 | |||
| Beltsville | Year | 0.165 | 0.08 | 0.377 | 0.05 |
| Treatment | 0.004 ** | 0.31 | 0.635 | 0.06 | |
| Year × Treatment | 0.121 | 0.25 | 0.498 | 0.15 | |
| n | 56 | 56 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sanderman, J.; Savage, K.; Dangal, S.R.S.; Duran, G.; Rivard, C.; Cavigelli, M.A.; Gollany, H.T.; Jin, V.L.; Liebig, M.A.; Omondi, E.C.; et al. Can Agricultural Management Induced Changes in Soil Organic Carbon Be Detected Using Mid-Infrared Spectroscopy? Remote Sens. 2021, 13, 2265. https://doi.org/10.3390/rs13122265
Sanderman J, Savage K, Dangal SRS, Duran G, Rivard C, Cavigelli MA, Gollany HT, Jin VL, Liebig MA, Omondi EC, et al. Can Agricultural Management Induced Changes in Soil Organic Carbon Be Detected Using Mid-Infrared Spectroscopy? Remote Sensing. 2021; 13(12):2265. https://doi.org/10.3390/rs13122265
Chicago/Turabian StyleSanderman, Jonathan, Kathleen Savage, Shree R. S. Dangal, Gabriel Duran, Charlotte Rivard, Michel A. Cavigelli, Hero T. Gollany, Virginia L. Jin, Mark A. Liebig, Emmanuel Chiwo Omondi, and et al. 2021. "Can Agricultural Management Induced Changes in Soil Organic Carbon Be Detected Using Mid-Infrared Spectroscopy?" Remote Sensing 13, no. 12: 2265. https://doi.org/10.3390/rs13122265
APA StyleSanderman, J., Savage, K., Dangal, S. R. S., Duran, G., Rivard, C., Cavigelli, M. A., Gollany, H. T., Jin, V. L., Liebig, M. A., Omondi, E. C., Rui, Y., & Stewart, C. (2021). Can Agricultural Management Induced Changes in Soil Organic Carbon Be Detected Using Mid-Infrared Spectroscopy? Remote Sensing, 13(12), 2265. https://doi.org/10.3390/rs13122265