Identifying Leaf Phenology of Deciduous Broadleaf Forests from PhenoCam Images Using a Convolutional Neural Network Regression Method
Abstract
:1. Introduction
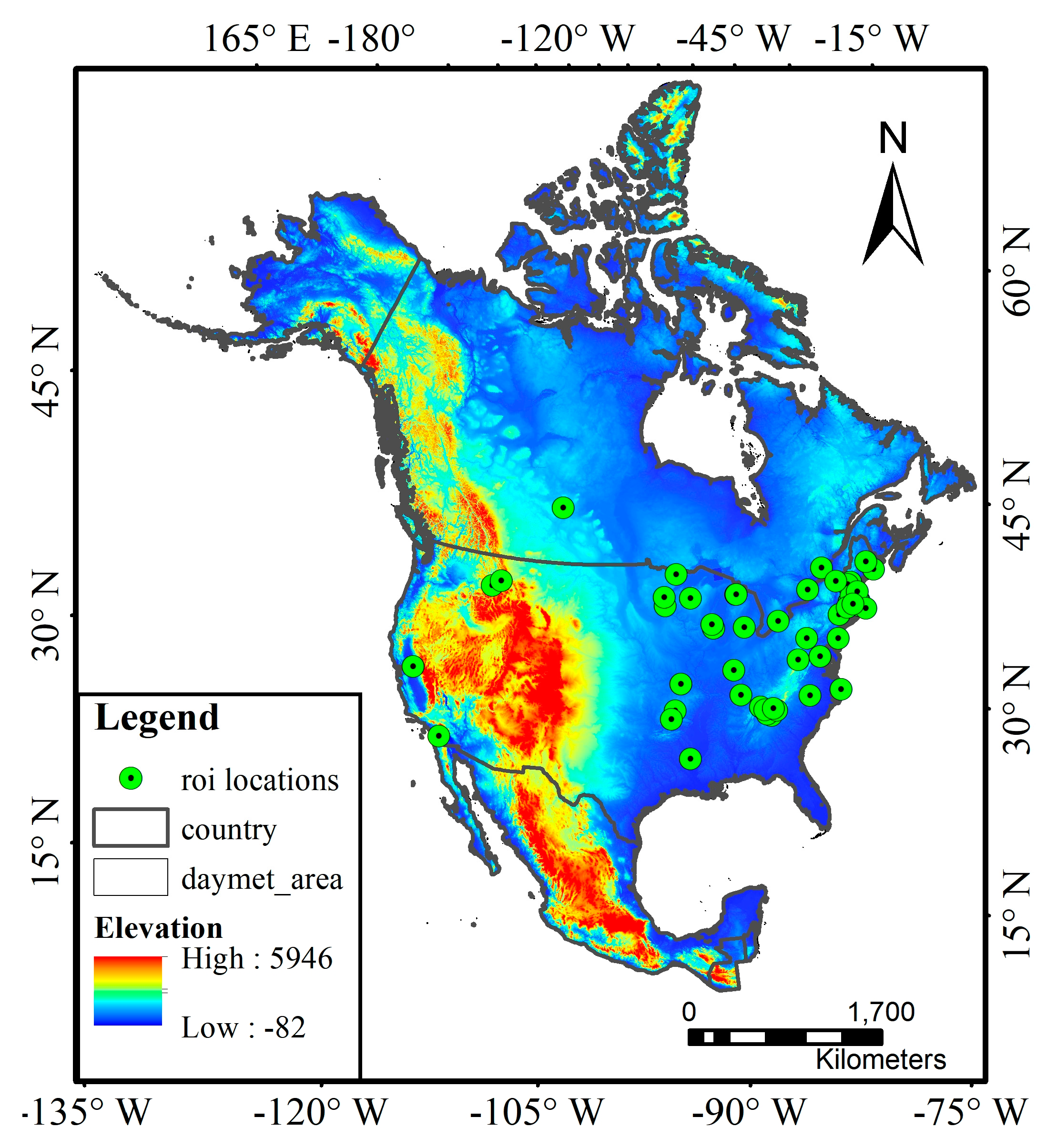
2. Study Materials
3. Methods
3.1. Data Preprocessing
3.2. Leaf Phenology Prediction
3.3. Leaf Phenology Prediction Using Detected ROI Images
3.4. Model Assessment
4. Results
4.1. Predicting Leaf Phenology Using the Entire Phenocam Images
4.2. Predicting Leaf Phenology Using Detected ROI Images
5. Discussion
6. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Site Name | Latitude (°) | Longitude (°) | Elevation (M) | SOS (Star-of-Season) | EOS (Star-of-Season) | Image Number and the Year |
|---|---|---|---|---|---|---|
| Acadia | 44.3769 | −68.2608 | 158 | 2016/5/8 | 2016/10/21 | 338/2016 |
| Alligatorriver | 35.7879 | −75.9038 | 1 | 2016/3/27 | 2016/10/16 | 285/2016 |
| Asa | 57.1645 | 14.7825 | 180 | 2015/5/23 | 2015/10/28 | 186/2015 |
| Bartlett | 44.0646 | −71.2881 | 268 | 2015/4/10 | 2015/11/6 | 297/2015 |
| Bartlettir | 44.0646 | −71.2881 | 268 | 2016/5/10 | 2016/11/26 | 353/2016 |
| Bitterootvalley | 46.5070 | −114.0910 | 1017 | 2016/5/9 | 2016/10/7 | 357/2016 |
| Bostoncommon | 42.3559 | −71.0641 | 10 | 2016/4/9 | 2016/10/27 | 157/2016 |
| Boundarywaters | 47.9467 | −91.4955 | 519 | 2016/4/25 | 2016/11/21 | 350/2016 |
| Bullshoals | 36.5628 | −93.0666 | 260 | 2016/5/19 | 2016/9/25 | 315/2016 |
| Canadaoa | 53.6289 | −106.1978 | 601 | 2016/4/1 | 2016/11/13 | 212/2016 |
| Caryinstitute | 41.7839 | −73.7341 | 127 | 2016/5/1 | 2016/6/4 | 347/2016 |
| Cedarcreek | 45.4019 | −93.2042 | 276 | 2016/5/1 | 2016/10/30 | 288/2016 |
| Columbiamissouri | 38.7441 | −92.1997 | 232 | 2009/4/11 | 2009/11/9 | 144/2009 |
| Coweeta | 35.0596 | −83.4280 | 680 | 2016/4/8 | 2016/10/20 | 291/2016 |
| Dollysods | 39.0995 | −79.4270 | 1133 | 2003/4/21 | 2003/11/2 | 230/2003 |
| Downerwoods | 43.0794 | −87.8808 | 213 | 2016/5/8 | 2016/10/21 | 339/2016 |
| Drippingsprings | 33.3000 | −116.8000 | 400 | 2006/4/2 | 2006/12/29 | 364/2006 |
| Dukehw | 35.9736 | −79.1004 | 400 | 2016/3/12 | 2016/10/31 | 329/2016 |
| Harvard | 42.5378 | −72.1715 | 340 | 2016/5/6 | 2016/10/27 | 359/2016 |
| Harvardbarn2 | 42.5353 | −72.1899 | 350 | 2016/5/9 | 2016/10/21 | 360/2016 |
| Harvardlph | 42.5420 | −72.1850 | 380 | 2016/5/10 | 2016/10/20 | 357/2016 |
| Howland2 | 45.2128 | −68.7418 | 79 | 2016/5/21 | 2016/10/4 | 206/2016 |
| Hubbardbrook | 43.9438 | −71.7010 | 253 | 2016/5/2 | 2016/11/6 | 351/2016 |
| Hubbardbrooknfws | 42.9580 | −71.7762 | 930 | 2016/5/21 | 2016/10/9 | 73/2016 |
| Joycekilmer | 35.2570 | −83.7950 | 1373 | 2016/4/26 | 2016/10/20 | 314/2016 |
| Laurentides | 45.9881 | −74.0055 | 350 | 2016/5/16 | 2016/10/8 | 354/2016 |
| Mammothcave | 37.1858 | −86.1019 | 226 | 2016/3/31 | 2016/10/29 | 336/2016 |
| Missouriozarks | 38.7441 | −92.2000 | 219 | 2016/4/15 | 2016/10/30 | 319/2016 |
| Monture | 47.0202 | −113.1283 | 1255 | 2007/4/29 | 2007/10/13 | 274/2007 |
| Morganmonroe | 39.3231 | −86.4131 | 275 | 2016/4/10 | 2016/11/3 | 345/2016 |
| Nationalcapital | 38.8882 | −77.0695 | 28 | 2016/3/19 | 2016/11/14 | 269/2016 |
| Northattleboroma | 41.9837 | −71.3106 | 60 | 2016/5/4 | 2016/10/18 | 349/2016 |
| Oakridge1 | 35.9311 | −84.3323 | 371 | 2016/3/26 | 2016/5/26 | 182/2016 |
| Oakridge2 | 35.9311 | −84.3323 | 371 | 2016/3/27 | 2016/5/28 | 182/2016 |
| Proctor | 44.5250 | −72.8660 | 403 | 2016/5/10 | 2016/10/18 | 353/2016 |
| Queens | 44.5650 | −76.3240 | 126 | 2016/5/8 | 2016/10/16 | 347/2016 |
| Readingma | 42.5304 | −71.1272 | 100 | 2016/4/22 | 2016/11/7 | 348/2016 |
| Russellsage | 32.4570 | −91.9743 | 20 | 2016/3/17 | 2016/11/29 | 320/2016 |
| Sanford | 42.7268 | −84.4645 | 268 | 2016/4/22 | 2016/11/2 | 364/2016 |
| Shalehillsczo | 40.6500 | −77.9000 | 310 | 2016/4/23 | 2016/10/28 | 278/2016 |
| Shiningrock | 35.3902 | −82.7750 | 1500 | 2006/5/12 | 2006/10/18 | 313/2006 |
| Silaslittle | 39.9137 | −74.5960 | 33 | 2016/12/6 | 2016/10/28 | 247/2016 |
| Smokylook | 35.6325 | −83.9431 | 801 | 2016/4/16 | 2016/10/22 | 333/2016 |
| Smokypurchase | 35.5900 | −83.0775 | 1550 | 2016/4/22 | 2016/10/18 | 342/2016 |
| Snakerivermn | 46.1206 | −93.2447 | 1181 | 2016/5/9 | 2016/10/4 | 263/2016 |
| Springfieldma | 42.1352 | −72.5860 | 56 | 2016/11/8 | 2016/11/1 | 318/2016 |
| Thompsonfarm2n | 43.1086 | −70.9505 | 23 | 2016/3/25 | 2016/12/4 | 322/2016 |
| Tonzi | 38.4309 | −120.9659 | 177 | 2016/2/28 | 2016/6/13 | 252/2016 |
| Turkeypointdbf | 42.6353 | −80.5576 | 211 | 2016/5/11 | 2016/10/18 | 298/2016 |
| Umichbiological | 45.5598 | −84.7138 | 230 | 2016/5/16 | 2016/10/21 | 347/2016 |
| Umichbiological2 | 45.5625 | −84.6976 | 240 | 2016/5/8 | 2016/10/19 | 336/2016 |
| Upperbuffalo | 35.8637 | −93.4932 | 777 | 2006/4/8 | 2006/10/19 | 257/2006 |
| Uwmfieldsta | 43.3871 | −88.0229 | 265 | 2016/5/5 | 2016/10/22 | 344/2016 |
| Willowcreek | 45.8060 | −90.0791 | 521 | 2016/5/7 | 2016/10/3 | 307/2016 |
| Woodshole | 41.5495 | −70.6432 | 10 | 2016/5/8 | 2016/11/1 | 288/2016 |
| Worcester | 42.2697 | −71.8428 | 185 | 2016/4/22 | 2016/10/20 | 353/2016 |



References
- Liu, Y.; Wu, C.Y.; Sonnentag, O.; Desai, A.R.; Wang, J. Using the red chromatic coordinate to characterize the phenology of forest canopy photosynthesis. Agric. For. Meteorol. 2020, 285–286, 107910. [Google Scholar] [CrossRef]
- Fitter, A.H.; Fitter, R.S.R. Rapid changes in flowering time in British plants. Science 2002, 296, 1689–1691. [Google Scholar] [CrossRef] [PubMed]
- Menzel, A. Phenology: Its importance to the global change community—An editorial comment. Clim. Change 2002, 54, 379–385. [Google Scholar] [CrossRef]
- Morisette, J.T.; Richardson, A.D.; Knapp, A.K.; Fisher, J.I.; Graham, E.A.; Abatzoglou, J.; Wilson, B.E.; Breshears, D.D.; Henebry, G.M.; Hanes, J.M.; et al. Tracking the rhythm of the seasons in the face of global change: Phenological research in the 21st century. Front. Ecol. Environ. 2009, 7, 253–260. [Google Scholar] [CrossRef] [Green Version]
- Richardson, A.D.; Klosterman, S.; Toomey, M. Near-Surface Sensor-Derived Phenology. Phenology: An Integrative Environmental Science; Springer: Berlin/Heidelberg, Germany, 2013; pp. 413–430. [Google Scholar]
- Hogg, E.H.; Price, D.T.; Black, T.A. Postulated feedbacks of deciduous forest phenology on seasonal climate patterns in the western Canadian interior. J. Clim. 2000, 13, 4229–4243. [Google Scholar] [CrossRef]
- Xin, Q.C.; Zhou, X.W.; Wei, N.; Yuan, H.; Ao, Z.R.; Dai, Y.J. A semiprognostic phenology model for simulating multidecadal dynamics of global vegetation leaf area index. J. Adv. Model. Earth Syst. 2020, 12, e2019MS001935. [Google Scholar] [CrossRef]
- Kikuzawa, K. Leaf phenology as an optimal strategy for carbon gain in plants. Can. J. Bot. 1995, 73, 158–163. [Google Scholar] [CrossRef]
- Luo, Y.P.; El-Madany, T.S.; Filippa, G.; Ma, X.L.; Ahrens, B.; Carrara, A.; Gonzalez-Cascon, R.; Cremonese, E.; Galvagno, M.; Hammer, T.W.; et al. Using Near-Infrared-Enabled Digital Repeat Photography to Track Structural and Physiological Phenology in Mediterranean Tree–Grass Ecosystems. Remote Sens. 2018, 10, 1293. [Google Scholar] [CrossRef] [Green Version]
- Fisher, J.I.; Mustard, J.F.; Vadeboncoeur, M.A. Green leaf phenology at Landsat resolution: Scaling from the field to the satellite. Remote Sens. Environ. 2006, 100, 265–279. [Google Scholar] [CrossRef]
- Sparks, T.H.; Menzel, A. Observed changes in seasons: An overview. Int. J. Climatol. 2002, 22, 1715–1725. [Google Scholar] [CrossRef]
- White, M.A.; Thornton, P.E.; Runnin, S.W. A continental phenology model for monitoring vegetation responses to interannual climatic variability. Glob. Biogeochem. Cycles 1997, 11, 217–234. [Google Scholar] [CrossRef]
- Justice, C.O.; Townshend, J.R.G.; Holben, B.N.; Tucker, C.J. Analysis of the phenology of global vegetation using meteorological satellite data. Int. J. Remote Sens. 1985, 6, 1271–1318. [Google Scholar] [CrossRef]
- Zhang, X.Y.; Friedl, M.A.; Schaaf, C.B.; Strahler, A.H.; Hodges, J.C.F.; Gao, F.; Reed, B.C.; Huete, A. Monitoring vegetation phenology using MODIS. Remote Sens. Environ. 2003, 84, 471–475. [Google Scholar] [CrossRef]
- Hilker, T.; Wulder, M.A.; Coops, N.C.; Linke, J.; McDermid, G.; Masek, J.G.; Gao, F.; White, J.C. A new data fusion model for high spatial- and temporal-resolution mapping of forest disturbance based on Landsat and MODIS. Remote Sens. Environ. 2009, 113, 1613–1627. [Google Scholar] [CrossRef]
- Badeck, F.W.; Bondeau, A.; Bottcher, K.; Doktor, D.; Lucht, W.; Schaber, J.; Sitch, S. Responses of spring phenology to climate change. New Phytol. 2004, 162, 295–309. [Google Scholar] [CrossRef]
- Richardson, A.D. Tracking seasonal rhythms of plants in diverse ecosystems with digital camera imagery. New Phytol. 2019, 222, 1742–1750. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Brown, T.B.; Hultine, K.R.; Steltzer, H.; Denny, E.G.; Denslow, M.W.; Granados, J.; Henderson, S.; Moore, D.; Nagai, S.; SanClements, M.; et al. Using phenocams to monitor our changing Earth: Toward a global phenocam network. Front. Ecol. Environ. 2016, 14, 84–93. [Google Scholar] [CrossRef] [Green Version]
- Bornez, K.; Richardson, A.D.; Verger, A.; Descals, A.; Penuelas, J. Evaluation of vegetation and proba-v phenology using phenocam and eddy covariance data. Remote Sens. 2020, 12, 3077. [Google Scholar] [CrossRef]
- Richardson, A.D.; Hufkens, K.; Milliman, T.; Frolking, S. Intercomparison of phenological transition dates derived from the PhenoCam Dataset V1.0 and MODIS satellite remote sensing. Sci. Rep. 2018, 8, 5679. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, H.S.; Jia, G.S.; Epstein, H.E.; Zhao, H.C.; Zhang, A.Z. Integrating a PhenoCam-derived vegetation index into a light use efficiency model to estimate daily gross primary production in a semi-arid grassland. Agric. For. Meteorol. 2020, 288–289, 107983. [Google Scholar] [CrossRef]
- Wu, J.; Rogers, A.; Albert, L.P.; Ely, K.; Prohaska, N.; Wolfe, B.T.; Oliveira, R.C.; Saleska, S.R.; Serbin, S.P. Leaf reflectance spectroscopy captures variation in carboxylation capacity across species, canopy environment and leaf age in lowland moist tropical forests. New Phytol. 2019, 224, 663–674. [Google Scholar] [CrossRef] [PubMed]
- Zhang, X.Y.; Jayavelu, S.; Liu, L.L.; Friedl, M.A.; Henebry, G.M.; Liu, Y.; Schaaf, C.B.; Richardson, A.D.; Gray, J. Evaluation of land surface phenology from VIIRS data using time series of PhenoCam imagery. Agric. For. Meteorol. 2018, 256, 137–149. [Google Scholar] [CrossRef]
- Zhang, S.K.; Butto, V.; Khare, S.; Deslauriers, A.; Morin, H.; Huang, J.G.; Ren, H.; Rossi, S. Calibrating PhenoCam Data with Phenological Observations of a Black Spruce Stand. Can. J. Remote Sens. 2020, 46, 154–165. [Google Scholar] [CrossRef]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Richardson, A.D.; Hufkens, K.; Milliman, T.; Aubrecht, D.M.; Chen, A.; Gray, J.M.; Johnston, M.R.; Keenan, T.F.; Klosterman, S.T.; Kosmala, M.; et al. Tracking vegetation phenology across diverse North American biomes using PhenoCam imagery. Sci. Data 2018, 5, 1–24. [Google Scholar] [CrossRef]
- Sonnentag, O.; Hufkens, K.; Teshera-Sterne, C.; Young, A.M.; Friedl, M.; Braswell, B.H.; Milliman, T.; O’Keefe, J.; Richardson, A.D. Digital repeat photography for phenological research in forest ecosystems. Agric. For. Meteorol. 2012, 152, 159–177. [Google Scholar] [CrossRef]
- Seyednasrollah, B.; Young, A.M.; Hufkens, K.; Milliman, T.; Friedl, M.A.; Frolking, S.; Richardson, A.D. Tracking vegetation phenology across diverse biomes using Version 2.0 of the PhenoCam Dataset. Sci. Data 2019, 6, 222. [Google Scholar] [CrossRef] [Green Version]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Alexnet Imagenet classification with deep convolutional neural networks. Adv. Neural Info Process. Syst. 2012, 25, 1097–1105. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Szegedy, C.; Liu, W.; Jia, Y.; Sermanet, P.; Reed, S.; Anguelov, D.; Erhan, D.; Vanhoucke, V.; Rabinovich, A. Going Deeper with Convolutions. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 1–9. [Google Scholar]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Las Vegas, NV, USA, 27–30 June 2016; pp. 770–778. [Google Scholar]
- Ma, L.; Shuai, R.J.; Ran, X.M.; Liu, W.J.; Ye, C. Combining DC-GAN with ResNet for blood cell image classification. Med. Biol. Eng. Comput. 2020, 58, 1251–1264. [Google Scholar] [CrossRef]
- Long, J.; Shelhamer, E.; Darrell, T. Fully convolutional networks for semantic segmentation. In Proceedings of the 2015 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Boston, MA, USA, 7–12 June 2015; pp. 3431–3440. [Google Scholar]
- Badrinarayanan, V.; Kendall, A.; Cipolla, R. Segnet: A deep convolutional encoder-decoder architecture for image segmentation. IEEE Trans. Pattern Analy. Mach. Intel. 2017, 39, 2481–2495. [Google Scholar] [CrossRef]
- Lalli, P.; Murphey, C.L.; Kucheryavaya, A.Y.; Bray, R.A. Evaluation of discrepant deceased donor hla typings reported in Unet. Hum. Immunol. 2017, 78, 92. [Google Scholar] [CrossRef]
- Chen, L.C.; Papandreou, G.; Kokkinos, I.; Murphy, K.; Yuille, A.L. Deeplab: Semantic image segmentation with deep convolutional nets, atrous convolution, and fully connected crfs. IEEE Trans. Pattern Analy. Mach. Intel. 2018, 40, 834–848. [Google Scholar] [CrossRef] [PubMed]
- Zhou, J.C.; Hao, M.L.; Zhang, D.H.; Zou, P.Y.; Zhang, W.S. Fusion PSPnet Image Segmentation Based Method for Multi-Focus Image Fusion. IEEE Photonics J. 2019, 11, 1–12. [Google Scholar] [CrossRef]
- Jiang, X.; Zhang, X.C.; Xin, Q.C.; Xi, X.; Zhang, P.C. Arbitrary-Shaped Building Boundary-Aware Detection with Pixel Aggregation Network. IEEE J. Sel. Top. App. Remote Sens. 2020. [Google Scholar] [CrossRef]
- Willmott, C.J.; Ackleson, S.G.; Davis, R.E.; Feddema, J.J.; Klink, K.M.; Legates, D.R.; O’Donnell, J.; Rowe, C.M. Statistics for the evaluation and comparison of models. J. Geophys. Res. 1985, 90, 8995–9005. [Google Scholar] [CrossRef] [Green Version]








| Model | Overall Accuracies | Precision | Recall | F1 | Mean IOU |
|---|---|---|---|---|---|
| Unet | 0.955 | 0.831 | 0.740 | 0.721 | 0.636 |
| PSPNet | 0.958 | 0.861 | 0.715 | 0.711 | 0.632 |
| DeepLabV3+ | 0.961 | 0.874 | 0.813 | 0.810 | 0.739 |
| FPN | 0.976 | 0.884 | 0.824 | 0.823 | 0.751 |
| BAPANet | 0.981 | 0.984 | 0.961 | 0.966 | 0.880 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Cao, M.; Sun, Y.; Jiang, X.; Li, Z.; Xin, Q. Identifying Leaf Phenology of Deciduous Broadleaf Forests from PhenoCam Images Using a Convolutional Neural Network Regression Method. Remote Sens. 2021, 13, 2331. https://doi.org/10.3390/rs13122331
Cao M, Sun Y, Jiang X, Li Z, Xin Q. Identifying Leaf Phenology of Deciduous Broadleaf Forests from PhenoCam Images Using a Convolutional Neural Network Regression Method. Remote Sensing. 2021; 13(12):2331. https://doi.org/10.3390/rs13122331
Chicago/Turabian StyleCao, Mengying, Ying Sun, Xin Jiang, Ziming Li, and Qinchuan Xin. 2021. "Identifying Leaf Phenology of Deciduous Broadleaf Forests from PhenoCam Images Using a Convolutional Neural Network Regression Method" Remote Sensing 13, no. 12: 2331. https://doi.org/10.3390/rs13122331
APA StyleCao, M., Sun, Y., Jiang, X., Li, Z., & Xin, Q. (2021). Identifying Leaf Phenology of Deciduous Broadleaf Forests from PhenoCam Images Using a Convolutional Neural Network Regression Method. Remote Sensing, 13(12), 2331. https://doi.org/10.3390/rs13122331









