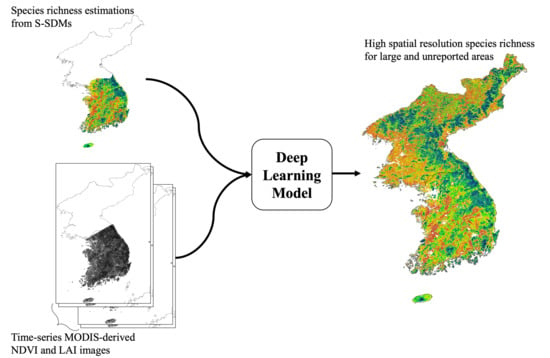
Mapping Potential Plant Species Richness over Large Areas with Deep Learning, MODIS, and Species Distribution Models
Abstract
:1. Introduction
2. Materials and Methods
2.1. Species Distribution Modeling
2.2. Species Richness (as a Response Variable)
2.3. MODIS Products: NDVI and LAI (as Input Variables)
2.4. Deep Learning-Based Species Richness Model
2.5. Independent Validation of Species Richness
3. Results
3.1. Species Richness Estimation from S-SDMs
3.2. Deep Learning-Based Species Richness Estimation Model Using Remote Sensing Data
3.3. Statistical Feature Importance
3.4. Independent Validation of Species Richness
4. Discussion
4.1. Deep Learning-Based Species Richness Estimation
4.2. Limitations and Recommendations
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Gotelli, N.J.; Colwell, R.K. Quantifying biodiversity: Procedures and pitfalls in the measurement and comparison of species richness. Ecol. Lett. 2001, 4, 379–391. [Google Scholar] [CrossRef] [Green Version]
- Pereira, H.M.; Ferrier, S.; Walters, M.; Geller, G.N.; Jongman, R.H.G.; Scholes, R.J.; Bruford, M.W.; Brummitt, N.; Butchart, S.H.M.; Cardoso, A.C.; et al. Essential biodiversity variables. Science 2013, 339, 277–278. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wich, S.A.; Koh, L.P. Conservation Drones: Mapping and Monitoring Biodiversity; Oxford University Press: Oxford, UK, 2018. [Google Scholar]
- La Sorte, F.A.; Aronson, M.F.; Lepczyk, C.A.; Horton, K.G. Area is the primary correlate of annual and seasonal patterns of avian species richness in urban green spaces. Landsc. Urban Plan. 2020, 203, 103892. [Google Scholar] [CrossRef]
- Rocchini, D.; Balkenhol, N.; Carter, G.A.; Foody, G.; Gillespie, T.W.; He, K.S.; Kark, S.; Levin, N.; Lucas, K.; Luoto, M.; et al. Remotely sensed spectral heterogeneity as a proxy of species diversity: Recent advances and open challenges. Ecol. Inform. 2010, 5, 318–329. [Google Scholar] [CrossRef]
- Gotelli, N.J.; Colwell, R.K. Estimating species richness. In Biological Diversity: Frontiers in Measurement and Assessment; Oxford University Press: Oxford, UK, 2011; Volume 12, pp. 39–54. [Google Scholar]
- Benito, B.M.; Cayuela, L.; Albuquerque, F.S. The impact of modelling choices in the predictive performance of richness maps derived from species-distribution models: Guidelines to build better diversity models. Methods Ecol. Evol. 2013, 4, 327–335. [Google Scholar] [CrossRef]
- Rocchini, D.; Marcantonio, M.; Ricotta, C. Measuring Rao’s Q diversity index from remote sensing: An open source solution. Ecol. Indic. 2017, 72, 234–238. [Google Scholar] [CrossRef]
- Scholes, R.J.; Gill, M.J.; Costello, M.J.; Sarantakos, G.; Walters, M. Working in networks to make biodiversity data more available. In The GEO Handbook on Biodiversity Observation Networks; Walters, M., Scholes, R.J., Eds.; Springer: Cham, Switzerland, 2017; pp. 1–17. [Google Scholar]
- Kühl, H.S.; Bowler, D.E.; Bösch, L.; Bruelheide, H.; Dauber, J.; Eichenberg, D.; Eisenhauer, N.; Fernández, N.; Guerra, C.A.; Henle, K.; et al. Effective biodiversity monitoring needs a culture of integration. One Earth 2020, 3, 462–474. [Google Scholar] [CrossRef]
- Guralnick, R.P.; Hill, A.W.; Lane, M. Towards a collaborative, global infrastructure for biodiversity assessment. Ecol. Lett. 2007, 10, 663–672. [Google Scholar] [CrossRef] [Green Version]
- Turner, W. Sensing biodiversity. Science 2014, 346, 301–302. [Google Scholar] [CrossRef]
- Schmeller, D.S.; Böhm, M.; Arvanitidis, C.; Barber-Meyer, S.; Brummitt, N.; Chandler, M.; Chatzinikolaou, E.; Costello, M.J.; Ding, H.; García-Moreno, J.; et al. Building capacity in biodiversity monitoring at the global scale. Biodivers. Conserv. 2017, 26, 2765–2790. [Google Scholar] [CrossRef] [Green Version]
- Amano, T.; Lamming, J.D.L.; Sutherland, W.J. Spatial gaps in global biodiversity information and the role of citizen science. Bioscience 2016, 66, 393–400. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Leathwick, J.R. Species distribution models: Ecological explanation and prediction across space and time. Annu. Rev. Ecol. Evol. Syst. 2009, 40, 677–697. [Google Scholar] [CrossRef]
- Grenié, M.; Violle, C.; Munoz, F. Is prediction of species richness from stacked species distribution models biased by habitat saturation? Ecol. Ind. 2020, 111, 105970. [Google Scholar] [CrossRef]
- Turner, W.; Rondinini, C.; Pettorelli, N.; Mora, B.; Leidner, A.; Szantoi, Z.; Buchanan, G.; Dech, S.; Dwyer, J.; Herold, M.; et al. Free and open-access satellite data are key to biodiversity conservation. Biol. Conserv. 2015, 182, 173–176. [Google Scholar] [CrossRef] [Green Version]
- Madonsela, S.; Cho, M.A.; Ramoelo, A.; Mutanga, O.; Naidoo, L. Estimating tree species diversity in the savannah using NDVI and woody canopy cover. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 106–115. [Google Scholar] [CrossRef] [Green Version]
- Pettorelli, N.; Safi, K.; Turner, W. Satellite remote sensing, biodiversity research and conservation of the future. Philos. Trans. R. Soc. B Biol. Sci. 2014, 369, 20130190. [Google Scholar] [CrossRef]
- Wu, J.; Liang, S. Developing an integrated remote sensing based biodiversity index for predicting animal species richness. Remote Sens. 2018, 10, 739. [Google Scholar] [CrossRef] [Green Version]
- Xu, H.; Wang, Y.; Guan, H.; Shi, T.; Hu, X. Detecting ecological changes with a remote sensing based ecological index (RSEI) produced time series and change vector analysis. Remote Sens. 2019, 11, 2345. [Google Scholar] [CrossRef] [Green Version]
- Randin, C.F.; Ashcroft, M.B.; Bolliger, J.; Cavender-Bares, J.; Coops, N.C.; Dullinger, S.; Dirnböck, T.; Eckert, S.; Ellis, E.; Fernández, N.; et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 2020, 239, 111626. [Google Scholar] [CrossRef]
- Hernandez-Stefanoni, J.L.; Gallardo-Cruz, J.A.; Meave, J.A.; Rocchini, D.; Bello-Pineda, J.; López-Martínez, J.O. Modeling α- and β-diversity in a tropical forest from remotely sensed and spatial data. Int. J. Appl. Earth Obs. Geoinf. 2012, 19, 359–368. [Google Scholar] [CrossRef]
- Zarnetske, P.L.; Read, Q.D.; Record, S.; Gaddis, K.D.; Pau, S.; Hobi, M.L.; Malone, S.L.; Costanza, J.; Dahlin, K.M.; Latimer, A.M.; et al. Towards connecting biodiversity and geodiversity across scales with satellite remote sensing. Glob. Ecol. Biogeogr. 2019, 28, 548–556. [Google Scholar] [CrossRef] [Green Version]
- Soto-Navarro, C.; Ravilious, C.; Arnell, A.; de Lamo, X.; Harfoot, M.; Hill, S.L.L.; Wearn, O.R.; Santoro, M.; Bouvet, A.; Mermoz, S.; et al. Mapping co-benefits for carbon storage and biodiversity to inform conservation policy and action. Philos. Trans. R Soc. B Biol. Sci. 2020, 375, 20190128. [Google Scholar] [CrossRef] [Green Version]
- LeCun, Y.; Bengio, Y.; Hinton, G. Deep learning. Nature 2015, 521, 436–444. [Google Scholar] [CrossRef]
- Zhang, L.; Zhang, L.; Du, B. Deep learning for remote sensing data: A technical tutorial on the state of the art. IEEE Geosci. Remote Sens. Mag. 2016, 4, 22–40. [Google Scholar] [CrossRef]
- Zhu, X.X.; Tuia, D.; Mou, L.; Xia, G.-S.; Zhang, L.; Xu, F.; Fraundorfer, F. Deep learning in remote sensing: A comprehensive review and list of resources. IEEE Geosci. Remote Sens. Mag. 2017, 5, 8–36. [Google Scholar] [CrossRef] [Green Version]
- Lopatin, J.; Dolos, K.; Hernández, H.; Galleguillos, M.; Fassnacht, F. Comparing generalized linear models and random forest to model vascular plant species richness using LiDAR data in a natural forest in central Chile. Remote Sens. Environ. 2016, 173, 200–210. [Google Scholar] [CrossRef]
- Hakkenberg, C.R.; Zhu, K.; Peet, R.K.; Song, C. Mapping multi-scale vascular plant richness in a forest landscape with inte-grated LiDAR and hyperspectral remote-sensing. Ecology 2018, 99, 474–487. [Google Scholar] [CrossRef]
- Sun, Y.; Huang, J.; Ao, Z.; Lao, D.; Xin, Q. Deep learning approaches for the mapping of tree species diversity in a tropical wetland using airborne LiDAR and high-spatial-resolution remote sensing images. Forests 2019, 10, 1047. [Google Scholar] [CrossRef] [Green Version]
- Kim, K.C. Preserving biodiversity in Korea’s demilitarized zone. Science 1997, 278, 242–243. [Google Scholar] [CrossRef]
- Choe, H.; Thorne, J.H.; Seo, C. Mapping national plant biodiversity patterns in South Korea with the MARS species distribution model. PLoS ONE 2016, 11, e0149511. [Google Scholar] [CrossRef]
- Ministry of Environment. The 3rd Master Plans for Protection of Wildlife (2016–2020); Ministry of Environment: Sejong-si, Korea, 2016. (In Korean)
- Dinerstein, E.; Olson, D.; Joshi, A.; Vynne, C.; Burgess, N.D.; Wikramanayake, E.; Hahn, N.; Palminteri, S.; Hedao, P.; Noss, R.; et al. An ecoregion-based approach to protecting half the terrestrial realm. Bioscience 2017, 67, 534–545. [Google Scholar] [CrossRef] [PubMed]
- Hernandez, P.A.; Graham, C.H.; Master, L.L.; Albert, D.L. The effect of sample size and species characteristics on performance of different species distribution modeling methods. Ecography 2006, 29, 773–785. [Google Scholar] [CrossRef]
- Van Proosdij, A.S.J.; Sosef, M.S.M.; Wieringa, J.J.; Raes, N. Minimum required number of specimen records to develop accurate species distribution models. Ecography 2016, 39, 542–552. [Google Scholar] [CrossRef]
- Hijmans, R.J.; Cameron, S.E.; Parra, J.L.; Jones, P.G.; Jarvis, A. Very high resolution interpolated climate surfaces for global land areas. Int. J. Climatol. J. R. Meteorol. Soc. 2005, 25, 1965–1978. [Google Scholar] [CrossRef]
- FAO; IIASA; ISRIC; ISSCAS; JRC. Harmonized World Soil Database (Version 1.2); FAO: Rome, Italy; IIASA: Laxenburg, Austria, 2012. [Google Scholar]
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-Filled Seamless SRTM Data V4, International Centre for Tropical Agri-culture (CIAT). 2008. Available online: http://srtm.csi.cgiar.org (accessed on 24 June 2021).
- Choe, H.; Thorne, J.H.; Joo, W.; Kwon, H. The biodiversity representation assessment in South Korea’s protected area network. J. Korea Soc. Environ. Restor. Technol. 2020, 23, 77–87. [Google Scholar]
- Dormann, C.F.; Elith, J.; Bacher, S.; Buchmann, C.; Carl, G.; Carré, G.; Marquéz, J.R.G.; Gruber, B.; Lafourcade, B.; Leitão, P.J.; et al. Collinearity: A review of methods to deal with it and a simulation study evaluating their performance. Ecography 2013, 36, 27–46. [Google Scholar] [CrossRef]
- Manzoor, S.A.; Griffiths, G.; Lukac, M. Species distribution model transferability and model grain size—Finer may not always be better. Sci. Rep. 2018, 8, 7168. [Google Scholar] [CrossRef] [Green Version]
- Phillips, S.J.; Anderson, R.P.; Schapire, R.E. Maximum entropy modeling of species geographic distributions. Ecol. Model. 2006, 190, 231–259. [Google Scholar] [CrossRef] [Green Version]
- Elith, J.; Graham, C.H.; Anderson, R.P.; Dudík, M.; Ferrier, S.; Guisan, A.; Hijmans, R.J.; Huettmann, F.; Leathwick, J.R.; Lehmann, A. Novel methods improve prediction of species’ distributions from occurrence data. Ecography 2006, 29, 129–151. [Google Scholar] [CrossRef] [Green Version]
- Aguirre-Gutiérrez, J.; Carvalheiro, L.G.; Polce, C.; van Loon, E.E.; Raes, N.; Reemer, M.; Biesmeijer, J.C. Fit-for-purpose: Species distribution model performance depends on evaluation criteria—Dutch hoverflies as a case study. PLoS ONE 2013, 8, e63708. [Google Scholar] [CrossRef] [Green Version]
- Lomba, A.; Pellissier, L.; Randin, C.; Vicente, J.; Moreira, F.; Honrado, J.; Guisan, A. Overcoming the rare species modelling paradox: A novel hierarchical framework applied to an Iberian endemic plant. Biol. Conserv. 2010, 143, 2647–2657. [Google Scholar] [CrossRef]
- Breiner, F.T.; Guisan, A.; Bergamini, A.; Nobis, M. Overcoming limitations of modelling rare species by using ensembles of small models. Methods Ecol. Evol. 2015, 6, 1210–1218. [Google Scholar] [CrossRef]
- Naimi, B.; Araújo, M.B. Sdm: A reproducible and extensible R platform for species distribution modelling. Ecography 2016, 39, 368–375. [Google Scholar] [CrossRef] [Green Version]
- D’Amen, M.; Dubuis, A.; Fernandes, R.F.; Pottier, J.; Pellissier, L.; Guisan, A. Using species richness and functional traits predictions to constrain assemblage predictions from stacked species distribution models. J. Biogeogr. 2015, 42, 1255–1266. [Google Scholar] [CrossRef]
- Mateo, R.G.; Felicisimo, A.M.; Pottier, J.; Guisan, A.; Muñoz, J. Do stacked species distribution models reflect altitudinal diversity patterns? PLoS ONE 2012, 7, e32586. [Google Scholar] [CrossRef] [Green Version]
- Sullivan, B.L.; Wood, C.L.; Iliff, M.J.; Bonney, R.E.; Fink, D.; Kelling, S. EBird: A citizen-based bird observation network in the biological sciences. Biol. Conserv. 2009, 142, 2282–2292. [Google Scholar] [CrossRef]
- Scherrer, D.; Mod, H.K.; Guisan, A. How to evaluate community predictions without thresholding? Methods Ecol. Evol. 2020, 11, 51–63. [Google Scholar] [CrossRef] [Green Version]
- Del Toro, I.; Ribbons, R.R.; Hayward, J.; Andersen, A.N. Are stacked species distribution models accurate at predicting multiple levels of diversity along a rainfall gradient? Austral. Ecol. 2019, 44, 105–113. [Google Scholar] [CrossRef] [Green Version]
- Justice, C.; Townshend, J.; Vermote, E.; Masuoka, E.; Wolfe, R.; Saleous, N.; Roy, D.; Morisette, J. An overview of MODIS land data processing and product status. Remote. Sens. Environ. 2002, 83, 3–15. [Google Scholar] [CrossRef]
- Huete, A.R. Vegetation indices, remote sensing and forest monitoring. Geogr. Compass 2012, 6, 513–532. [Google Scholar] [CrossRef]
- Jensen, J.R. Remote Sensing of the Environment: An Earth Resource Perspective; Pearson Prentice Hall: Upper Saddle River, NJ, USA, 2007. [Google Scholar]
- Didan, K.; Munoz, A.B.; Solano, R.; Huete, A. MODIS Vegetation Index User’s Guide (MOD13 Series). Version 3.00; Vegetation Index and Phenology Lab, University of Arizona: Tucson, AZ, USA, 2015. [Google Scholar]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Campillo, C.; García, M.; Daza, C.; Prieto, M. Study of a non-destructive method for estimating the leaf area index in vegetable crops using digital images. HortScience 2010, 45, 1459–1463. [Google Scholar] [CrossRef]
- Myneni, R.; Knyazikhin, Y.; Park, T. MCD15A2H MODIS/Terra + Aqua Leaf Area Index/FPAR 8-day L4 Global 500 m SIN Grid V006; NASA EOSDIS Land Processes DAAC: Sioux Falls, SD, USA, 2015. [CrossRef]
- Gardner, M.W.; Dorling, S.R. Artificial neural networks (the multilayer perceptron)—A review of applications in the at-mospheric sciences. Atmos. Environ. 1998, 32, 2627–2636. [Google Scholar] [CrossRef]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Commun. ACM 2017, 60, 84–90. [Google Scholar] [CrossRef]
- Tieleman, T.; Hinton, G. Lecture 6.5-rmsprop: Divide the gradient by a running average of its recent magnitude. COURSERA Neural Netw. Mach. Learn. 2012, 4, 26–31. [Google Scholar]
- Chollet, F. Deep Learning with R; Manning Publications: Shelter Island, NY, USA, 2018. [Google Scholar]
- Srivastava, N.; Hinton, G.; Krizhevsky, A.; Sutskever, I.; Salakhutdinov, R. Dropout: A simple way to prevent neural networks from overfitting. J. Mach. Learn. Res. 2014, 15, 1929–1958. [Google Scholar]
- Bhatnagar, S.; Gill, L.; Ghosh, B. Drone image segmentation using machine and deep learning for mapping raised bog vegetation communities. Remote Sens. 2020, 12, 2602. [Google Scholar] [CrossRef]
- Kim, Y.J.; Kim, H.-C.; Han, D.; Lee, S.; Im, J. Prediction of monthly Arctic sea ice concentrations using satellite and reanalysis data based on convolutional neural networks. Cryosphere 2020, 14, 1083–1104. [Google Scholar] [CrossRef] [Green Version]
- Korotcov, A.; Tkachenko, V.; Russo, D.P.; Ekins, S. Comparison of deep learning with multiple machine learning methods and metrics using diverse drug discovery data sets. Mol. Pharm. 2017, 14, 4462–4475. [Google Scholar] [CrossRef]
- Joharestani, M.Z.; Cao, C.; Ni, X.; Bashir, B.; Talebiesfandarani, S. PM2.5 prediction based on random forest, XGBoost, and deep learning using multisource remote sensing data. Atmosphere 2019, 10, 373. [Google Scholar] [CrossRef] [Green Version]
- Fernández-Delgado, M.; Cernadas, E.; Barro, S.; Amorim, D. Do we need hundreds of classifiers to solve real world classifi-cation problems? J. Mach. Learn. Res. 2014, 15, 3133–3181. [Google Scholar]
- Korea Forest Service. The 6th National Forest Inventory and Monitoring; Korea Forest Service: Daejeon, Korea, 2016. [Google Scholar]
- Choe, H.; Thorne, J.H.; Huber, P.R.; Lee, D.; Quinn, J.F. Assessing shortfalls and complementary conservation areas for national plant biodiversity in South Korea. PLoS ONE 2018, 13, e0190754. [Google Scholar] [CrossRef]
- Malhi, Y.; Franklin, J.; Seddon, N.; Solan, M.; Turner, M.G.; Field, C.B.; Knowlton, N. Climate change and ecosystems: Threats, opportunities and solutions. Philos. Trans. R. Soc. B Biol. Sci. 2020, 375, 20190104. [Google Scholar] [CrossRef] [Green Version]
- Pau, S.; Gillespie, T.W.; Wolkovich, E.M. Dissecting NDVI-species richness relationships in Hawaiian dry forests. J. Biogeogr. 2012, 39, 1678–1686. [Google Scholar] [CrossRef]
- Pausas, J.G.; Austin, M.P. Patterns of plant species richness in relation to different environments: An appraisal. J. Veg. Sci. 2001, 12, 153–166. [Google Scholar] [CrossRef]
- Kang, J.; Suh, M.; Kwak, C. Classification of land cover over the Korean peninsula using MODIS data. Atmosphere 2009, 19, 169–182. [Google Scholar]
- Camathias, L.; Küchler, M.; Stofer, S.; Baltensweiler, A.; Bergamini, A. High-resolution remote sensing data improves models of species richness. Appl. Veg. Sci. 2013, 16, 539–551. [Google Scholar] [CrossRef]
- Cord, A.F.; Klein, D.; Gernandt, D.S.; de la Rosa, J.A.P.; Dech, S. Remote sensing data can improve predictions of species richness by stacked species distribution models: A case study for Mexican pines. J. Biogeogr. 2014, 41, 736–748. [Google Scholar] [CrossRef]
- Choe, H.; Thorne, J.H.; Hijmans, R.; Seo, C. Integrating the Rabinowitz rarity framework with a National Plant Inventory in South Korea. Ecol. Evol. 2019, 9, 1353–1363. [Google Scholar] [CrossRef]
- Ince, T.; Kiranyaz, S.; Eren, L.; Askar, M.; Gabbouj, M. Real-time motor fault detection by 1-D convolutional neural networks. IEEE Trans. Ind. Electron. 2016, 63, 7067–7075. [Google Scholar] [CrossRef]
- Peng, D.; Liu, Z.; Wang, H.; Qin, Y.; Jia, L. A novel deeper one-dimensional CNN with residual learning for fault diagnosis of wheelset bearings in high-speed trains. IEEE Access 2019, 7, 10278–10293. [Google Scholar] [CrossRef]
- Sak, H.; Senior, A.; Beaufays, F. Long short-term memory based recurrent neural network architectures for large vocabulary speech recognition. arXiv 2014, arXiv:1402.1128. [Google Scholar]
- Hong, D.; Gao, L.; Yao, J.; Zhang, B.; Plaza, A.; Chanussot, J. Graph convolutional networks for hyperspectral image classification. IEEE Trans. Geosci. Remote Sens. 2020, 1–13. [Google Scholar] [CrossRef]
- Ledig, C.; Theis, L.; Huszar, F.; Caballero, J.; Cunningham, A.; Acosta, A.; Aitken, A.; Tejani, A.; Totz, J.; Wang, Z.; et al. Photo-realistic single image super-resolution using a generative adversarial network. In Proceedings of the 2017 IEEE Conference on Computer Vision and Pattern Recognition (CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5892–5900. [Google Scholar]
- GEO BON. Global Biodiversity Change Indicators. Version 1.2; Group on Earth Observations Biodiversity Observation Network Secretariat: Leipzig, Germany, 2015; 20p. [Google Scholar]








| Model Types | MAE (Mean Absolute Error) | RMSE (Root Mean Square Error) | Bias | Correlation |
|---|---|---|---|---|
| Random Forest | 61.1028 | 78.9512 | 0.5656 | 0.8843 |
| Deep Learning | 28.8105 | 38.5759 | 10.2055 | 0.9752 |
| DOY (Day of Year) | LAI (Leaf Area Index) | NDVI (Normalized Difference Vegetation Index) | |
|---|---|---|---|
| Spring | 97–177 | 46.46 (0.50) | 49.59 (0.61) |
| Summer | 193–257 | 38.51 (0.38) | 35.06 (0.38) |
| Fall | 273–321 | 28.19 (0.21) | 43.50 (0.49) |
| Winter | 1–81; 337–353 | 20.84 (0.01) | 40.83 (0.41) |
| Average | 32.64 (0.25) | 42.32 (0.47) |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Choe, H.; Chi, J.; Thorne, J.H. Mapping Potential Plant Species Richness over Large Areas with Deep Learning, MODIS, and Species Distribution Models. Remote Sens. 2021, 13, 2490. https://doi.org/10.3390/rs13132490
Choe H, Chi J, Thorne JH. Mapping Potential Plant Species Richness over Large Areas with Deep Learning, MODIS, and Species Distribution Models. Remote Sensing. 2021; 13(13):2490. https://doi.org/10.3390/rs13132490
Chicago/Turabian StyleChoe, Hyeyeong, Junhwa Chi, and James H. Thorne. 2021. "Mapping Potential Plant Species Richness over Large Areas with Deep Learning, MODIS, and Species Distribution Models" Remote Sensing 13, no. 13: 2490. https://doi.org/10.3390/rs13132490
APA StyleChoe, H., Chi, J., & Thorne, J. H. (2021). Mapping Potential Plant Species Richness over Large Areas with Deep Learning, MODIS, and Species Distribution Models. Remote Sensing, 13(13), 2490. https://doi.org/10.3390/rs13132490