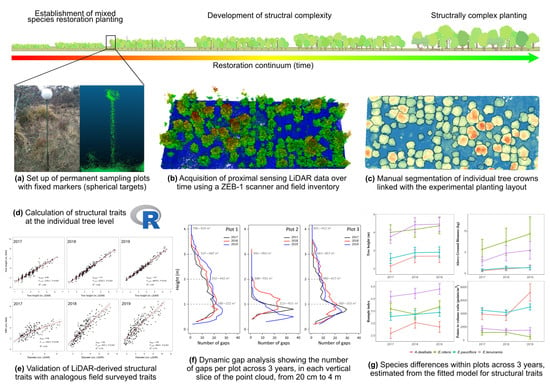
Handheld Laser Scanning Detects Spatiotemporal Differences in the Development of Structural Traits among Species in Restoration Plantings
Abstract
:1. Introduction
2. Materials and Methods
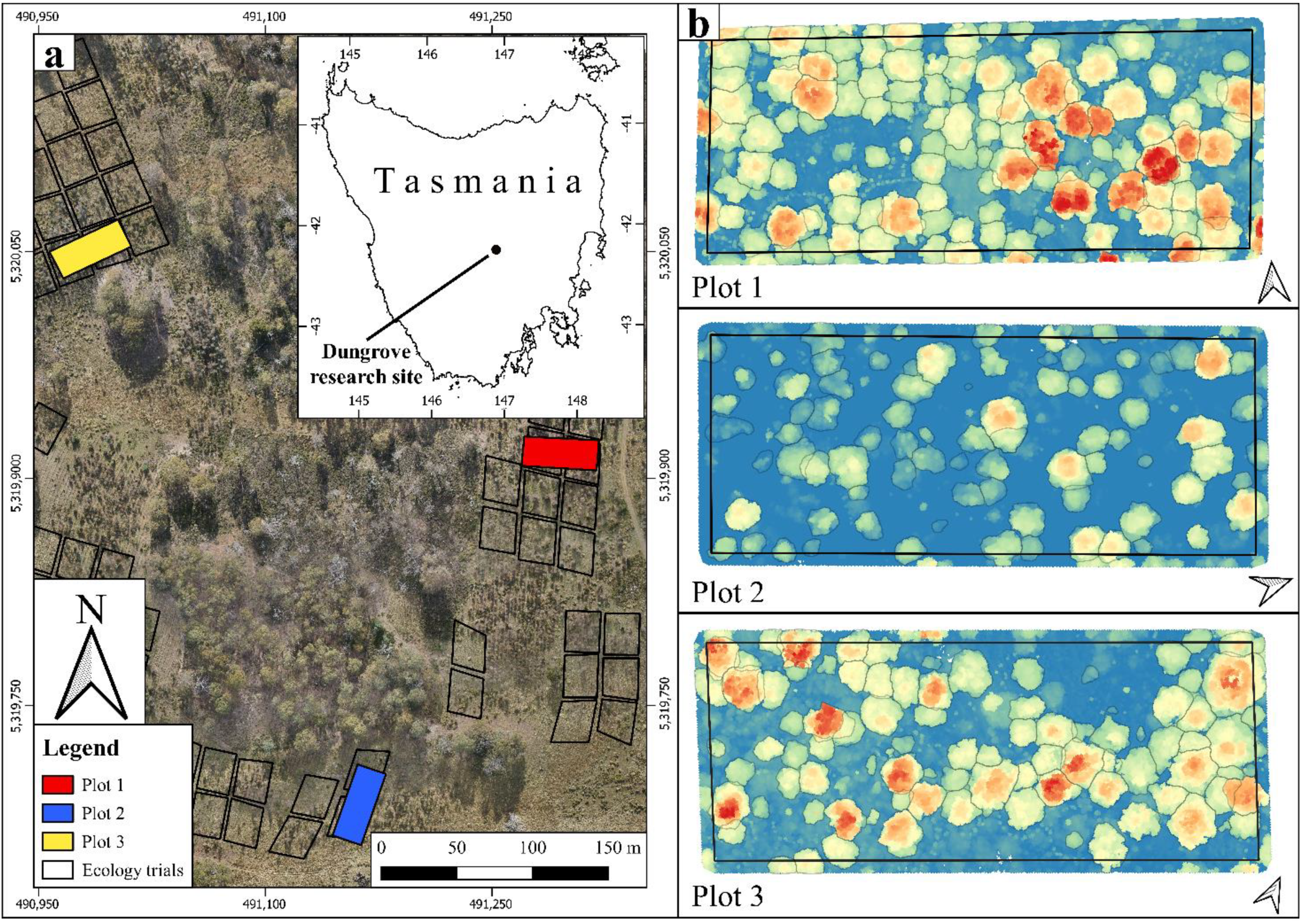
2.1. Study Site
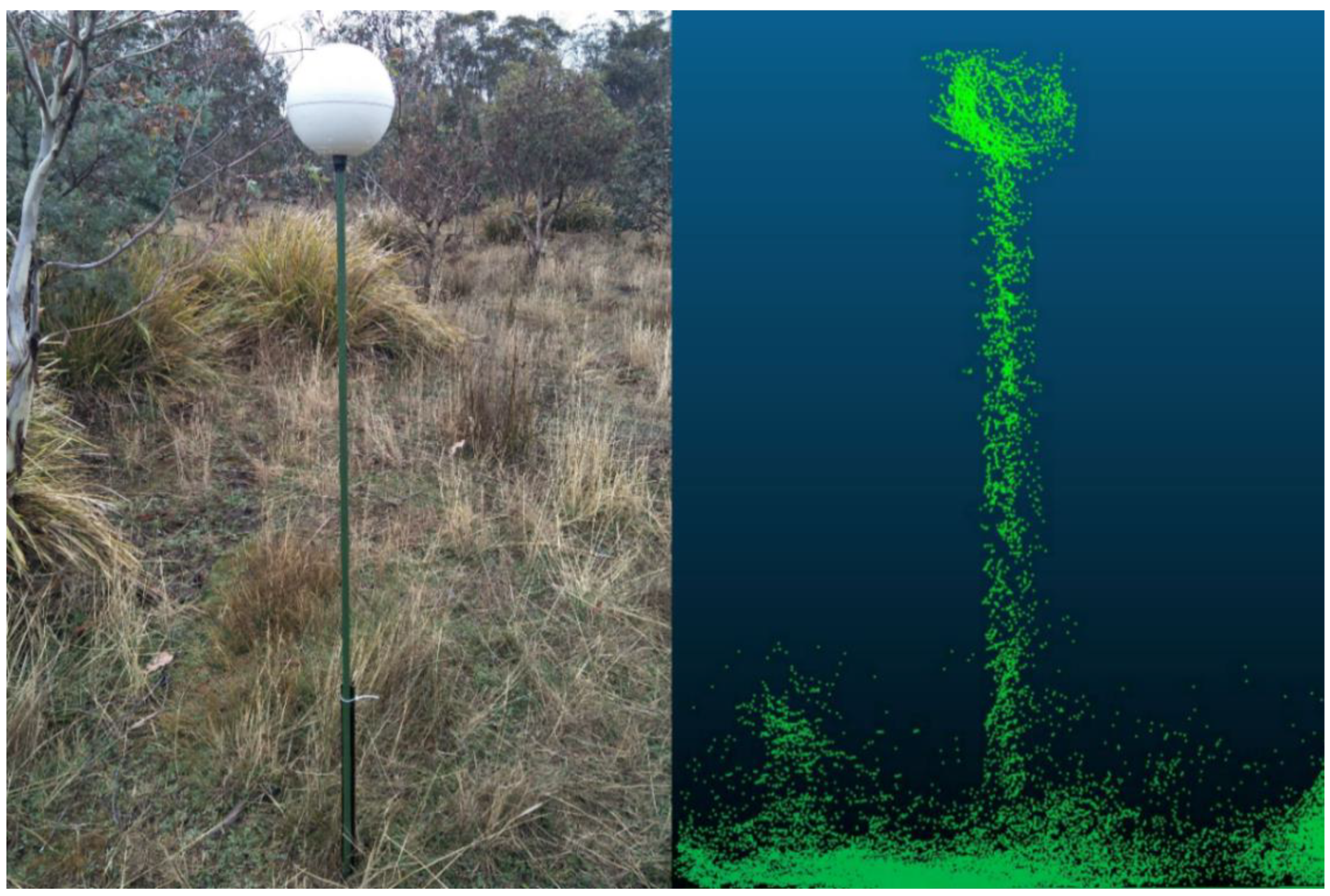
2.2. ZEB1 Data Acquisition and Co-Registration
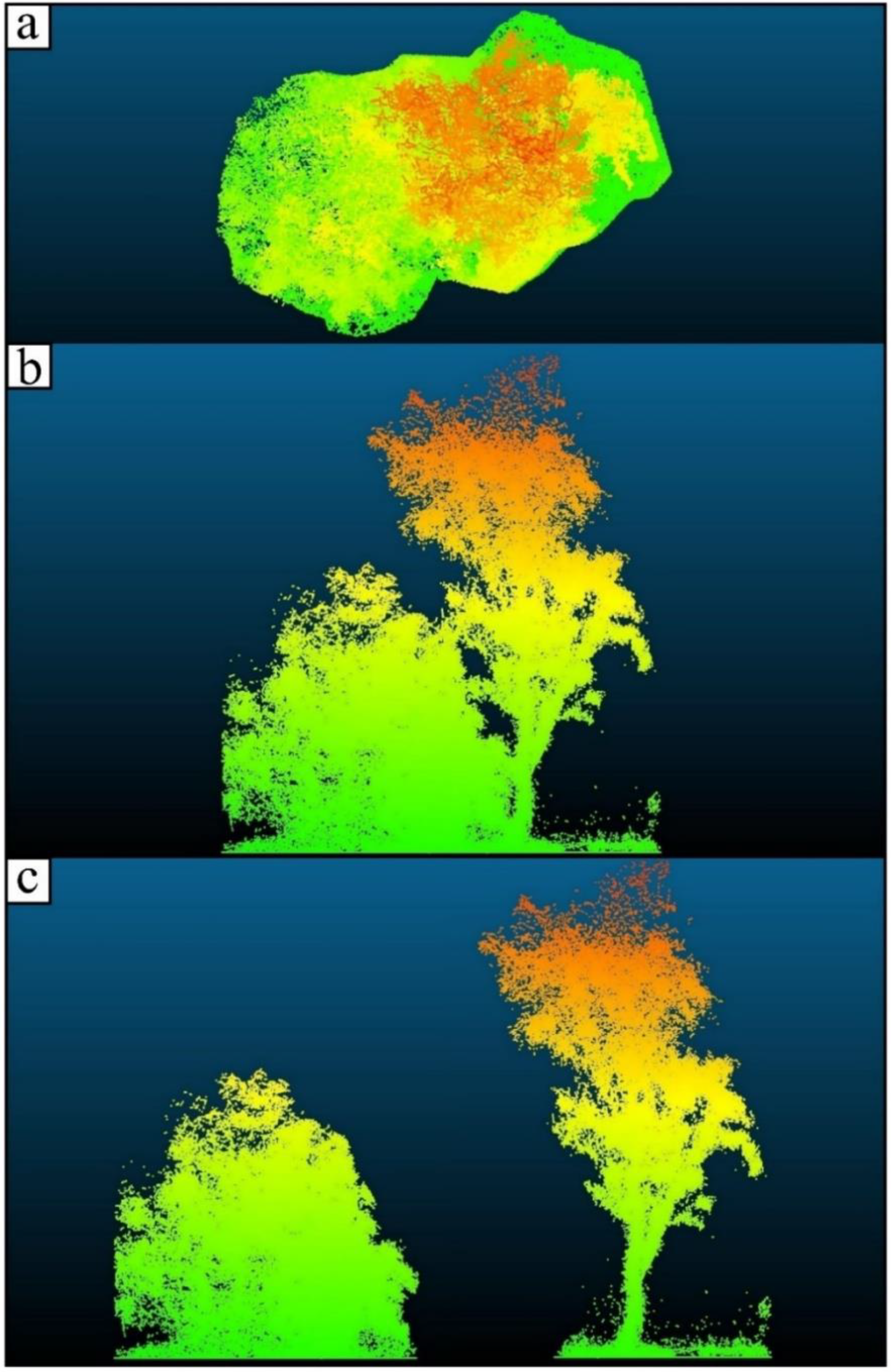
2.3. Individual Tree and Plot-Level Structural Trait Extraction
2.4. Ground-Based Measurements
2.5. Statistical Analysis
3. Results
4. Discussion
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Perring, M.P.; Standish, R.J.; Price, J.N.; Craig, M.D.; Erickson, T.E.; Ruthrof, K.X.; Whiteley, A.S.; Valentine, L.E.; Hobbs, R.J. Advances in restoration ecology: Rising to the challenges of the coming decades. Ecosphere 2015, 6, art131. [Google Scholar] [CrossRef] [Green Version]
- Maginel, C.J.; Knapp, B.O.; Kabrick, J.M.; Olson, E.K.; Muzika, R.-M. Floristic quality index for woodland ground flora restoration: Utility and effectiveness in a fire-managed landscape. Ecol. Indic. 2016, 67, 58–67. [Google Scholar] [CrossRef]
- Noss, R.F. Indicators for monitoring biodiversity: A hierarchical approach. Conserv. Biol. 1990, 4, 355–364. [Google Scholar] [CrossRef]
- McElhinny, C.; Gibbons, P.; Brack, C.; Bauhus, J. Forest and woodland stand structural complexity: Its definition and measurement. For. Ecol. Manag. 2005, 218, 1–24. [Google Scholar] [CrossRef]
- Kormann, U.; Scherber, C.; Tscharntke, T.; Klein, N.; Larbig, M.; Valente, J.J.; Hadley, A.S.; Betts, M.G. Corridors restore animal-mediated pollination in fragmented tropical forest landscapes. Proc. R. Soc. B Biol. Sci. 2016, 283, 20152347. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Chazdon, R.L. Beyond Deforestation: Restoring Forests and Ecosystem Services on Degraded Lands. Science 2008, 320, 1458–1460. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- MacArthur, R.H.; MacArthur, J.W. On bird species diversity. Ecology 1961, 42, 594–598. [Google Scholar] [CrossRef]
- Davies, A.B.; Ancrenaz, M.; Oram, F.; Asner, G.P. Canopy structure drives orangutan habitat selection in disturbed Bornean forests. Proc. Natl. Acad. Sci. USA 2017, 114, 8307–8312. [Google Scholar] [CrossRef] [Green Version]
- Camarretta, N.; Harrison, P.A.; Bailey, T.; Potts, B.; Lucieer, A.; Davidson, N.; Hunt, M. Monitoring forest structure to guide adaptive management of forest restoration: A review of remote sensing approaches. New For. 2019, 1–24. [Google Scholar] [CrossRef]
- Cordell, S.; Questad, E.J.; Asner, G.P.; Kinney, K.M.; Thaxton, J.M.; Uowolo, A.; Brooks, S.; Chynoweth, M.W. Remote sensing for restoration planning: How the big picture can inform stakeholders. Restor. Ecol. 2017, 25, S147–S154. [Google Scholar] [CrossRef]
- Camarretta, N.; A Harrison, P.; Lucieer, A.; M Potts, B.; Davidson, N.; Hunt, M. From Drones to Phenotype: Using UAV-LiDAR to Detect Species and Provenance Variation in Tree Productivity and Structure. Remote Sens. 2020, 12, 3184. [Google Scholar] [CrossRef]
- Vepakomma, U.; Cormier, D. Potential of multi-temporal UAV-borne lidar in assessing effectiveness of silvicultural treatments. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. ISPRS Arch. 2017, 42, 393–397. [Google Scholar] [CrossRef] [Green Version]
- Næsset, E.; Gobakken, T. Estimation of above- and below-ground biomass across regions of the boreal forest zone using airborne laser. Remote Sens. Environ. 2008, 112, 3079–3090. [Google Scholar] [CrossRef]
- Price, O.F.; Gordon, C.E. The potential for LiDAR technology to map fire fuel hazard over large areas of Australian forest. J. Environ. Manag. 2016, 181, 663–673. [Google Scholar] [CrossRef]
- Muir, J.; Phinn, S.; Eyre, T.; Scarth, P. Measuring plot scale woodland structure using terrestrial laser scanning. Remote Sens. Ecol. Conserv. 2018, 4, 320–338. [Google Scholar] [CrossRef] [Green Version]
- Marselis, S.M.; Yebra, M.; Jovanovic, T.; van Dijk, A. Deriving comprehensive forest structure information from mobile laser scanning observations using automated point cloud classification. Environ. Model. Softw. 2016, 82, 142–151. [Google Scholar] [CrossRef]
- Bauwens, S.; Bartholomeus, H.; Calders, K.; Lejeune, P. Forest inventory with terrestrial LiDAR: A comparison of static and hand-held mobile laser scanning. Forests 2016, 7, 127. [Google Scholar] [CrossRef] [Green Version]
- Ryding, J.; Williams, E.; Smith, M.; Eichhorn, M. Assessing handheld mobile laser scanners for forest surveys. Remote Sens. 2015, 7, 1095. [Google Scholar] [CrossRef] [Green Version]
- Jaakkola, A.; Hyyppä, J.; Yu, X.; Kukko, A.; Kaartinen, H.; Liang, X.; Hyyppä, H.; Wang, Y. Autonomous collection of forest field reference—The outlook and a first step with UAV laser scanning. Remote Sens. 2017, 9, 785. [Google Scholar] [CrossRef] [Green Version]
- Bailey, T.G.; Harrison, P.A.; Davidson, N.; Weller-Wong, A.; Tilyard, P.; Steane, D.; Vaillancourt, R.E.; Potts, B.M. Embedding genetics experiments in restoration to assist in plant choice for a degraded landscape with a changing climate. Ecol. Manag. Restor. 2021, 999, 999. [Google Scholar]
- Camarretta, N.; Harrison, P.A.; Bailey, T.; Davidson, N.; Lucieer, A.; Hunt, M.; Potts, B.M. Stability of species and provenance performance when translocated into different community assemblages. Restor. Ecol. 2020, 28, 447–458. [Google Scholar] [CrossRef]
- GEOSLAM. ZEB1 User Guide; v3.0.1.; GeoSLAM Ltd.: Nottingham, UK, 2015; v3.0.1. [Google Scholar]
- R Core Team. R: A Language and Environment for Statistical Computing. Available online: https://www.r-project.org (accessed on 9 January 2017).
- Jucker, T.; Caspersen, J.; Chave, J.; Antin, C.; Barbier, N.; Bongers, F.; Dalponte, M.; van Ewijk, K.Y.; Forrester, D.I.; Haeni, M.; et al. Allometric equations for integrating remote sensing imagery into forest monitoring programmes. Glob. Chang. Biol. 2017, 23, 177–190. [Google Scholar] [CrossRef] [PubMed]
- Silva, C.A.; Valbuena, R.; Pinagé, E.R.; Mohan, M.; de Almeida, D.R.A.; North Broadbent, E.; Jaafar, W.S.W.M.; de Almeida Papa, D.; Cardil, A.; Klauberg, C. ForestGapR: An r Package for forest gap analysis from canopy height models. Methods Ecol. Evol. 2019, 10, 1347–1356. [Google Scholar] [CrossRef] [Green Version]
- Wolfinger, R.D. Heterogeneous variance-covariance structures for repeated measures. J. Agric. Biol. Environ. Stat. 1996, 1, 205–230. [Google Scholar] [CrossRef]
- Pinheiro, J.C.; Bates, D.M. Unconstrained parametrizations for variance-covariance matrices. Stat. Comput. 1996, 6, 289–296. [Google Scholar] [CrossRef]
- Zuur, A.F.; Ieno, E.N. A protocol for conducting and presenting results of regression-type analyses. Methods Ecol. Evol. 2016, 7, 636–645. [Google Scholar] [CrossRef]
- Del Perugia, B.; Giannetti, F.; Chirici, G.; Travaglini, D. Influence of scan density on the estimation of single-tree attributes by hand-held mobile laser scanning. Forests 2019, 10, 277. [Google Scholar] [CrossRef] [Green Version]
- Tompalski, P.; Rakofsky, J.; Coops, N.C.; White, J.C.; Graham, A.N.V.; Rosychuk, K. Challenges of multi-temporal and multi-sensor forest growth analyses in a highly disturbed boreal mixedwood forests. Remote Sens. 2019, 11, 2102. [Google Scholar] [CrossRef] [Green Version]
- Hyyppä, E.; Yu, X.; Kaartinen, H.; Hakala, T.; Kukko, A.; Vastaranta, M.; Hyyppä, J. Comparison of backpack, handheld, under-canopy UAV, and above-canopy UAV laser scanning for field reference data collection in boreal forests. Remote Sens. 2020, 12, 3327. [Google Scholar] [CrossRef]
- Puletti, N.; Grotti, M.; Scotti, R. Evaluating the Eccentricities of Poplar Stem Profiles with Terrestrial Laser Scanning. Forests 2019, 10, 239. [Google Scholar] [CrossRef] [Green Version]
- Krisanski, S.; Taskhiri, M.S.; Turner, P. Enhancing methods for under-canopy unmanned aircraft system based photogrammetry in complex forests for tree diameter measurement. Remote Sens. 2020, 12, 1652. [Google Scholar] [CrossRef]
- Harikumar, A.; Bovolo, F.; Bruzzone, L. A local projection-based approach to individual tree detection and 3-d crown delineation in multistoried coniferous forests using high-density airborne LiDAR data. IEEE Trans. Geosci. Remote Sens. 2019, 57, 1168–1182. [Google Scholar] [CrossRef]
- Dalponte, M.; Frizzera, L.; Gianelle, D. How to map forest structure from aircraft, one tree at a time. Ecol. Evol. 2018, 8, 5611–5618. [Google Scholar] [CrossRef] [PubMed]
- Kaartinen, H.; Hyyppä, J.; Yu, X.; Vastaranta, M.; Hyyppä, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An international comparison of individual tree detection and extraction using airborne laser scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef] [Green Version]
- Millikan, P.H.K.; Silva, C.A.; Rodriguez, L.C.E.; Oliveira, T.M.; Carvalho, M.P.L.C.; Carvalho, S.P.C. Automated individual tree detection in Amazon tropical forest from airborne laser scanning data. CERNE 2019, 25, 273–282. [Google Scholar] [CrossRef]
- Hastings, J.H.; Ollinger, S.V.; Ouimette, A.P.; Sanders-DeMott, R.; Palace, M.W.; Ducey, M.J.; Sullivan, F.B.; Basler, D.; Orwig, D.A. Tree species traits determine the success of LiDAR-based crown mapping in a mixed temperate forest. Remote Sens. 2020, 12, 309. [Google Scholar] [CrossRef] [Green Version]
- Jaskierniak, D.; Lucieer, A.; Kuczera, G.; Turner, D.; Lane, P.N.J.; Benyon, R.G.; Haydon, S. Individual tree detection and crown delineation from Unmanned Aircraft System (UAS) LiDAR in structurally complex mixed species eucalypt forests. ISPRS J. Photogramm. Remote Sens. 2021, 171, 171–187. [Google Scholar] [CrossRef]
- Dalponte, M.; Coomes, D.A. Tree-centric mapping of forest carbon density from airborne laser scanning and hyperspectral data. Methods Ecol. Evol. 2016, 7, 1236–1245. [Google Scholar] [CrossRef] [Green Version]
- Hamilton, M.; Joyce, K.; Williams, D.; Dutkowski, G.; Potts, B. Achievements in forest tree improvement in Australia and New Zealand 9. Genetic improvement of Eucalyptus nitens in Australia. Aust. For. 2008, 71, 82–93. [Google Scholar] [CrossRef]
- Close, D.C.; Davidson, N.J.; Churchill, K.C.; Grosser, P. Evaluation of establishment techniques on Eucalyptus nitens and E. pauciflora in the Midlands of Tasmania. Ecol. Manag. Restor. 2005, 6, 149–151. [Google Scholar] [CrossRef]
- Close, D.C.; Davidson, N.J.; Churchill, K.C.; Corkrey, R. Establishment of native Eucalyptus pauciflora and exotic Eucalyptus nitens on former grazing land. New For. 2010, 40, 143–152. [Google Scholar] [CrossRef]
- Whitehead, D.; Beadle, C.L. Physiological regulation of productivity and water use in Eucalyptus: A review. For. Ecol. Manag. 2004, 193, 113–140. [Google Scholar] [CrossRef]
- White, D.A.; Crombie, D.S.; Kinal, J.; Battaglia, M.; McGrath, J.F.; Mendham, D.S.; Walker, S.N. Managing productivity and drought risk in Eucalyptus globulus plantations in south-western Australia. For. Ecol. Manag. 2009, 259, 33–44. [Google Scholar] [CrossRef]
- Wardlaw, T. A Climate Analysis of the Current and Potential Future Eucalyptus Nitens and E. globulus Plantation Estate on Tasmanian State Forest. Tasforests 2017, 19, 17–27. [Google Scholar]
- Tomlinson, K.W.; Sterck, F.J.; Bongers, F.; da Silva, D.A.; Barbosa, E.R.M.; Ward, D.; Bakker, F.T.; van Kaauwen, M.; Prins, H.H.T.; de Bie, S.; et al. Biomass partitioning and root morphology of savanna trees across a water gradient. J. Ecol. 2012, 100, 1113–1121. [Google Scholar] [CrossRef] [Green Version]
- Müller, J.; Bae, S.; Röder, J.; Chao, A.; Didham, R.K. Airborne LiDAR reveals context dependence in the effects of canopy architecture on arthropod diversity. For. Ecol. Manag. 2014, 312, 129–137. [Google Scholar] [CrossRef]
- Setiawan, N.N.; Vanhellemont, M.; Baeten, L.; Gobin, R.; De Smedt, P.; Proesmans, W.; Ampoorter, E.; Verheyen, K. Does neighbourhood tree diversity affect the crown arthropod community in saplings? Biodivers. Conserv. 2016, 25, 169–185. [Google Scholar] [CrossRef]
- Müller, J.; Stadler, J.; Brandl, R. Composition versus physiognomy of vegetation as predictors of bird assemblages: The role of lidar. Remote Sens. Environ. 2010, 114, 490–495. [Google Scholar] [CrossRef]
- Munro, N.T.; Fischer, J.; Barrett, G.; Wood, J.; Leavesley, A.; Lindenmayer, D.B. Bird’s response to revegetation of different structure and floristics-Are “restoration plantings” restoring bird communities? Restor. Ecol. 2011, 19, 223–235. [Google Scholar] [CrossRef]
- North, M.P.; Kane, J.T.; Kane, V.R.; Asner, G.P.; Berigan, W.; Churchill, D.J.; Conway, S.; Gutiérrez, R.J.; Jeronimo, S.; Keane, J.; et al. Cover of tall trees best predicts California spotted owl habitat. For. Ecol. Manag. 2017, 405, 166–178. [Google Scholar] [CrossRef]
- Froidevaux, J.S.P.; Zellweger, F.; Bollmann, K.; Jones, G.; Obrist, M.K. From field surveys to LiDAR: Shining a light on how bats respond to forest structure. Remote Sens. Environ. 2016, 175, 242–250. [Google Scholar] [CrossRef] [Green Version]
- Aitken, S.N.; Whitlock, M.C. Assisted gene flow to facilitate local adaptation to climate change. Annu. Rev. Ecol. Evol. Syst. 2013, 44, 367–388. [Google Scholar] [CrossRef]
- Prober, S.M.; Byrne, M.; McLean, E.H.; Steane, D.A.; Potts, B.M.; Vaillancourt, R.E.; Stock, W.D. Climate-adjusted provenancing: A strategy for climate-resilient ecological restoration. Front. Ecol. Evol. 2015, 3, 40–53. [Google Scholar] [CrossRef] [Green Version]
- Hanzelka, J.; Reif, J. Effects of vegetation structure on the diversity of breeding bird communities in forest stands of non-native black pine (Pinus nigra A.) and black locust (Robinia pseudoacacia L.) in the Czech Republic. For. Ecol. Manag. 2016, 379, 102–113. [Google Scholar] [CrossRef]
- Chen, S.; Liu, H.; Feng, Z.; Shen, C.; Chen, P. Applicability of personal laser scanning in forestry inventory. PLoS ONE 2019, 14, e211392. [Google Scholar] [CrossRef] [PubMed]
- Vatandaşlar, C.; Zeybek, M. Application of handheld laser scanning technology for forest inventory purposes in the NE Turkey. Turkish J. Agric. For. 2020, 229–242. [Google Scholar] [CrossRef]

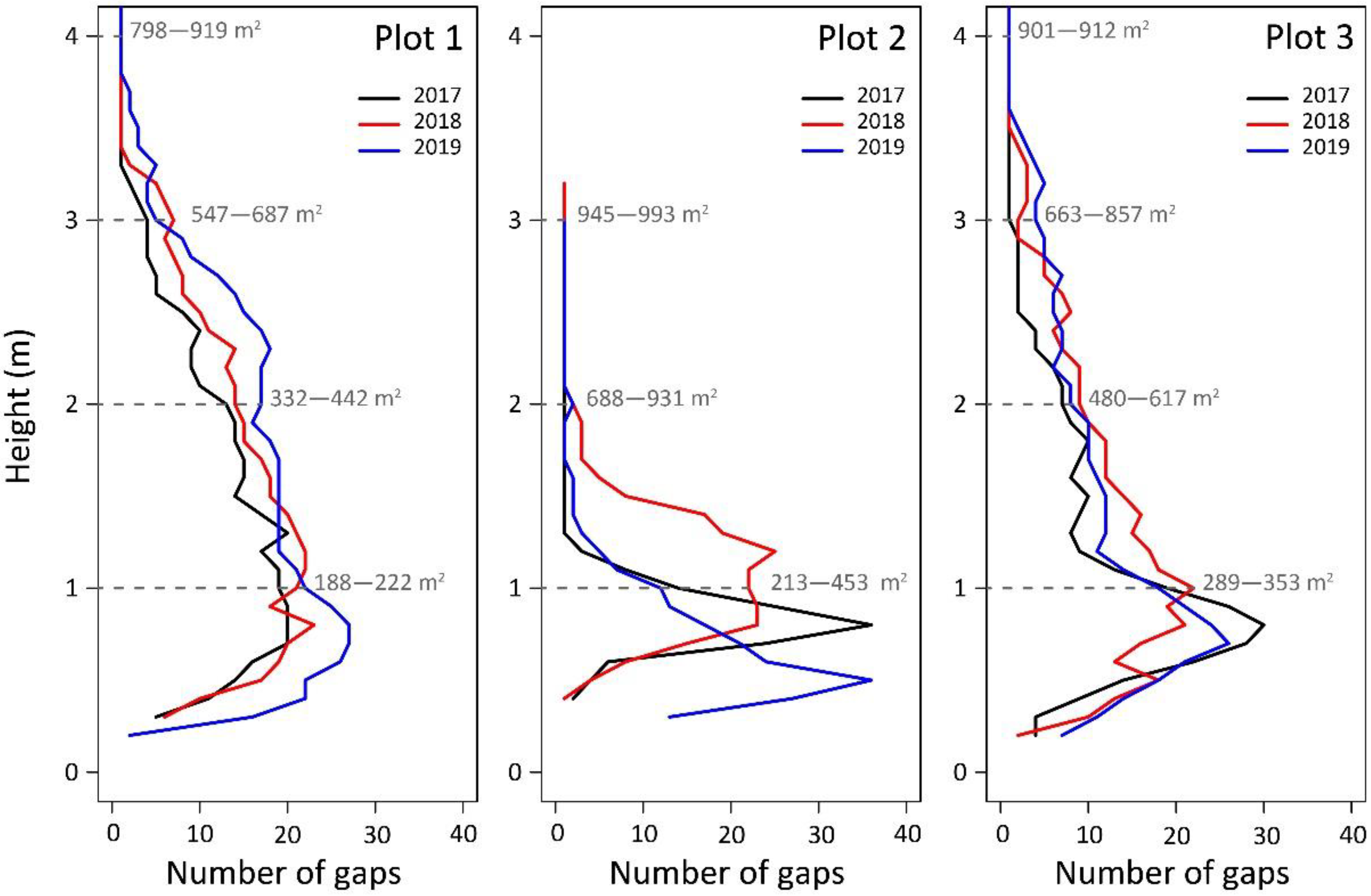
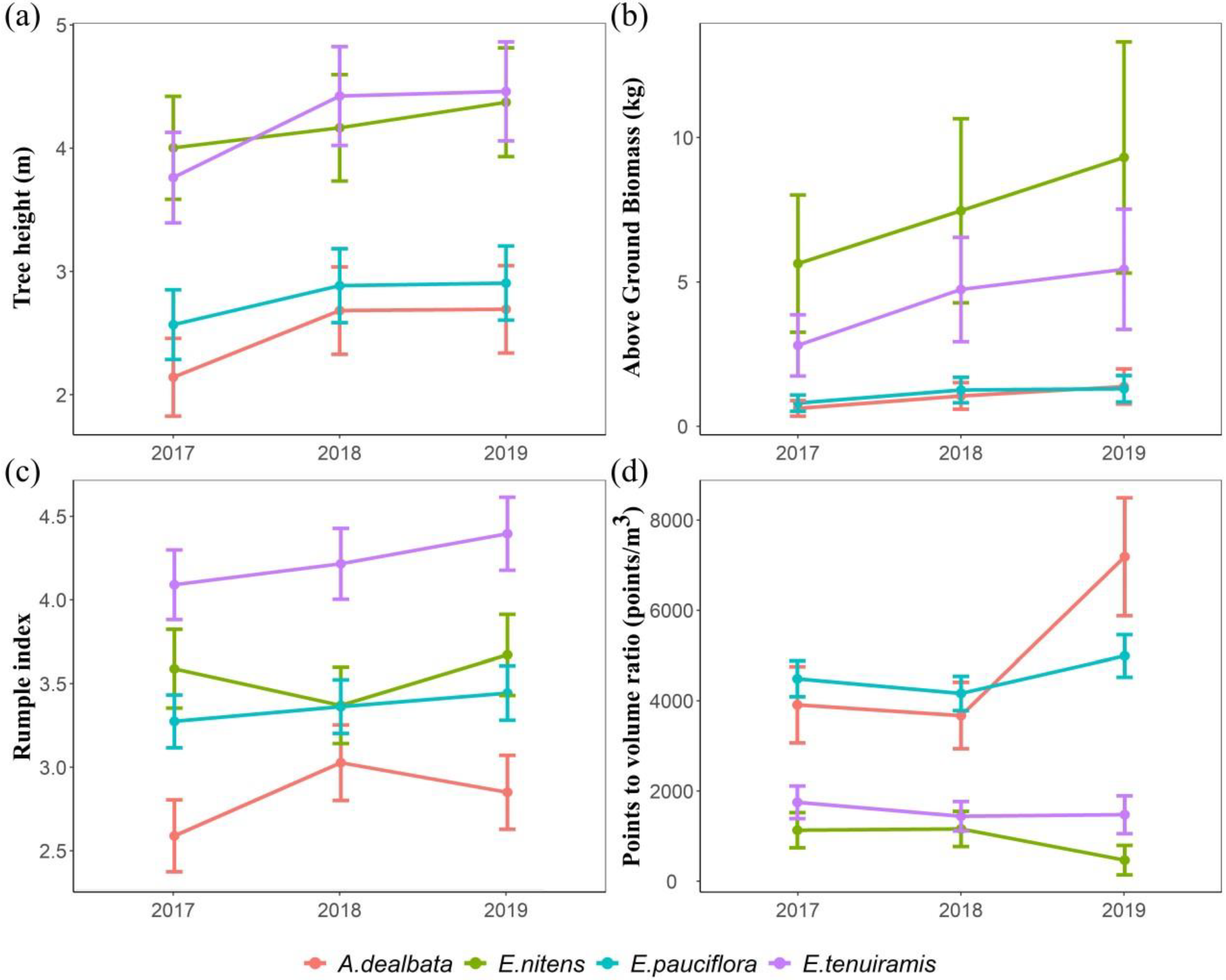
| Plot | Acquisition Date | Registration RMSE (m) | Points/m2 | Mean Height (m) | Max Height (m) | Standard Deviation (m) |
|---|---|---|---|---|---|---|
| 21 February 2017 | 0.14 a | 12,822.6 | 2.27 | 7.38 | 0.77 | |
| 1 | 15 May 2018 | 0.20 a | 15,522.2 | 2.34 | 8.35 | 0.84 |
| 2 April 2019 | 0.19 a | 14,543.2 | 2.24 | 8.93 | 0.82 | |
| 20 February 2017 | 0.11 | 6149.3 | 1.96 | 5.40 | 0.52 | |
| 2 | 15 May 2018 | 0.29 | 11,543.0 | 2.18 | 6.48 | 0.66 |
| 10 May 2019 | 0.20 | 14,159.6 | 2.07 | 6.42 | 0.65 | |
| 21 February 2017 | 0.15 | 7919.7 | 2.28 | 7.52 | 0.79 | |
| 3 | 15 May 2018 b | 0.13 a | 13,502.2 | 2.33 | 7.94 | 0.85 |
| 2 April 2019 | 0.16 | 8085.1 | 2.19 | 8.22 | 0.74 |
| Structural Traits | Description of LiDAR-Derived Traits |
|---|---|
| Height P99 (m) | 99th percentile of height within point cloud. |
| Crown skewness | Skewness of the height distribution within each tree (median − Q1)/(Q3 − median). |
| Height of widest cross-section (m) | Height of the crown at its widest cross-section. |
| Max crown diameter (m) | The widest cross-section of the crown in any given direction. |
| Crown volume (convex hull-m3) | Crown volume of a 3D convex hull calculated from the point cloud defined above crown insertion. It is calculated using the “convhulln” function of the R geometry package. |
| Crown surface area (m2) | The surface area of a 3D convex hull calculated using the point cloud defined above the canopy insertion point. |
| Crown projected area (m2) | The area of the projected polygon describing the crown ground cover. |
| Height to area ratio (m/m2) | The ratio of tree height to crown surface area. It represents the total height of the tree per unit of area. |
| Height to volume ratio (m/m3) | The ratio of tree height to crown volume. It represents the total height of the tree per unit of volume. |
| Points to area ratio (points/m2) | The ratio of number of points in the crown to crown surface area, representing a proxy for crown density. |
| Points to volume ratio (points/m3) | The ratio of number of points in the crown to crown volume, representing a proxy for crown density. |
| Above Ground Biomass (AGB-kg) | AGB estimated through the allometric Equation (1) developed by Jucker et al. [24], using Tree height and Max crown width. Measured as dry weight. |
| Diameter at breast height (cm) | Diameter at 1.3m, derived from a general allometric Equation (2) using total tree height and maximum crown width [24]. |
| Rumple index | Calculated as the ratio between crown surface area on ground surface area, this index reflects crown structural complexity. Calculated using the “rumple_index” function from the lidR package. |
| Area (3D:2D) | The ratio between crown volume calculated from the point cloud (i.e., convex hull) and the crown area obtained from the projected polygon. |
| Structural Traits | Transformation | Time (df = 2) | Species (df = 3) | Time Species (df = 6) | |||
|---|---|---|---|---|---|---|---|
| F | Pr | F | Pr | F | Pr | ||
| Height P99 (m) | sqrt | 18.9 | <0.001 | 32.4 | <0.001 | 6.0 | <0.001 |
| Crown skewness | ln | 2.5 | 0.082 | 9.8 | <0.001 | 1.3 | 0.250 |
| Height of widest cross-section (m) | 7.4 | <0.001 | 6.8 | <0.001 | 1.0 | 0.441 | |
| Max crown diameter (m) | 54.1 | <0.001 | 25.1 | <0.001 | 5.6 | <0.001 | |
| Crown volume (convex hull-m3) | sqrt | 15.9 | <0.001 | 31.4 | <0.001 | 5.7 | <0.001 |
| Crown surface area (m2) | sqrt | 18.0 | <0.001 | 27.6 | <0.001 | 4.3 | <0.001 |
| Crown projected area (m2) | sqrt | 18.1 | <0.001 | 27.1 | <0.001 | 7.7 | <0.001 |
| Height to area ratio (m/m2) | ln | 9.8 | <0.001 | 17.9 | <0.001 | 2.5 | 0.021 |
| Height to volume ratio (m/m3) | ln | 14.4 | <0.001 | 22.3 | <0.001 | 2.8 | 0.010 |
| Points to area ratio (points/m2) | sqrt | 5.3 | <0.001 | 16.0 | 0.005 | 4.4 | <0.001 |
| Points to volume ratio (points/m3) | sqrt | 1.5 | 0.226 | 25.4 | <0.001 | 2.4 | 0.027 |
| Above Ground Biomass (AGB-kg) | ln | 17.9 | <0.001 | 29.1 | <0.001 | 3.7 | 0.001 |
| Diameter at breast height (m) | sqrt | 15.6 | <0.001 | 32.5 | <0.001 | 6.0 | <0.001 |
| Rumple index | sqrt | 7.7 | <0.001 | 19.0 | <0.001 | 5.1 | <0.001 |
| Area (3D:2D) | 25.3 | <0.001 | 16.1 | <0.001 | 3.0 | 0.007 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Camarretta, N.; Harrison, P.A.; Lucieer, A.; Potts, B.M.; Davidson, N.; Hunt, M. Handheld Laser Scanning Detects Spatiotemporal Differences in the Development of Structural Traits among Species in Restoration Plantings. Remote Sens. 2021, 13, 1706. https://doi.org/10.3390/rs13091706
Camarretta N, Harrison PA, Lucieer A, Potts BM, Davidson N, Hunt M. Handheld Laser Scanning Detects Spatiotemporal Differences in the Development of Structural Traits among Species in Restoration Plantings. Remote Sensing. 2021; 13(9):1706. https://doi.org/10.3390/rs13091706
Chicago/Turabian StyleCamarretta, Nicolò, Peter A. Harrison, Arko Lucieer, Brad M. Potts, Neil Davidson, and Mark Hunt. 2021. "Handheld Laser Scanning Detects Spatiotemporal Differences in the Development of Structural Traits among Species in Restoration Plantings" Remote Sensing 13, no. 9: 1706. https://doi.org/10.3390/rs13091706
APA StyleCamarretta, N., Harrison, P. A., Lucieer, A., Potts, B. M., Davidson, N., & Hunt, M. (2021). Handheld Laser Scanning Detects Spatiotemporal Differences in the Development of Structural Traits among Species in Restoration Plantings. Remote Sensing, 13(9), 1706. https://doi.org/10.3390/rs13091706