Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging
Abstract
:1. Introduction
2. Materials
2.1. Study Area and Experimental Setup
2.2. Canopy Spectra Measurements
2.3. UAV-Based Hyperspectral Data Acquisition and Processing
3. Methods
3.1. Radiative Transfer Model
3.2. Look-Up Table Generation-Based SLC Model
- (1)
- Initialize the number of canopy simulations (n = 17,280) and the number of correlated variables with their normal distributions.
- (2)
- Generate the Latin Hypercube Samples (Z) with the size of n × 3, considering the number of canopy simulations (n) and correlated variables (3) that divide into samples (N), with the same probability of 1/N, and selecting one sampling value of these samples in each partition randomly [76].
- (3)
- Define the correlation matrices between three measured variables (M) and between the generated values of LHS (m), following the size of the correlated variables.
- (4)
- Calculate the non-singular lower triangular matrix (L) of the measured variables by using the Cholesky decomposition method (LU) for the correlation or covariance matrix (M), which satisfies:where M(3 × 3) is a Hermitian positive-definite matrix, which is decomposed into lower triangular (L) and upper triangular (LT) matrices; R is the correlation coefficient used in the correlation matrix M; i is the standard deviation of the variable xi; i,j is the covariance between xi and j; a, c, and d could be positive, negative or zero values of off-diagonal values; and b and e cannot be equal to zero.
- (5)
- Calculate the non-singular lower triangular matrix (Q) from the correlation matrix of the LHS realizations (m(3 × 3)):
- (6)
- Simulate the correlated random variate, which is based on transforming the realization matrix of LHS (Z) to a new matrix, denoted Z1, with size n × 3.
- (7)
- Convert the uniform correlated variables of Z1 to the normal distribution function, as defined before for three variables (Step 1). Then, each product of Z1 represents (Z1i) for LAI, (Z1j) for Cv, and (Z1k) for LCC.
| Parameter | Unit | Range | Distribution | Fixed Value | Reference | |
|---|---|---|---|---|---|---|
| Min | Max | |||||
| Leaf Parameter (PROSPECT-4) | ||||||
| Internal leaf structure, N | Unitless | 1 | 2.5 | Uniform | [78,79] | |
| Chlorophyll content, LCC | (g cm) | 40 | 90 | Gaussian | From field measurement | |
| = 65.36, = 9.38 | ||||||
| Water content, Cw | (cm) | 0.0317 | [5] | |||
| Dry matter content, Cm | (g cm) | 0.005 | [79] | |||
| Senescence material, Cs | Unitless | 0 | From field experience | |||
| Canopy Parameter (4SAIL2) | ||||||
| Leaf area index, LAI | (m m) | 0.05 | 7 | Gaussian | From field measurement | |
| = 2.85, = 1.17 | ||||||
| Leaf inclination distribution functions (LIDFa and LIDFb) | Unitless | LIDFa (0.66), LIDFb (−0.04) | [30] | |||
| Hotspot coefficient, hot | (m m) | 0.05 | [80] | |||
| Vertical crown cover, Cv | Unitless | 0.05 | 1 | Gaussian | [30] | |
| = 0.71, = 0.23 | ||||||
| Tree shape factor, zeta | Unitless | 1 | From field experience | |||
| Layer dissociation factor, D | Unitless | 1 | From field experience | |||
| Fraction of brown canopy area, fB | Unitless | 0 | From field experience | |||
| Soil parameters (Hapke) | ||||||
| Hapke_b | Unitless | 0.84 | [66,72] | |||
| Hapke_c | Unitless | 0.68 | - | |||
| Hapke_h | Unitless | 0.23 | - | |||
| Hapke_B0 | Unitless | 0.3 | - | |||
| Soil moisture, SM | Unitless | 15 | From field experience | |||
3.3. Retrieval Strategies
3.3.1. Physically Based Method
3.3.2. Hybrid Method
3.3.3. Statistical Method Using the Exposure Time
| Algorithm | Brief Description | References |
|---|---|---|
| Non-Linear Non-Parametric Regression | ||
| Random Forest (Tree Bagger) | RF is an extension over bagging trees. In particular, random selection is applied to construct different subsets of training data sets, as well as their features, to grow trees instead of using all features. This leads to a consensus prediction. | [51] |
| Conical Correlation Forest | CCF is a member of the decision tree ensemble family. Conical correlation analysis is used to find feature projections, wherein a voting rule combines the predictions of individual conical correlation trees to make a final prediction for unknown samples. | [85,86] |
| Gaussian Process Regression | GPR, as one of the kernel-based regression methods, is a stochastic probability distribution-based process of estimation by providing the mean and covariance. Consequently, the confidence interval around the mean predictions can be provided to assess the uncertainties. | [87] |
3.4. Model Validation
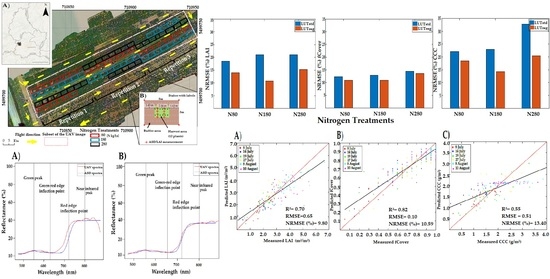
4. Results
4.1. Descriptive Statistics of Field Measurements
4.2. LUTreg- and LUTstd-Based Inversion
4.3. Hybrid Methods Based on ML
4.4. Retrieval Strategies under Illumination Variation and Crop Developments over Time
5. Discussion
5.1. The Use of Correlated Variables in LUTreg Inversion
5.2. Evaluation of the Retrieval Methods at Different Observation Dates
6. Conclusions
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| ASD FieldSpec3 | Analytical Spectral Devices |
| ARTMO | Automated Radiative Transfer Models Operator |
| C.V. | Coefficient of Variation |
| DART | Discrete Anisotropic Radiative Transfer |
| DN | Digital number |
| ENVI | Environment for Visualizing Images |
| exp | exposure time |
| ELC | empirical line calibration |
| FWHM | Full Width at Half Maximum |
| FOV | field of view |
| FV2000 | File Viewer software |
| GPS | Global Position System |
| INFORM | INvertible FOrest Reflectance Model |
| LIDFa | the average leaf slope |
| LIDFb | the distribution’s bimodality |
| LIDF | Leaf Inclination Distribution Function |
| NIR | Near-Infrared Range of spectrum |
| OSAVI | Optimized Soil-Adjusted Vegetation Index |
| PROSAIL | PROSPECT (leaf optical PROperties SPECTra model) and SAIL |
| (Scattering by Arbitrarily Inclined Leaves) | |
| PCA | Plant Canopy Analyzer |
| RTK-GPS | Real-Time Kinematic Global Positioning System |
| RGB | Red-Green-Blue |
| SLC | Soil–Leaf–Canopy |
| SCOPE | Soil Canopy Observation, Photochemistry, and Energy fluxes |
| SPAD | Soil Plant Analysis Development |
| SZA | solar zenith angle |
| SAA | solar azimuth angle |
| UAV | unmanned aerial vehicle |
| UTM31N | Universal Transverse Mercator Grid Zones 31North |
| VIS | Visible range of spectrum |
| VNIR | Visible and Near-Infrared Ranges |
| WDRVI | Wide Dynamic Range Vegetation Index |
| WGS84 | World Geodetic System 1984 |
Appendix A
| Observation Dates | 8 July | 14 July | 19 July | 27 July | 5 August | 10 August | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Day1 | Day2 | Day3 | Day4 | Day5 | Day6 | ||||||||
| Band No. | Band | R2 | RMSE | R2 | RMSE | R2 | RMSE | R2 | RMSE | R2 | RMSE | RMSE | |
| (nm) | |||||||||||||
| 1 | 474 | 0.99 | 0.0094 | 0.792 | 0.0422 | 0.9977 | 0.0044 | 0.9581 | 0.0183 | 0.982 | 0.0126 | 0.9634 | 0.018 |
| 2 | 483 | 0.9871 | 0.0106 | 0.8407 | 0.0367 | 0.999 | 0.003 | 0.9579 | 0.0181 | 0.9816 | 0.0127 | 0.9629 | 0.018 |
| 3 | 492 | 0.9842 | 0.0117 | 0.9236 | 0.0254 | 0.9997 | 0.0022 | 0.9633 | 0.0166 | 0.9814 | 0.0127 | 0.9619 | 0.018 |
| 4 | 501 | 0.9829 | 0.0121 | 0.9775 | 0.0142 | 0.9998 | 0.0029 | 0.9792 | 0.0121 | 0.9821 | 0.0125 | 0.9621 | 0.018 |
| 5 | 509 | 0.9838 | 0.0118 | 0.9914 | 0.009 | 0.9989 | 0.0051 | 0.9942 | 0.0067 | 0.9832 | 0.0122 | 0.9633 | 0.0179 |
| 6 | 518 | 0.9855 | 0.0112 | 0.9926 | 0.0081 | 0.9961 | 0.008 | 0.9883 | 0.0109 | 0.9842 | 0.0118 | 0.9649 | 0.0175 |
| 7 | 527 | 0.9868 | 0.0107 | 0.9925 | 0.0079 | 0.9905 | 0.0111 | 0.9588 | 0.0192 | 0.9848 | 0.0116 | 0.9661 | 0.0172 |
| 8 | 536 | 0.9871 | 0.0105 | 0.9921 | 0.008 | 0.9854 | 0.0134 | 0.9257 | 0.0252 | 0.9849 | 0.0114 | 0.9669 | 0.017 |
| 9 | 545 | 0.9869 | 0.0105 | 0.9913 | 0.0083 | 0.9835 | 0.014 | 0.9101 | 0.0273 | 0.9851 | 0.0113 | 0.9674 | 0.0168 |
| 10 | 554 | 0.9868 | 0.0106 | 0.99 | 0.0089 | 0.9853 | 0.013 | 0.9167 | 0.0257 | 0.9855 | 0.0111 | 0.9679 | 0.0166 |
| 11 | 569 | 0.9863 | 0.0106 | 0.9874 | 0.01 | 0.9897 | 0.0109 | 0.9322 | 0.0227 | 0.9856 | 0.0109 | 0.9682 | 0.0163 |
| 12 | 582 | 0.9857 | 0.0106 | 0.9838 | 0.0112 | 0.9935 | 0.009 | 0.9415 | 0.021 | 0.9855 | 0.0108 | 0.9681 | 0.0161 |
| 13 | 596 | 0.9854 | 0.0106 | 0.9785 | 0.0128 | 0.9963 | 0.0071 | 0.944 | 0.0205 | 0.9854 | 0.0107 | 0.9683 | 0.0158 |
| 14 | 610 | 0.9852 | 0.0106 | 0.9697 | 0.0152 | 0.9985 | 0.0048 | 0.9447 | 0.0203 | 0.9856 | 0.0106 | 0.969 | 0.0155 |
| 15 | 624 | 0.9845 | 0.0108 | 0.957 | 0.018 | 0.9995 | 0.0026 | 0.9444 | 0.0201 | 0.9855 | 0.0105 | 0.9697 | 0.0152 |
| 16 | 638 | 0.9836 | 0.011 | 0.9474 | 0.0197 | 0.9986 | 0.0026 | 0.9386 | 0.0208 | 0.9853 | 0.0105 | 0.9703 | 0.015 |
| 17 | 651 | 0.9811 | 0.0117 | 0.9461 | 0.0198 | 0.9979 | 0.0032 | 0.9447 | 0.0195 | 0.9855 | 0.0103 | 0.9711 | 0.0148 |
| 18 | 665 | 0.9762 | 0.0131 | 0.9518 | 0.0186 | 0.9986 | 0.003 | 0.967 | 0.0147 | 0.9867 | 0.0098 | 0.9725 | 0.0142 |
| 19 | 674 | 0.9706 | 0.0144 | 0.9619 | 0.0164 | 0.9996 | 0.0023 | 0.9838 | 0.01 | 0.9878 | 0.0093 | 0.9739 | 0.0137 |
| 20 | 682 | 0.968 | 0.0149 | 0.9723 | 0.0139 | 0.9996 | 0.0021 | 0.9889 | 0.0083 | 0.9875 | 0.0092 | 0.9741 | 0.0135 |
| 21 | 691 | 0.9829 | 0.038 | 0.9978 | 0.0136 | 0.9988 | 0.0093 | 0.998 | 0.0128 | 0.9976 | 0.0146 | 0.996 | 0.0187 |
| 22 | 699 | 0.9868 | 0.0335 | 0.9984 | 0.0117 | 0.9986 | 0.0096 | 0.9982 | 0.0115 | 0.9983 | 0.0119 | 0.9972 | 0.0157 |
| 23 | 708 | 0.9902 | 0.0287 | 0.9987 | 0.0105 | 0.9992 | 0.0077 | 0.9965 | 0.0171 | 0.9984 | 0.0117 | 0.9969 | 0.0163 |
| 24 | 716 | 0.9922 | 0.0248 | 0.9988 | 0.01 | 0.9996 | 0.0065 | 0.9877 | 0.0322 | 0.9984 | 0.0113 | 0.9965 | 0.0168 |
| 25 | 725 | 0.9882 | 0.0294 | 0.9988 | 0.0092 | 0.9998 | 0.0053 | 0.9338 | 0.0722 | 0.9982 | 0.0113 | 0.9956 | 0.0173 |
| 26 | 743 | 0.9845 | 0.0305 | 0.9981 | 0.0097 | 0.9995 | 0.0055 | 0.8446 | 0.1088 | 0.998 | 0.0111 | 0.9949 | 0.0156 |
| 27 | 761 | 0.9884 | 0.0237 | 0.998 | 0.0109 | 0.9995 | 0.0056 | 0.878 | 0.0937 | 0.9977 | 0.0111 | 0.9952 | 0.0135 |
| 28 | 779 | 0.987 | 0.0264 | 0.9988 | 0.0102 | 0.9996 | 0.0076 | 0.8976 | 0.087 | 0.9975 | 0.012 | 0.996 | 0.0131 |
| 29 | 797 | 0.9684 | 0.048 | 0.9988 | 0.0092 | 0.9997 | 0.0037 | 0.7959 | 0.1276 | 0.9981 | 0.0111 | 0.9961 | 0.0146 |
| 30 | 815 | 0.9747 | 0.0441 | 0.9984 | 0.0103 | 0.9999 | 0.0028 | 0.8049 | 0.126 | 0.9984 | 0.0102 | 0.9965 | 0.0144 |
| 31 | 825 | 0.9902 | 0.0275 | 0.9985 | 0.0102 | 0.9995 | 0.0073 | 0.9358 | 0.0726 | 0.9986 | 0.0094 | 0.9972 | 0.0129 |
| 32 | 835 | 0.9892 | 0.0293 | 0.9988 | 0.0092 | 0.9993 | 0.0081 | 0.9426 | 0.0683 | 0.9985 | 0.01 | 0.9972 | 0.0133 |
| 33 | 845 | 0.979 | 0.0406 | 0.9984 | 0.0105 | 0.9997 | 0.0061 | 0.862 | 0.1055 | 0.9978 | 0.0121 | 0.9965 | 0.0154 |
| 34 | 855 | 0.9851 | 0.0341 | 0.9984 | 0.0106 | 0.9996 | 0.0065 | 0.98 | 0.0417 | 0.9978 | 0.0124 | 0.9966 | 0.0156 |
| 35 | 865 | 0.992 | 0.0251 | 0.9987 | 0.0091 | 0.9997 | 0.0065 | 0.9906 | 0.0253 | 0.9981 | 0.0116 | 0.9973 | 0.014 |
| 36 | 875 | 0.9882 | 0.0309 | 0.9987 | 0.0089 | 0.9994 | 0.0082 | 0.986 | 0.0333 | 0.9986 | 0.0099 | 0.9979 | 0.0122 |
| 37 | 885 | 0.9882 | 0.0306 | 0.9978 | 0.0124 | 0.9985 | 0.0118 | 0.9921 | 0.0247 | 0.9985 | 0.0102 | 0.9976 | 0.0138 |
| 38 | 895 | 0.9887 | 0.0297 | 0.9975 | 0.0135 | 0.9997 | 0.0039 | 0.9687 | 0.0505 | 0.9973 | 0.0138 | 0.9971 | 0.0145 |
| 39 | 905 | 0.993 | 0.0229 | 0.9979 | 0.012 | 0.9989 | 0.0106 | 0.9428 | 0.0676 | 0.9982 | 0.0116 | 0.9966 | 0.0151 |
| 40 | 915 | 0.9943 | 0.0204 | 0.9987 | 0.01 | 0.9995 | 0.0058 | 0.968 | 0.0495 | 0.9986 | 0.0103 | 0.9972 | 0.0122 |
| Total mean | 0.985 | 0.0215 | 0.9777 | 0.014 | 0.9974 | 0.0066 | 0.9446 | 0.041 | 0.9914 | 0.0113 | 0.9821 | 0.0155 | |


| MLRAs | RF | CCF | GPR | |||
|---|---|---|---|---|---|---|
| Samples | CV | GV | CV | GV | CV | GV |
| 100 | 7.07 g | 15.96 i | 7.38 f | 12.58 e | 6.40 e | 9.80 a |
| 200 | 7.56 h | 14.20 g | 7.58 g | 12.21 b | 7.23 f | 10.99 d |
| 250 | 7.02 g | 14.54 h | 6.88 e | 13.97 g | 6.53 f | 12.99 d |
| 500 | 6.20 f | 13.13 f | 6.55 d | 12.56 d | 6.20 c | 10.33 b |
| 1000 | 5.18 e | 12.01 e | 6.52 d | 12.94 g | 6.20 c | 15.39 h |
| 2000 | 4.63 d | 11.46 d | 6.36 c | 12.30 c | 6.09 b | 13.28 f |
| 2500 | 4.48 c | 10.59 a | 6.53 d | 11.59 a | 6.29 d | 12.10 d |
| 3000 | 4.38 b | 11.42 d | 6.50 d | 12.85 f | 6.21 c | 14.06 g |
| 4000 | 4.06 a | 10.90 c | 6.15 a | 12.83 f | 5.86 a | 13.00 e |
| 5000 | 3.98 a | 10.70 b | 6.24 b | 12.94 g | 5.96 b | 10.83 c |
| MLRAs | RF | CCF | GPR | |||
|---|---|---|---|---|---|---|
| Samples | CV | GV | CV | GV | CV | GV |
| 100 | 2.70 g | 12.58 f | 1.49 d | 17.03 e | 3.72 e | 17.58 b |
| 200 | 2.23 f | 11.51 e | 2.58 f | 16.65 b | 1.45 d | 17.49 a |
| 250 | 2.26 f | 11.16 c | 2.24 f | 16.95 d | 1.37 c | 18.20 d |
| 500 | 1.97 e | 10.59 a | 1.90 e | 16.58 a | 1.41 d | 17.96 c |
| 1000 | 1.67 d | 10.83 a | 1.61 | 16.84 c | 1.32 b | 18.48 e |
| 2000 | 1.54 c | 11.22 d | 1.46 d | 17.33 g | 1.30 b | 21.29 i |
| 2500 | 1.51 c | 11.06 b | 1.42 c | 17.13 f | 1.30 b | 20.73 h |
| 3000 | 1.46 b | 12.07 | 1.40 c | 17.08 d | 1.29 b | 18.90 f |
| 4000 | 1.41 a | 11.01 b | 1.37 a | 17.29 h | 1.25 a | 20.37 g |
| 5000 | 1.42 b | 11.29 d | 1.38 b | 17.14 e | 1.29 b | 18.24 d |
| MLRAs | RF | CCF | GPR | |||
|---|---|---|---|---|---|---|
| Samples | CV | GV | CV | GV | CV | GV |
| 100 | 7.85 i | 30.45 i | 8.00 g | 14.20 f | 7.63 h | 18.21 b |
| 200 | 8.93 j | 26.85 h | 8.58 h | 15.94 g | 8.17 i | 29.90 i |
| 250 | 7.77 h | 27.84 g | 7.69 f | 13.85 e | 7.25 g | 30.47 j |
| 500 | 6.57 g | 23.17 f | 6.93 c | 14.91 g | 6.60 c | 17.26 a |
| 1000 | 5.78 f | 22.61 e | 7.13 | 13.40 a | 6.86 f | 20.99 h |
| 2000 | 5.01 e | 20.87 d | 6.99 d | 13.49 b | 6.69 d | 19.92 e |
| 2500 | 4.81 d | 15.06 a | 7.09 e | 17.13 i | 6.83 f | 20.73 f |
| 3000 | 4.66 c | 19.07 b | 7.06 e | 13.44 a | 6.75 e | 19.86 d |
| 4000 | 4.40 b | 20.05 d | 6.77 a | 13.66 d | 6.45 a | 20.91 g |
| 5000 | 4.32 a | 19.92 c | 6.89 b | 13.53 c | 6.59 b | 19.84 c |
References
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Long, H.; Yue, J.; Li, Z.; Yang, G.; Yang, X.; Fan, L. Estimation of crop growth parameters using UAV-based hyperspectral remote sensing data. Sensors 2020, 20, 1296. [Google Scholar] [CrossRef] [Green Version]
- Cilia, C.; Panigada, C.; Rossini, M.; Meroni, M.; Busetto, L.; Amaducci, S.; Boschetti, M.; Picchi, V.; Colombo, R. Nitrogen status assessment for variable rate fertilization in maize through hyperspectral imagery. Remote Sens. 2014, 6, 6549–6565. [Google Scholar] [CrossRef] [Green Version]
- Verger, A.; Martínez, B.; Camacho-de Coca, F.; García-Haro, F. Accuracy assessment of fraction of vegetation cover and leaf area index estimates from pragmatic methods in a cropland area. Int. J. Remote Sens. 2009, 30, 2685–2704. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33. [Google Scholar] [CrossRef] [Green Version]
- Clevers, J.G.; Kooistra, L. Using hyperspectral remote sensing data for retrieving canopy chlorophyll and nitrogen content. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2011, 5, 574–583. [Google Scholar] [CrossRef]
- Hoeppner, J.M.; Skidmore, A.K.; Darvishzadeh, R.; Heurich, M.; Chang, H.C.; Gara, T.W. Mapping canopy chlorophyll content in a temperate forest using airborne hyperspectral data. Remote Sens. 2020, 12, 3573. [Google Scholar] [CrossRef]
- Cheng, T.; Lu, N.; Wang, W.; Zhang, Q.; Li, D.; YAO, X.; Tian, Y.; Zhu, Y.; Cao, W.; Baret, F. Estimation of nitrogen nutrition status in winter wheat from unmanned aerial vehicle based multi-angular multispectral imagery. Front. Plant Sci. 2019, 10, 1601. [Google Scholar]
- Shang, J.; McNairn, H.; Schulthess, U.; Fernandes, R.; Storie, J. Estimation of crop ground cover and leaf area index (LAI) of wheat using RapidEye satellite data: Prelimary study. In Proceedings of the 2012 First International Conference on Agro-Geoinformatics (Agro-Geoinformatics); IEEE: New York, NY, USA, 2012; pp. 1–5. [Google Scholar]
- Zhu, W.; Sun, Z.; Huang, Y.; Lai, J.; Li, J.; Zhang, J.; Yang, B.; Li, B.; Li, S.; Zhu, K. Improving Field-Scale Wheat LAI Retrieval Based on UAV Remote-Sensing Observations and Optimized VI-LUTs. Remote Sens. 2019, 11, 2456. [Google Scholar] [CrossRef] [Green Version]
- Tian, J.; Wang, L.; Li, X.; Gong, H.; Shi, C.; Zhong, R.; Liu, X. Comparison of UAV and WorldView-2 imagery for mapping leaf area index of mangrove forest. Int. J. Appl. Earth Obs. Geoinf. 2017, 61, 22–31. [Google Scholar] [CrossRef]
- Stuart, M.B.; McGonigle, A.J.S.; Willmott, J.R. Hyperspectral Imaging in Environmental Monitoring: A Review of Recent Developments and Technological Advances in Compact Field Deployable Systems. Sensors 2019, 19, 3071. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kanning, M.; Kühling, I.; Trautz, D.; Jarmer, T. High-resolution UAV-based hyperspectral imagery for LAI and chlorophyll estimations from wheat for yield prediction. Remote Sens. 2018, 10, 2000. [Google Scholar] [CrossRef] [Green Version]
- Tian, M.; Ban, S.; Chang, Q.; You, M.; Luo, D.; Wang, L.; Wang, S. Use of hyperspectral images from UAV-based imaging spectroradiometer to estimate cotton leaf area index. Trans. Chin. Soc. Agric. Eng. 2016, 32, 102–108. [Google Scholar]
- Yue, J.; Feng, H.; Jin, X.; Yuan, H.; Li, Z.; Zhou, C.; Yang, G.; Tian, Q. A comparison of crop parameters estimation using images from UAV-mounted snapshot hyperspectral sensor and high-definition digital camera. Remote Sens. 2018, 10, 1138. [Google Scholar] [CrossRef] [Green Version]
- Kalisperakis, I.; Stentoumis, C.; Grammatikopoulos, L.; Karantzalos, K. Leaf area index estimation in vineyards from UAV hyperspectral data, 2D image mosaics and 3D canopy surface models. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2015, 40, 299. [Google Scholar] [CrossRef] [Green Version]
- Roosjen, P.P.J.; Brede, B.; Suomalainen, J.M.; Bartholomeus, H.M.; Kooistra, L.; Clevers, J.G.P.W. Improved estimation of leaf area index and leaf chlorophyll content of a potato crop using multi-angle spectral data-potential of unmanned aerial vehicle imagery. Int. J. Appl. Earth Obs. Geoinf. 2018, 66, 14–26. [Google Scholar] [CrossRef]
- Duan, S.B.; Li, Z.L.; Wu, H.; Tang, B.H.; Ma, L.; Zhao, E.; Li, C. Inversion of the PROSAIL model to estimate leaf area index of maize, potato, and sunflower fields from unmanned aerial vehicle hyperspectral data. Int. J. Appl. Earth Obs. Geoinf. 2014, 26, 12–20. [Google Scholar] [CrossRef]
- Riihimäki, H.; Luoto, M.; Heiskanen, J. Estimating fractional cover of tundra vegetation at multiple scales using unmanned aerial systems and optical satellite data. Remote Sens. Environ. 2019, 224, 119–132. [Google Scholar] [CrossRef]
- Sankey, T.T.; McVay, J.; Swetnam, T.L.; McClaran, M.P.; Heilman, P.; Nichols, M. UAV hyperspectral and lidar data and their fusion for arid and semi-arid land vegetation monitoring. Remote Sens. Ecol. Conserv. 2018, 4, 20–33. [Google Scholar] [CrossRef]
- Bian, J.; Li, A.; Zhang, Z.; Zhao, W.; Lei, G.; Yin, G.; Jin, H.; Tan, J.; Huang, C. Monitoring fractional green vegetation cover dynamics over a seasonally inundated alpine wetland using dense time series HJ-1A/B constellation images and an adaptive endmember selection LSMM model. Remote Sens. Environ. 2017, 197, 98–114. [Google Scholar] [CrossRef]
- Paul, S.; Poliyapram, V.; İmamoğlu, N.; Uto, K.; Nakamura, R.; Kumar, D.N. Canopy Averaged Chlorophyll Content Prediction of Pear Trees Using Convolutional Autoencoder on Hyperspectral Data. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2020, 13, 1426–1437. [Google Scholar] [CrossRef]
- Li, D.; Zheng, H.; Xu, X.; Lu, N.; Yao, X.; Jiang, J.; Wang, X.; Tian, Y.; Zhu, Y.; Cao, W.; et al. BRDF effect on the estimation of canopy chlorophyll content in paddy rice from UAV-based hyperspectral imagery. In Proceedings of the IGARSS 2018-2018 IEEE International Geoscience and Remote Sensing Symposium; IEEE: New York, NY, USA, 2018; pp. 6464–6467. [Google Scholar]
- Vanbrabant, Y.; Tits, L.; Delalieux, S.; Pauly, K.; Verjans, W.; Somers, B. Multitemporal chlorophyll mapping in pome fruit orchards from remotely piloted aircraft systems. Remote Sens. 2019, 11, 1468. [Google Scholar] [CrossRef] [Green Version]
- Gewali, U.B.; Monteiro, S.T.; Saber, E. Gaussian Processes for Vegetation Parameter Estimation from Hyperspectral Data with Limited Ground Truth. Remote Sens. 2019, 11, 1614. [Google Scholar] [CrossRef] [Green Version]
- Combal, B.; Baret, F.; Weiss, M.; Trubuil, A.; Macé, D.; Pragnère, A.; Myneni, R.; Knyazikhin, Y.; Wang, L. Retrieval of canopy biophysical variables from bidirectional reflectance: Using prior information to solve the ill-posed inverse problem. Remote Sens. Environ. 2003, 84, 1–15. [Google Scholar] [CrossRef]
- Atzberger, C. Object-based retrieval of biophysical canopy variables using artificial neural nets and radiative transfer models. Remote Sens. Environ. 2004, 93, 53–67. [Google Scholar] [CrossRef]
- Atzberger, C.; Richter, K. Spatially constrained inversion of radiative transfer models for improved LAI mapping from future Sentinel-2 imagery. Remote Sens. Environ. 2012, 120, 208–218. [Google Scholar] [CrossRef]
- Zurita-Milla, R.; Laurent, V.; van Gijsel, J. Visualizing the ill-posedness of the inversion of a canopy radiative transfer model: A case study for Sentinel-2. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 7–18. [Google Scholar] [CrossRef]
- Verrelst, J.; Vicent, J.; Rivera-Caicedo, J.P.; Lumbierres, M.; Morcillo-Pallarés, P.; Moreno, J. Global sensitivity analysis of leaf-canopy-atmosphere RTMs: Implications for biophysical variables retrieval from top-of-atmosphere radiance data. Remote Sens. 2019, 11, 1923. [Google Scholar] [CrossRef] [Green Version]
- Abdelbaki, A.; Schlerf, M.; Verhoef, W.; Udelhoven, T. Introduction of Variable Correlation for the Improved Retrieval of Crop Traits Using Canopy Reflectance Model Inversion. Remote Sens. 2019, 11, 2681. [Google Scholar] [CrossRef] [Green Version]
- Quan, X.; He, B.; Li, X. A Bayesian Network-Based Method to Alleviate the Ill-Posed Inverse Problem: A Case Study on Leaf Area Index and Canopy Water Content Retrieval. IEEE Trans. Geosci. Remote Sens. 2015, 53, 6507–6517. [Google Scholar] [CrossRef]
- Verrelst, J.; Malenovský, Z.; Van der Tol, C.; Camps-Valls, G.; Gastellu-Etchegorry, J.P.; Lewis, P.; North, P.; Moreno, J. Quantifying vegetation biophysical variables from imaging spectroscopy data: A review on retrieval methods. Surv. Geophys. 2019, 40, 589–629. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Camps-Valls, G.; Muñoz-Marí, J.; Rivera, J.P.; Veroustraete, F.; Clevers, J.G.P.W.; Moreno, J. Optical remote sensing and the retrieval of terrestrial vegetation bio-geophysical properties-A review. ISPRS J. Photogramm. Remote Sens. 2015, 108, 273–290. [Google Scholar] [CrossRef]
- Caicedo, J.P.R.; Verrelst, J.; Muñoz-Marí, J.; Moreno, J.; Camps-Valls, G. Toward a semiautomatic machine learning retrieval of biophysical parameters. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 1249–1259. [Google Scholar] [CrossRef]
- Belda, S.; Pipia, L.; Morcillo-Pallarés, P.; Verrelst, J. Optimizing Gaussian Process Regression for Image Time Series Gap-Filling and Crop Monitoring. Agronomy 2020, 10, 618. [Google Scholar] [CrossRef]
- Atzberger, C.; Darvishzadeh, R.; Immitzer, M.; Schlerf, M.; Skidmore, A.; le Maire, G. Comparative analysis of different retrieval methods for mapping grassland leaf area index using airborne imaging spectroscopy. Int. J. Appl. Earth Obs. Geoinf. 2015, 43, 19–31. [Google Scholar] [CrossRef] [Green Version]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; Heurich, M. Machine learning methods’ performance in radiative transfer model inversion to retrieve plant traits from Sentinel-2 data of a mixed mountain forest. Int. J. Digit. Earth 2020, 14, 106–120. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Skidmore, A.; Schlerf, M.; Atzberger, C.; Corsi, F.; Cho, M. LAI and chlorophyll estimation for a heterogeneous grassland using hyperspectral measurements. ISPRS J. Photogramm. Remote Sens. 2008, 63, 409–426. [Google Scholar] [CrossRef]
- Liang, L.; Di, L.; Zhang, L.; Deng, M.; Qin, Z.; Zhao, S.; Lin, H. Estimation of crop LAI using hyperspectral vegetation indices and a hybrid inversion method. Remote Sens. Environ. 2015, 165, 123–134. [Google Scholar] [CrossRef]
- Upreti, D.; Huang, W.; Kong, W.; Pascucci, S.; Pignatti, S.; Zhou, X.; Ye, H.; Casa, R. A comparison of hybrid machine learning algorithms for the retrieval of wheat biophysical variables from sentinel-2. Remote Sens. 2019, 11, 481. [Google Scholar] [CrossRef] [Green Version]
- De Grave, C.; Verrelst, J.; Morcillo-Pallarés, P.; Pipia, L.; Rivera-Caicedo, J.P.; Amin, E.; Belda, S.; Moreno, J. Quantifying vegetation biophysical variables from the Sentinel-3/FLEX tandem mission: Evaluation of the synergy of OLCI and FLORIS data sources. Remote Sens. Environ. 2020, 251, 112101. [Google Scholar] [CrossRef]
- Zheng, H.; Li, W.; Jiang, J.; Liu, Y.; Cheng, T.; Tian, Y.; Zhu, Y.; Cao, W.; Zhang, Y.; Yao, X. A comparative assessment of different modeling algorithms for estimating leaf nitrogen content in winter wheat using multispectral images from an unmanned aerial vehicle. Remote Sens. 2018, 10, 2026. [Google Scholar] [CrossRef] [Green Version]
- Luo, S.; He, Y.; Li, Q.; Jiao, W.; Zhu, Y.; Zhao, X. Nondestructive estimation of potato yield using relative variables derived from multi-period LAI and hyperspectral data based on weighted growth stage. Plant Methods 2020, 16, 1–14. [Google Scholar] [CrossRef] [PubMed]
- Li, S.; Yuan, F.; Ata-UI-Karim, S.T.; Zheng, H.; Cheng, T.; Liu, X.; Tian, Y.; Zhu, Y.; Cao, W.; Cao, Q. Combining color indices and textures of UAV-based digital imagery for rice LAI estimation. Remote Sens. 2019, 11, 1763. [Google Scholar] [CrossRef] [Green Version]
- Verrelst, J.; Dethier, S.; Rivera, J.P.; Munoz-Mari, J.; Camps-Valls, G.; Moreno, J. Active Learning Methods for Efficient Hybrid Biophysical Variable Retrieval. IEEE Geosci. Remote Sens. Lett. 2016, 13, 1012–1016. [Google Scholar] [CrossRef]
- Verrelst, J.; Rivera, J.P.; Gitelson, A.; Delegido, J.; Moreno, J.; Camps-Valls, G. Spectral band selection for vegetation properties retrieval using Gaussian processes regression. Int. J. Appl. Earth Obs. Geoinf. 2016, 52, 554–567. [Google Scholar] [CrossRef]
- Pasolli, L.; Melgani, F.; Blanzieri, E. Gaussian Process Regression for Estimating Chlorophyll Concentration in Subsurface Waters From Remote Sensing Data. IEEE Geosci. Remote Sens. Lett. 2010, 7, 464–468. [Google Scholar] [CrossRef]
- Lu, B.; He, Y. Evaluating empirical regression, machine learning, and radiative transfer modelling for estimating vegetation chlorophyll content using bi-seasonal hyperspectral images. Remote Sens. 2019, 11, 1979. [Google Scholar] [CrossRef] [Green Version]
- Liu, D.; Yang, L.; Jia, K.; Liang, S.; Xiao, Z.; Wei, X.; Yao, Y.; Xia, M.; Li, Y. Global fractional vegetation cover estimation algorithm for VIIRS reflectance data based on machine learning methods. Remote Sens. 2018, 10, 1648. [Google Scholar] [CrossRef] [Green Version]
- Danner, M.; Berger, K.; Wocher, M.; Mauser, W.; Hank, T. Efficient RTM-based training of machine learning regression algorithms to quantify biophysical & biochemical traits of agricultural crops. ISPRS J. Photogramm. Remote Sens. 2021, 173, 278–296. [Google Scholar]
- Breiman, L. Random forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Yang, J.; Liu, X.; Du, L.; Shi, S.; Sun, J.; Chen, B. Estimation of Multi-Species Leaf Area Index Based on Chinese GF-1 Satellite Data Using Look-Up Table and Gaussian Process Regression Methods. Sensors 2020, 20, 2460. [Google Scholar] [CrossRef]
- Xie, R.; Darvishzadeh, R.; Skidmore, A.K.; Heurich, M.; Holzwarth, S.; Gara, T.W.; Reusen, I. Mapping leaf area index in a mixed temperate forest using Fenix airborne hyperspectral data and Gaussian processes regression. Int. J. Appl. Earth Obs. Geoinf. 2021, 95, 102242. [Google Scholar] [CrossRef]
- Pourshamsi, M.; Xia, J.; Yokoya, N.; Garcia, M.; Lavalle, M.; Pottier, E.; Balzter, H. Tropical forest canopy height estimation from combined polarimetric SAR and LiDAR using machine-learning. ISPRS J. Photogramm. Remote Sens. 2021, 172, 79–94. [Google Scholar] [CrossRef]
- Colkesen, I.; Kavzoglu, T. Ensemble-based canonical correlation forest (CCF) for land use and land cover classification using sentinel-2 and Landsat OLI imagery. Remote Sens. Lett. 2017, 8, 1082–1091. [Google Scholar] [CrossRef]
- Näsi, R.; Viljanen, N.; Kaivosoja, J.; Alhonoja, K.; Hakala, T.; Markelin, L.; Honkavaara, E. Estimating biomass and nitrogen amount of barley and grass using UAV and aircraft based spectral and photogrammetric 3D features. Remote Sens. 2018, 10, 1082. [Google Scholar] [CrossRef] [Green Version]
- Nevalainen, O.; Honkavaara, E.; Tuominen, S.; Viljanen, N.; Hakala, T.; Yu, X.; Hyyppä, J.; Saari, H.; Pölönen, I.; Imai, N.N.; et al. Individual Tree Detection and Classification with UAV-Based Photogrammetric Point Clouds and Hyperspectral Imaging. Remote Sens. 2017, 9, 185. [Google Scholar] [CrossRef] [Green Version]
- Stow, D.; Nichol, C.J.; Wade, T.; Assmann, J.J.; Simpson, G.; Helfter, C. Illumination geometry and flying height influence surface reflectance and NDVI derived from multispectral UAS imagery. Drones 2019, 3, 55. [Google Scholar] [CrossRef] [Green Version]
- Assmann, J.J.; Kerby, J.T.; Cunliffe, A.M.; Myers-Smith, I.H. Vegetation monitoring using multispectral sensors-Best practices and lessons learned from high latitudes. J. Unmanned Veh. Syst. 2018, 7, 54–75. [Google Scholar] [CrossRef] [Green Version]
- Wendel, A.; Underwood, J. Illumination compensation in ground based hyperspectral imaging. ISPRS J. Photogramm. Remote Sens. 2017, 129, 162–178. [Google Scholar] [CrossRef]
- Danner, M.; Locherer, M.; Hank, T.; Richter, K.; EnMAP Consortium. Measuring Leaf Area Index (LAI) with the LI-Cor LAI 2200C or LAI-2200 (+ 2200Clear Kit)—Theory, Measurement, Problems, Interpretation. 2015. Available online: http://doi.org/10.2312/enmap.2015.009 (accessed on 22 October 2015).
- Nackaerts, K.; Coppin, P.; Muys, B.; Hermy, M. Sampling methodology for LAI measurements with LAI-2000 in small forest stands. Agric. For. Meteorol. 2000, 101, 247–250. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Inversion of a forest reflectance model to estimate structural canopy variables from hyperspectral remote sensing data. Remote Sens. Environ. 2006, 100, 281–294. [Google Scholar] [CrossRef]
- Uddling, J.; Gelang-Alfredsson, J.; Piikki, K.; Pleijel, H. Evaluating the relationship between leaf chlorophyll concentration and SPAD-502 chlorophyll meter readings. Photosynth. Res. 2007, 91, 37–46. [Google Scholar] [CrossRef] [PubMed]
- Smith, G.M.; Milton, E.J. The use of the empirical line method to calibrate remotely sensed data to reflectance. Int. J. Remote Sens. 1999, 20, 2653–2662. [Google Scholar] [CrossRef]
- Verhoef, W.; Bach, H. Coupled soil-leaf-canopy and atmosphere radiative transfer modeling to simulate hyperspectral multi-angular surface reflectance and TOA radiance data. Remote Sens. Environ. 2007, 109, 166–182. [Google Scholar] [CrossRef]
- Jacquemoud, S.; Baret, F. PROSPECT: A model of leaf optical properties spectra. Remote Sens. Environ. 1990, 34, 75–91. [Google Scholar] [CrossRef]
- Hapke, B. Bidirectional reflectance spectroscopy: 1. Theory. J. Geophys. Res. Solid Earth 1981, 86, 3039–3054. [Google Scholar] [CrossRef]
- Gastellu-Etchegorry, J.P.; Demarez, V.; Pinel, V.; Zagolski, F. Modeling radiative transfer in heterogeneous 3-D vegetation canopies. Remote Sens. Environ. 1996, 58, 131–156. [Google Scholar] [CrossRef] [Green Version]
- Levashova, N.; Lukyanenko, D.; Mukhartova, Y.; Olchev, A. Application of a three-dimensional radiative transfer model to retrieve the species composition of a mixed forest stand from canopy reflected radiation. Remote Sens. 2018, 10, 1661. [Google Scholar] [CrossRef] [Green Version]
- Baret, F.; Jacquemoud, S.; Guyot, G.; Leprieur, C. Modeled analysis of the biophysical nature of spectral shifts and comparison with information content of broad bands. Remote Sens. Environ. 1992, 41, 133–142. [Google Scholar] [CrossRef]
- Mousivand, A.; Menenti, M.; Gorte, B.; Verhoef, W. Multi-temporal, multi-sensor retrieval of terrestrial vegetation properties from spectral-directional radiometric data. Remote Sens. Environ. 2015, 158, 311–330. [Google Scholar] [CrossRef]
- Sacks, J.; Welch, W.J.; Mitchell, T.J.; Wynn, H.P. Design and analysis of computer experiments. Stat. Sci. 1989, 4, 409–423. [Google Scholar] [CrossRef]
- Sallaberry, C.J.; Helton, J.C.; Hora, S.C. Extension of Latin hypercube samples with correlated variables. Reliab. Eng. Syst. Saf. 2008, 93, 1047–1059. [Google Scholar] [CrossRef] [Green Version]
- Ha, N.T.; Manley-Harris, M.; Pham, T.D.; Hawes, I. A Comparative Assessment of Ensemble-Based Machine Learning and Maximum Likelihood Methods for Mapping Seagrass Using Sentinel-2 Imagery in Tauranga Harbor, New Zealand. Remote. Sens. 2020, 12, 355. [Google Scholar] [CrossRef] [Green Version]
- Zhang, Y.; Pinder, G. Latin hypercube lattice sample selection strategy for correlated random hydraulic conductivity fields. Water Resour. Res. 2003, 39. [Google Scholar] [CrossRef]
- Ali, A.M.; Darvishzadeh, R.; Skidmore, A.; Gara, T.W.; O’Connor, B.; Roeoesli, C.; Heurich, M.; Paganini, M. Comparing methods for mapping canopy chlorophyll content in a mixed mountain forest using Sentinel-2 data. Int. J. Appl. Earth Obs. Geoinf. 2020, 87, 102037. [Google Scholar] [CrossRef]
- Kooistra, L.; Clevers, J.G.P.W. Estimating potato leaf chlorophyll content using ratio vegetation indices. Remote Sens. Lett. 2016, 7, 611–620. [Google Scholar] [CrossRef] [Green Version]
- Botha, E.J.; Leblon, B.; Zebarth, B.; Watmough, J. Non-destructive estimation of potato leaf chlorophyll from canopy hyperspectral reflectance using the inverted PROSAIL model. Int. J. Appl. Earth Obs. Geoinf. 2007, 9, 360–374. [Google Scholar] [CrossRef]
- Casa, R.; Jones, H.G. Retrieval of crop canopy properties: A comparison between model inversion from hyperspectral data and image classification. Int. J. Remote Sens. 2004, 25, 1119–1130. [Google Scholar] [CrossRef]
- Darvishzadeh, R.; Atzberger, C.; Skidmore, A.; Schlerf, M. Mapping grassland leaf area index with airborne hyperspectral imagery: A comparison study of statistical approaches and inversion of radiative transfer models. ISPRS J. Photogramm. Remote Sens. 2011, 66, 894–906. [Google Scholar] [CrossRef]
- Weiss, M.; Baret, F. Evaluation of canopy biophysical variable retrieval performances from the accumulation of large swath satellite data. Remote Sens. Environ. 1999, 70, 293–306. [Google Scholar] [CrossRef]
- Brown, L.A.; Ogutu, B.O.; Dash, J. Estimating Forest Leaf Area Index and Canopy Chlorophyll Content with Sentinel-2: An Evaluation of Two Hybrid Retrieval Algorithms. Remote. Sens. 2019, 11, 1752. [Google Scholar] [CrossRef] [Green Version]
- Li, Z.; Wang, J.; Tang, H.; Huang, C.; Yang, F.; Chen, B.; Wang, X.; Xin, X.; Ge, Y. Predicting grassland leaf area index in the meadow steppes of northern china: A comparative study of regression approaches and hybrid geostatistical methods. Remote Sens. 2016, 8, 632. [Google Scholar] [CrossRef] [Green Version]
- Rainforth, T.; Wood, F. Canonical correlation forests. arXiv 2015, arXiv:1507.05444. [Google Scholar]
- Xia, J.; Yokoya, N.; Iwasaki, A. Hyperspectral image classification with canonical correlation forests. IEEE Trans. Geosci. Remote Sens. 2016, 55, 421–431. [Google Scholar] [CrossRef]
- Williams, C.K.I.; Rasmussen, C.E. Gaussian Processes for Machine Learning; MIT Press: Cambridge, MA, USA, 2006; Volume 2. [Google Scholar]
- Demšar, J. Statistical comparisons of classifiers over multiple data sets. J. Mach. Learn. Res. 2006, 7, 1–30. [Google Scholar]
- D’Urso, G.; Dini, L.; Vuolo, F.; Alonso, L.; Guanter, L. Retrieval of leaf area index by inverting hyperspectral multiangular CHRIS PROBA data from SPARC 2003. In Proceedings of the 2nd CHRIS Proba Workshop, Frascati, Italy, 28–30 April 2004; Volume 28. [Google Scholar]
- Weiss, M.; Baret, F.; Myneni, R.; Pragnère, A.; Knyazikhin, Y. Investigation of a model inversion technique to estimate canopy biophysical variables from spectral and directional reflectance data. Agronomie 2000, 20, 3–22. [Google Scholar] [CrossRef]
- Domingues Franceschini, M.H.; Bartholomeus, H.; Van Apeldoorn, D.; Suomalainen, J.; Kooistra, L. Intercomparison of unmanned aerial vehicle and ground-based narrow band spectrometers applied to crop trait monitoring in organic potato production. Sensors 2017, 17, 1428. [Google Scholar] [CrossRef]
- Gitelson, A.A. Remote estimation of crop fractional vegetation cover: The use of noise equivalent as an indicator of performance of vegetation indices. Int. J. Remote Sens. 2013, 34, 6054–6066. [Google Scholar] [CrossRef]
- Liang, S. Recent developments in estimating land surface biogeophysical variables from optical remote sensing. Prog. Phys. Geogr. 2007, 31, 501–516. [Google Scholar] [CrossRef] [Green Version]
- Houborg, R.; McCabe, M.F. A hybrid training approach for leaf area index estimation via Cubist and random forests machine-learning. ISPRS J. Photogramm. Remote Sens. 2018, 135, 173–188. [Google Scholar] [CrossRef]
- Dovey, S.B.; Du Toit, B. Calibration of LAI-2000 canopy analyser with leaf area index in a young eucalypt stand. Trees 2006, 20, 273–277. [Google Scholar] [CrossRef]
- Chmura, D.; Salachna, A. The errors in visual estimation of plants cover in the context of education of phytosociology. Chem. Didact. Ecol. Metrol. 2016, 21, 75–82. [Google Scholar] [CrossRef] [Green Version]
- Ling, Q.; Huang, W.; Jarvis, P. Use of a SPAD-502 meter to measure leaf chlorophyll concentration in Arabidopsis thaliana. Photosynth. Res. 2011, 107, 209–214. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Atzberger, C. Development of an invertible forest reflectance model: The INFOR-Model. A decade of trans-european remote sensing cooperation. In Proceedings of the 20th EARSeL Symposium Dresden, Dresden, Germany, 14–16 June 2000; Volume 14, pp. 39–44. [Google Scholar]
- Tol, C.; Verhoef, W.; Timmermans, J.; Verhoef, A.; Su, Z. An integrated model of soil-canopy spectral radiances, photosynthesis, fluorescence, temperature and energy balance. Biogeosciences 2009, 6, 3109–3129. [Google Scholar]







| Date | Growth Stage | Flight Time | SZA | SAA | Illumination | exp (Second) | |
|---|---|---|---|---|---|---|---|
| VIS | NIR | ||||||
| 8-July | Tuber bulking | 12:00 | 28 | 165.58 | Partial cloud cover | 1/840 | 1/1135 |
| 14-July | Tuber bulking and flowering | 12:30 | 29 | 165.51 | Partial cloud cover | 1/840 | 1/1135 |
| 19-July | Tuber bulking and flowering | 12:15 | 30 | 165.62 | Clear/ sunny | 1/840 | 1/1135 |
| 27-July | Maturity | 12:15 | 31 | 166.09 | Partial cloud cover | 1/496 | 1/840 |
| 5-August | Maturity | 11:25 | 33 | 177.94 | Full cloud cover | 1/328 | 1/716 |
| 10-August | Maturity | 11:46 | 35 | 162.3 | Full cloud cover | 1/328 | 1/552 |
| Var. | Stats. | 8 July | 14 July | 19 July | 27 July | 05 August | 10 August | All Data |
|---|---|---|---|---|---|---|---|---|
| Tuber Bulking | Tuber Bulking and Flowering | Tuber Bulking and Flowering | Maturity | Maturity | Maturity | |||
| LAI (m2/m2) | Mean | 1.91 | 2.19 | 2.22 | 2.98 | 3.94 | 3.69 | 2.85 |
| Min | 0.19 | 0.06 | 0.56 | 0.92 | 1.64 | 2.35 | 0.06 | |
| Max | 2.84 | 3.74 | 4.04 | 5.25 | 6.67 | 5.46 | 6.67 | |
| Stdev | 0.62 | 0.91 | 0.86 | 0.99 | 1.05 | 0.77 | 1.17 | |
| C.V. | 0.32 | 0.42 | 0.39 | 0.33 | 0.27 | 0.21 | 0.41 | |
| fCover | Mean | 0.47 | 0.58 | 0.62 | 0.77 | 0.91 | 0.88 | 0.71 |
| Min | 0.05 | 0.15 | 0.1 | 0.35 | 0.55 | 0.7 | 0.05 | |
| Max | 0.65 | 0.85 | 0.95 | 0.95 | 1 | 1 | 1 | |
| Stdev | 0.17 | 0.22 | 0.25 | 0.14 | 0.12 | 0.09 | 0.23 | |
| C.V. | 0.36 | 0.38 | 0.41 | 0.19 | 0.13 | 0.11 | 0.33 | |
| CCC (g/m2) | Mean | 1.37 | 1.48 | 1.66 | 1.93 | 2.27 | 2.22 | 1.84 |
| Min | 0.15 | 0.05 | 0.38 | 0.48 | 0.81 | 1.18 | 0.05 | |
| Max | 2.1 | 2.89 | 3.3 | 3.62 | 3.85 | 3.63 | 3.85 | |
| Stdev | 0.47 | 0.68 | 0.77 | 0.78 | 0.69 | 0.6 | 0.75 | |
| C.V. | 0.35 | 0.46 | 0.46 | 0.4 | 0.31 | 0.27 | 0.41 |
| Methods | Stats. | LAI(m2/m2) | fCover | CCC(g/m2) |
|---|---|---|---|---|
| RF | R2 | 0.77 | 0.82 | 0.81 |
| NRMSE(%) | 10.59 | 10.59 | 15.06 | |
| CCF | R2 | 0.59 | 0.65 | 0.55 |
| NRMSE(%) | 11.59 | 16.58 | 13.40 | |
| GPR | R2 | 0.70 | 0.68 | 0.60 |
| NRMSE(%) | 9.80 | 17.58 | 17.26 |
| Estimations | Growth Seasons | Illumination | Different Retrieval Strategies | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| Hybrid | LUTreg | RF | RFexp | |||||||
| R2 | NRMSE | R2 | NRMSE | R2 | NRMSE | R2 | NRMSE | |||
| LAI(m2/m2) | 8 July (Tuber bulking) | Partial cloud cover | 0.56 | 23.23 | 0.73 | 13.81 | 0.70 | 12.64 | 0.8 | 12.27 |
| 14 July (Tuber bulking and flowering) | Partial cloud cover | 0.64 | 15.26 | 0.71 | 14.83 | 0.65 | 16.23 | 0.76 | 14.69 | |
| 19 July (Tuber bulking and flowering) | Clear/Sunny | 0.83 | 16.66 | 0.73 | 13.87 | 0.87 | 9.33 | 0.88 | 8.11 | |
| 27 July (Maturity) | Partial cloud cover | 0.52 | 17.35 | 0.59 | 14.57 | 0.58 | 14.54 | 0.71 | 11.59 | |
| 5 August (Maturity) | Full cloud cover | 0.61 | 15.15 | 0.70 | 12.09 | 0.46 | 15.50 | 0.63 | 11.81 | |
| 10 August (Maturity) | Full cloud cover | 0.16 | 33.95 | 0.26 | 24.53 | 0.25 | 25.27 | 0.43 | 14.25 | |
| All data | - | 0.70 | 9.80 | 0.77 | 9.18 | 0.80 | 5.51 | 0.83 | 5.36 | |
| fCover | 8 July (Tuber bulking) | Partial cloud cover | 0.41 | 22.92 | 0.75 | 14.37 | 0.7 | 15.61 | 0.76 | 13.82 |
| 14 July(Tuber bulking and flowering) | Partial cloud cover | 0.64 | 19.92 | 0.77 | 17.12 | 0.72 | 17.5 | 0.79 | 13.71 | |
| 19 July (Tuber bulking and flowering) | Clear/Sunny | 0.71 | 14.35 | 0.74 | 14.99 | 0.77 | 14.46 | 0.80 | 13.14 | |
| 27 July (Maturity) | Partial cloud cover | 0.55 | 13.97 | 0.74 | 12.80 | 0.740 | 12.53 | 0.86 | 8.03 | |
| 5 August (Maturity) | Full cloud cover | 0.38 | 13.96 | 0.71 | 13.76 | 0.66 | 21.89 | 0.91 | 8.81 | |
| 10 August (Maturity) | Full cloud cover | 0.11 | 33.42 | 0.12 | 33.06 | 0.45 | 36.56 | 0.71 | 10.93 | |
| All data | - | 0.82 | 10.59 | 0.83 | 10.46 | 0.85 | 6.23 | 0.86 | 5.87 | |
| CCC(g/m2) | 8 July (Tuber bulking) | Partial cloud cover | 0.64 | 26.12 | 0.6 | 18.05 | 0.61 | 17.20 | 0.68 | 15.49 |
| 14 July(Tuber bulking and flowering) | Partial cloud cover | 0.68 | 16.83 | 0.6 | 17.09 | 0.65 | 15.59 | 0.75 | 13.35 | |
| 19 July (Tuber bulking and flowering) | Clear/Sunny | 0.79 | 15.83 | 0.64 | 16.11 | 0.80 | 17.32 | 0.85 | 13.66 | |
| 27 July (Maturity) | Partial cloud cover | 0.52 | 18.25 | 0.62 | 16.92 | 0.32 | 18.15 | 0.52 | 15.04 | |
| 5 August (Maturity) | Full cloud cover | 0.55 | 15.59 | 0.54 | 14.49 | 0.47 | 19.28 | 0.56 | 14.53 | |
| 10 August (Maturity) | Full cloud cover | 0.08 | 30.86 | 0.09 | 23.33 | 0.11 | 22.34 | 0.44 | 15.12 | |
| All data | - | 0.55 | 13.40 | 0.62 | 12.16 | 0.65 | 16.21 | 0.61 | 15.01 | |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Abdelbaki, A.; Schlerf, M.; Retzlaff, R.; Machwitz, M.; Verrelst, J.; Udelhoven, T. Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging. Remote Sens. 2021, 13, 1748. https://doi.org/10.3390/rs13091748
Abdelbaki A, Schlerf M, Retzlaff R, Machwitz M, Verrelst J, Udelhoven T. Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging. Remote Sensing. 2021; 13(9):1748. https://doi.org/10.3390/rs13091748
Chicago/Turabian StyleAbdelbaki, Asmaa, Martin Schlerf, Rebecca Retzlaff, Miriam Machwitz, Jochem Verrelst, and Thomas Udelhoven. 2021. "Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging" Remote Sensing 13, no. 9: 1748. https://doi.org/10.3390/rs13091748
APA StyleAbdelbaki, A., Schlerf, M., Retzlaff, R., Machwitz, M., Verrelst, J., & Udelhoven, T. (2021). Comparison of Crop Trait Retrieval Strategies Using UAV-Based VNIR Hyperspectral Imaging. Remote Sensing, 13(9), 1748. https://doi.org/10.3390/rs13091748