Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve
Abstract
:1. Introduction
- (1)
- a set of S1-derived parameters and 14 cloud-free S2 scenes were classified individually and in combination;
- (2)
- the monotemporal S2 scenes were classified separately as well as paired with S1 data;
- (3)
- the accuracies obtained from the monotemporal S2 scenes were used to determine the most- and least-accurate S2 scenes from spring, summer, and autumn seasons. Combinations of the least- and most-accurate seasonal S2 scenes were classified with and without S1 data.
2. Materials and Methods
2.1. Study Site, Reference Data
2.2. Sentinel-2 Data
2.3. Sentinel-1 Data
2.3.1. Backscatter Averages and Ratios
2.3.2. Phenological Parameters
2.3.3. Harmonic Parameters
2.4. Classification Approach
3. Results
3.1. Full S1/S2 Dataset Comparison
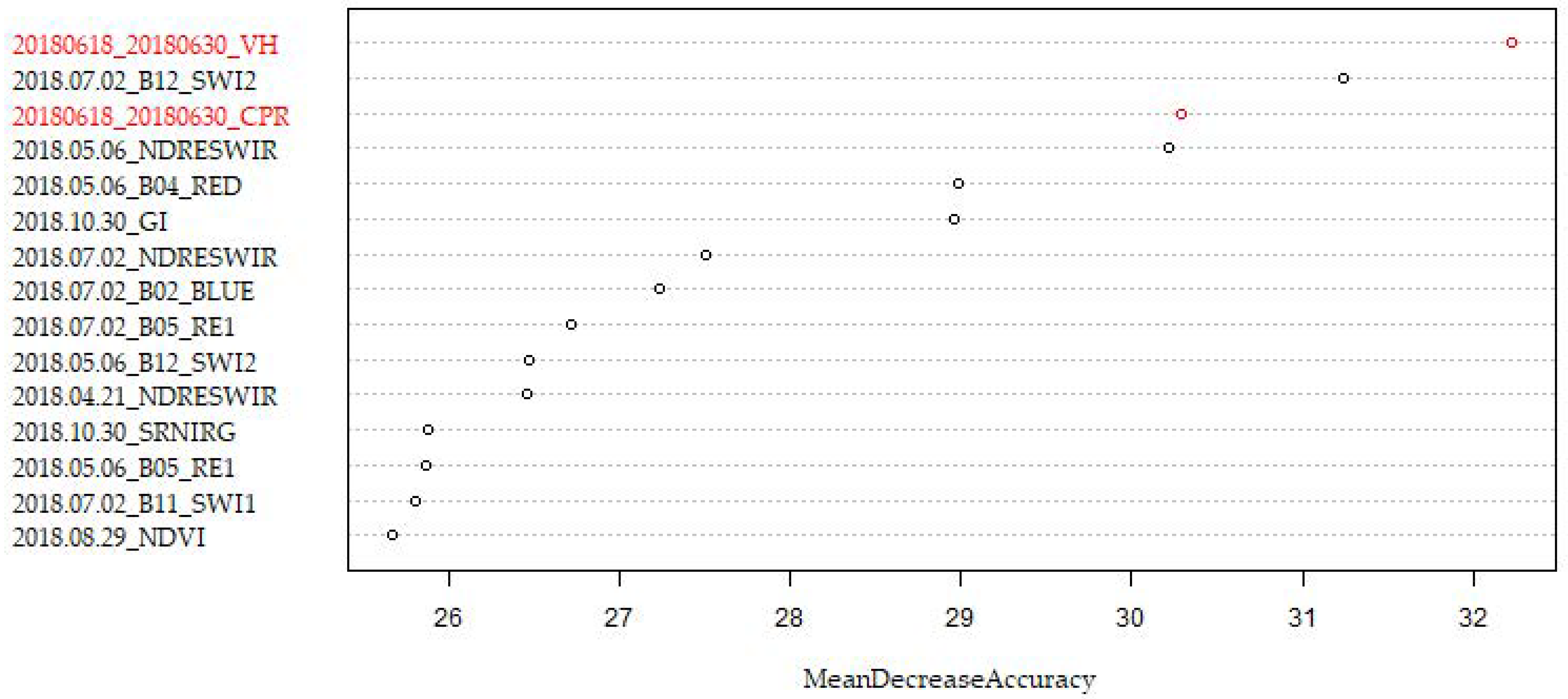
3.2. Added Value of S1 on Monotemporal S2 Datasets
3.3. Added Value of S1 on Multitemporal S2 Dataset
4. Discussion
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Index-Name | Formula | Reference |
|---|---|---|
| Built-up Area Index (BAI) | [49] | |
| Chlorophyll Green Index (CGI) | [50] | |
| Greenness Index (GI) | [51] | |
| Green Normalized-Difference Vegetation Index (gNDVI) | [52] | |
| Leaf Chlorophyll Content Index (LCCI) | [53] | |
| Moisture Stress Index (MSI) | [54] | |
| Normalized-Difference Red-Edge and SWIR2(NDRESWIR) | [55] | |
| Normalized-Difference Tillage Index (NDTI) | [56] | |
| Normalized-Difference Vegetation Index (NDVI) | [57] | |
| Red-Edge Normalized-Difference Vegetation Index(reNDVI) | [52] | |
| Normalized-Difference Water Index 1 (NDWI1) | [58] | |
| Normalized-Difference Water Index 2 (NDWI2) | [52] | |
| Normalized Humidity Index (NHI) | [59] | |
| Red-Edge Peak Area (REPA) | [55,60] | |
| Red SWIR1 Difference (DIRESWIR) | [61] | |
| Red-Edge Triangular Vegetation Index (RETVI) | [62] | |
| Soil Adjusted Vegetation Index (SAVI) | [63] | |
| Blue and RE1 Ratio (SRBRE1) | [51] | |
| Blue and RE2 Ratio (SRBRE2) | [64] | |
| Blue and RE3 Ratio (SRBRE3) | [55] | |
| NIR and Blue Ratio (SRNIRB) | [65] | |
| NIR and Green Ratio (SRNIRG) | [51] | |
| NIR and Red Ratio (SRNIRR) | [65] | |
| NIR and RE1 Ratio (SRNIRRE1) | [50] | |
| NIR and RE2 Ratio (SRNIRRE2) | [55] | |
| NIR and RE3 Ratio (SRNIRRE3) | [55] | |
| Soil Tillage Index (STI) | [56] | |
| Water Body Index (WBI) | [66] |
References
- Secretariat of the Convention on Biological Diversity. Global Biodiversity Outlook 5; Secretariat of the Convention on Biological Diversity: Montreal, Canada, 2020.
- Breidenbach, J.; Waser, L.T.; Debella-Gilo, M.; Schumacher, J.; Rahlf, J.; Hauglin, M.; Puliti, S.; Astrup, R. National Mapping and Estimation of Forest Area by Dominant Tree Species Using Sentinel-2 Data. Can. J. For. Res. 2021, 51, 365–379. [Google Scholar] [CrossRef]
- Immitzer, M.; Vuolo, F.; Atzberger, C. First Experience with Sentinel-2 Data for Crop and Tree Species Classifications in Central Europe. Remote Sens. 2016, 8, 166. [Google Scholar] [CrossRef]
- Maschler, J.; Atzberger, C.; Immitzer, M. Individual Tree Crown Segmentation and Classification of 13 Tree Species Using Airborne Hyperspectral Data. Remote Sens. 2018, 10, 1218. [Google Scholar] [CrossRef] [Green Version]
- Grybas, H.; Congalton, R.G. A Comparison of Multi-Temporal RGB and Multispectral UAS Imagery for Tree Species Classification in Heterogeneous New Hampshire Forests. Remote Sens. 2021, 13, 2631. [Google Scholar] [CrossRef]
- Immitzer, M.; Neuwirth, M.; Böck, S.; Brenner, H.; Vuolo, F.; Atzberger, C. Optimal Input Features for Tree Species Classification in Central Europe Based on Multi-Temporal Sentinel-2 Data. Remote Sens. 2019, 11, 2599. [Google Scholar] [CrossRef] [Green Version]
- Bjerreskov, K.S.; Nord-Larsen, T.; Fensholt, R. Classification of Nemoral Forests with Fusion of Multi-Temporal Sentinel-1 and 2 Data. Remote Sens. 2021, 13, 950. [Google Scholar] [CrossRef]
- Hościło, A.; Lewandowska, A. Mapping Forest Type and Tree Species on a Regional Scale Using Multi-Temporal Sentinel-2 Data. Remote Sens. 2019, 11, 929. [Google Scholar] [CrossRef] [Green Version]
- Kohrs, R.A.; Lazzara, M.A.; Robaidek, J.O.; Santek, D.A.; Knuth, S.L. Global Satellite Composites—20 Years of Evolution. Atmospheric Res. 2014, 135–136, 8–34. [Google Scholar] [CrossRef] [Green Version]
- Vuolo, F.; Ng, W.-T.; Atzberger, C. Smoothing and Gap-Filling of High Resolution Multi-Spectral Time Series: Example of Landsat Data. Int. J. Appl. Earth Obs. Geoinf. 2017, 57, 202–213. [Google Scholar] [CrossRef]
- Moreno-Martínez, Á.; Izquierdo-Verdiguier, E.; Maneta, M.P.; Camps-Valls, G.; Robinson, N.; Muñoz-Marí, J.; Sedano, F.; Clinton, N.; Running, S.W. Multispectral High Resolution Sensor Fusion for Smoothing and Gap-Filling in the Cloud. Remote Sens. Environ. 2020, 247, 111901. [Google Scholar] [CrossRef]
- Griffiths, P.; Nendel, C.; Hostert, P. Intra-Annual Reflectance Composites from Sentinel-2 and Landsat for National-Scale Crop and Land Cover Mapping. Remote Sens. Environ. 2019, 220, 135–151. [Google Scholar] [CrossRef]
- Bauer-Marschallinger, B.; Cao, S.; Navacchi, C.; Freeman, V.; Reuß, F.; Geudtner, D.; Rommen, B.; Vega, F.C.; Snoeij, P.; Attema, E.; et al. The Normalised Sentinel-1 Global Backscatter Model, Mapping Earth’s Land Surface with C-Band Microwaves. Sci. Data 2021, 8, 277. [Google Scholar] [CrossRef] [PubMed]
- Dostálová, A.; Lang, M.; Ivanovs, J.; Waser, L.T.; Wagner, W. European Wide Forest Classification Based on Sentinel-1 Data. Remote Sens. 2021, 13, 337. [Google Scholar] [CrossRef]
- Waser, L.T.; Rüetschi, M.; Psomas, A.; Small, D.; Rehush, N. Mapping Dominant Leaf Type Based on Combined Sentinel-1/-2 Data—Challenges for Mountainous Countries. ISPRS J. Photogramm. Remote Sens. 2021, 180, 209–226. [Google Scholar] [CrossRef]
- Rüetschi, M.; Schaepman, M.; Small, D. Using Multitemporal Sentinel-1 C-Band Backscatter to Monitor Phenology and Classify Deciduous and Coniferous Forests in Northern Switzerland. Remote Sens. 2017, 10, 55. [Google Scholar] [CrossRef] [Green Version]
- Udali, A.; Lingua, E.; Persson, H.J. Assessing Forest Type and Tree Species Classification Using Sentinel-1 C-Band SAR Data in Southern Sweden. Remote Sens. 2021, 13, 3237. [Google Scholar] [CrossRef]
- Tran, A.T.; Nguyen, K.A.; Liou, Y.A.; Le, M.H.; Vu, V.T.; Nguyen, D.D. Classification and Observed Seasonal Phenology of Broadleaf Deciduous Forests in a Tropical Region by Using Multitemporal Sentinel-1A and Landsat 8 Data. Forests 2021, 12, 235. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Mrkvicka, A.; Drozdowski, I.; Brenner, H. Kernzonen im Biosphärenpark Wienerwald—Urwälder von morgen. Wiss. Mitt. Aus Niederösterreichischen Landesmus. 2014, 25, 41–88. [Google Scholar]
- Nkosi, S.E.; Adam, E.; Barrett, A.S.; Brown, L.R. Mapping the Spatial Distribution of Tree Species Selected by Elephants (Loxodonta Africana) in Venetia-Limpopo Nature Reserve Using Sentinel-2 Imagery. Appl. Geomat. 2021, 13, 701–713. [Google Scholar] [CrossRef]
- Louis, J.; Debaecker, V.; Pflug, B.; Main-Knorn, M.; Bieniarz, J.; Mueller-Wilm, U.; Cadau, E.; Gascon, F. Sentinel-2 Sen2Cor: L2A Processor for Users. In Proceedings of the Living Planet Symposium 2016, Prague, Czech Republic, 9–13 May 2016; Spacebooks Online. 2016; pp. 1–8. [Google Scholar]
- Myneni, R.B.; Nemani, R.R.; Shabanov, N.V.; Knyazikhin, Y.; Morisette, J.T.; Privette, J.L.; Running, S.W. LAI and FPAR. Available online: https://cce.nasa.gov/mtg2008_ab_presentations/LAI-FPAR_Myneni_whitepaper.pdf (accessed on 4 May 2022).
- Watson, D.J. Comparative Physiological Studies on the Growth of Field Crops: I. Variation in Net Assimilation Rate and Leaf Area between Species and Varieties, and within and between Years. Ann. Bot. 1947, 11, 41–76. [Google Scholar] [CrossRef]
- EODC GmbH Austrian Data Cube. Available online: https://acube.eodc.eu/ (accessed on 1 April 2022).
- Le Toan, T.; Beaudoin, A.; Riom, J.; Guyon, D. Relating Forest Biomass to SAR Data. IEEE Trans. Geosci. Remote Sens. 1992, 30, 403–411. [Google Scholar] [CrossRef]
- Vreugdenhil, M.; Wagner, W.; Bauer-Marschallinger, B.; Pfeil, I.; Teubner, I.; Rüdiger, C.; Strauss, P. Sensitivity of Sentinel-1 Backscatter to Vegetation Dynamics: An Austrian Case Study. Remote Sens. 2018, 10, 1396. [Google Scholar] [CrossRef] [Green Version]
- Dostálová, A.; Wagner, W.; Milenković, M.; Hollaus, M. Annual Seasonality in Sentinel-1 Signal for Forest Mapping and Forest Type Classification. Int. J. Remote Sens. 2018, 39, 7738–7760. [Google Scholar] [CrossRef]
- Frison, P.-L.; Fruneau, B.; Kmiha, S.; Soudani, K.; Dufrêne, E.; Toan, T.L.; Koleck, T.; Villard, L.; Mougin, E.; Rudant, J.-P. Potential of Sentinel-1 Data for Monitoring Temperate Mixed Forest Phenology. Remote Sens. 2018, 10, 2049. [Google Scholar] [CrossRef] [Green Version]
- Ahern, F.J.; Leckie, D.J.; Drieman, J.A. Seasonal Changes in Relative C-Band Backscatter of Northern Forest Cover Types. IEEE Trans. Geosci. Remote Sens. 1993, 31, 668–680. [Google Scholar] [CrossRef]
- Zeileis, A.; Leisch, F.; Hornik, K.; Kleiber, C. Strucchange: An R Package for Testing for Structural Change in Linear Regression Models. J. Stat. Softw. 2002, 7, 1–38. [Google Scholar] [CrossRef] [Green Version]
- Schlaffer, S.; Matgen, P.; Hollaus, M.; Wagner, W. Flood Detection from Multi-Temporal SAR Data Using Harmonic Analysis and Change Detection. Int. J. Appl. Earth Obs. Geoinf. 2015, 38, 15–24. [Google Scholar] [CrossRef]
- Einzmann, K.; Immitzer, M.; Böck, S.; Bauer, O.; Schmitt, A.; Atzberger, C. Windthrow Detection in European Forests with Very High-Resolution Optical Data. Forests 2017, 8, 21. [Google Scholar] [CrossRef] [Green Version]
- Immitzer, M.; Böck, S.; Einzmann, K.; Vuolo, F.; Pinnel, N.; Wallner, A.; Atzberger, C. Fractional Cover Mapping of Spruce and Pine at 1 Ha Resolution Combining Very High and Medium Spatial Resolution Satellite Imagery. Remote Sens. Environ. 2018, 204, 690–703. [Google Scholar] [CrossRef] [Green Version]
- Guyon, I.; Weston, J.; Barnhill, S. Gene Selection for Cancer Classification Using Support Vector Machines. Mach. Learn. 2002, 46, 389–422. [Google Scholar] [CrossRef]
- Wessel, M.; Brandmeier, M.; Tiede, D. Evaluation of Different Machine Learning Algorithms for Scalable Classification of Tree Types and Tree Species Based on Sentinel-2 Data. Remote Sens. 2018, 10, 1419. [Google Scholar] [CrossRef] [Green Version]
- Ma, M.; Liu, J.; Liu, M.; Zeng, J.; Li, Y. Tree Species Classification Based on Sentinel-2 Imagery and Random Forest Classifier in the Eastern Regions of the Qilian Mountains. Forests 2021, 12, 1736. [Google Scholar] [CrossRef]
- Kollert, A.; Bremer, M.; Löw, M.; Rutzinger, M. Exploring the Potential of Land Surface Phenology and Seasonal Cloud Free Composites of One Year of Sentinel-2 Imagery for Tree Species Mapping in a Mountainous Region. Int. J. Appl. Earth Obs. Geoinf. 2021, 94, 102208. [Google Scholar] [CrossRef]
- Persson, M.; Lindberg, E.; Reese, H. Tree Species Classification with Multi-Temporal Sentinel-2 Data. Remote Sens. 2018, 10, 1794. [Google Scholar] [CrossRef] [Green Version]
- Mngadi, M.; Odindi, J.; Peerbhay, K.; Mutanga, O. Examining the Effectiveness of Sentinel-1 and 2 Imagery for Commercial Forest Species Mapping. Geocarto Int. 2021, 36, 1–12. [Google Scholar] [CrossRef]
- Grabska, E.; Hostert, P.; Pflugmacher, D.; Ostapowicz, K. Forest Stand Species Mapping Using the Sentinel-2 Time Series. Remote Sens. 2019, 11, 1197. [Google Scholar] [CrossRef] [Green Version]
- Grabska, E.; Frantz, D.; Ostapowicz, K. Evaluation of Machine Learning Algorithms for Forest Stand Species Mapping Using Sentinel-2 Imagery and Environmental Data in the Polish Carpathians. Remote Sens. Environ. 2020, 251, 112103. [Google Scholar] [CrossRef]
- Karasiak, N.; Fauvel, M.; Dejoux, J.-F.; Monteil, C.; Sheeren, D. Optimal Dates for Deciduous Tree Species Mapping Using Full Years Sentinel-2 Time Series in South West France. ISPRS Ann. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, V-3-2020, 469–476. [Google Scholar] [CrossRef]
- Hemmerling, J.; Pflugmacher, D.; Hostert, P. Mapping Temperate Forest Tree Species Using Dense Sentinel-2 Time Series. Remote Sens. Environ. 2021, 267, 112743. [Google Scholar] [CrossRef]
- Xie, B.; Cao, C.; Xu, M.; Duerler, R.S.; Yang, X.; Bashir, B.; Chen, Y.; Wang, K. Analysis of Regional Distribution of Tree Species Using Multi-Seasonal Sentinel-1&2 Imagery within Google Earth Engine. Forests 2021, 12, 565. [Google Scholar] [CrossRef]
- Schlerf, M.; Atzberger, C. Inversion of a Forest Reflectance Model to Estimate Structural Canopy Variables from Hyperspectral Remote Sensing Data. Remote Sens. Environ. 2006, 100, 281–294. [Google Scholar] [CrossRef]
- ESA Sentinel-2 L1C Data Quality Report. Available online: https://sentinel.esa.int/documents/247904/685211/Sentinel-2_L1C_Data_Quality_Report (accessed on 2 May 2022).
- Ghassemi, B.; Dujakovic, A.; Żółtak, M.; Immitzer, M.; Atzberger, C.; Vuolo, F. Designing a European-Wide Crop Type Mapping Approach Based on Machine Learning Algorithms Using LUCAS Field Survey and Sentinel-2 Data. Remote Sens. 2022, 14, 541. [Google Scholar] [CrossRef]
- Shahi, K.; Shafri, H.Z.M.; Taherzadeh, E.; Mansor, S.; Muniandy, R. A Novel Spectral Index to Automatically Extract Road Networks from WorldView-2 Satellite Imagery. Egypt. J. Remote Sens. Space Sci. 2015, 18, 27–33. [Google Scholar] [CrossRef] [Green Version]
- Datt, B. A New Reflectance Index for Remote Sensing of Chlorophyll Content in Higher Plants: Tests Using Eucalyptus Leaves. J. Plant Physiol. 1999, 154, 30–36. [Google Scholar] [CrossRef]
- Le Maire, G.; François, C.; Dufrêne, E. Towards Universal Broad Leaf Chlorophyll Indices Using PROSPECT Simulated Database and Hyperspectral Reflectance Measurements. Remote Sens. Environ. 2004, 89, 1–28. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a Green Channel in Remote Sensing of Global Vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Wulf, H.; Stuhler, S. Sentinel-2: Land Cover, Preliminary User Feedback on Sentinel-2a Data. In Proceedings of the Sentinel-2A Expert Users Technical Meeting, Frascati, Italy, 27 February–1 March 2015; pp. 29–30. [Google Scholar]
- Vogelmann, J.E.; Rock, B.N. Spectral Characterization of Suspected Acid Deposition Damage in Red Spruce (Picea Rubens) Stands from Vermont. In Proceedings of the Airborne Imaging Spectrometer Data Anal. Workshop, Pasadena, CA, USA, 8–10 April 1985. [Google Scholar]
- Radoux, J.; Chomé, G.; Jacques, D.; Waldner, F.; Bellemans, N.; Matton, N.; Lamarche, C.; d’Andrimont, R.; Defourny, P. Sentinel-2’s Potential for Sub-Pixel Landscape Feature Detection. Remote Sens. 2016, 8, 488. [Google Scholar] [CrossRef] [Green Version]
- Van Deventer, A.P.; Ward, A.D.; Gowda, P.H.; Lyon, J.G. Using Thematic Mapper Data to Identify Contrasting Soil Plains and Tillage Practices. Photogramm. Eng. Remote Sens. 1997, 63, 87–93. [Google Scholar]
- Tucker, C.J. Red and Photographic Infrared Linear Combinations for Monitoring Vegetation. Remote Sens. Environ. 1979, 8, 127–150. [Google Scholar] [CrossRef] [Green Version]
- Gao, B. NDWI—A Normalized Difference Water Index for Remote Sensing of Vegetation Liquid Water from Space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Lacaux, J.P.; Tourre, Y.M.; Vignolles, C.; Ndione, J.A.; Lafaye, M. Classification of Ponds from High-Spatial Resolution Remote Sensing: Application to Rift Valley Fever Epidemics in Senegal. Remote Sens. Environ. 2007, 106, 66–74. [Google Scholar] [CrossRef]
- Filella, I.; Penuelas, J. The Red Edge Position and Shape as Indicators of Plant Chlorophyll Content, Biomass and Hydric Status. Int. J. Remote Sens. 1994, 15, 1459–1470. [Google Scholar] [CrossRef]
- Jacques, D.C.; Kergoat, L.; Hiernaux, P.; Mougin, E.; Defourny, P. Monitoring Dry Vegetation Masses in Semi-Arid Areas with MODIS SWIR Bands. Remote Sens. Environ. 2014, 153, 40–49. [Google Scholar] [CrossRef]
- Chen, P.-F.; Nicolas, T.; Wang, J.-H.; Philippe, V.; Huang, W.-J.; Li, B.-G. New Index for Crop Canopy Fresh Biomass Estimation. Spectrosc. Spectr. Anal. 2010, 30, 512–517. [Google Scholar]
- Huete, A.R. A Soil-Adjusted Vegetation Index (SAVI). Remote Sens. Environ. 1988, 25, 295–309. [Google Scholar] [CrossRef]
- Lichtenthaler, H.K.; Lang, M.; Sowinska, M.; Heisel, F.; Miehé, J.A. Detection of Vegetation Stress Via a New High Resolution Fluorescence Imaging System. J. Plant Physiol. 1996, 148, 599–612. [Google Scholar] [CrossRef]
- Blackburn, G.A. Quantifying Chlorophylls and Caroteniods at Leaf and Canopy Scales. Remote Sens. Environ. 1998, 66, 273–285. [Google Scholar] [CrossRef]
- Domenech, E.; Mallet, C. Change Detection in High Resolution Land Use/Land Cover Geodatabases (at Object Level); Official Publication No. 64; EuroSDR: Leuven, Belgium, 2014. [Google Scholar]



| S2 Satellite | Date | Orbit | Sun Zenith Angle | Sun Azimuth Angle |
|---|---|---|---|---|
| B | 8 April 2018 | 79 | 43.02 | 157.29 |
| B | 21 April 2018 | 122 | 37.72 | 160.39 |
| A | 6 May 2018 | 122 | 33.06 | 159.37 |
| A | 2 July 2018 | 79 | 28.23 | 147.73 |
| B | 9 August 2018 | 122 | 34.38 | 155.97 |
| A | 21 August 2018 | 79 | 38.49 | 154.81 |
| B | 29 August 2018 | 122 | 40.40 | 160.44 |
| A | 13 September 2018 | 122 | 45.59 | 163.93 |
| B | 18 September 2018 | 122 | 47.41 | 164.98 |
| B | 28 September 2018 | 122 | 51.10 | 166.96 |
| A | 30 September 2018 | 79 | 52.25 | 164.21 |
| B | 5 October 2018 | 79 | 54.08 | 165.12 |
| A | 10 October 2018 | 79 | 55.90 | 165.94 |
| A | 30 October 2018 | 79 | 62.82 | 168.14 |
| Temporal Average Backscatter | Leaf-Off | Leaf-On |
|---|---|---|
| VH | 20180314_20180326_VH | 20180618_20180630_VH |
| VV | 20180314_20180326_VV | 20180618_20180630_VV |
| VH/VV | 20180618_20180630_CPR |
| Backscatter ratio | VH or VV |
| Leaf-on/Leaf-off | Rat_Leaf_on_off |
| Phenology | VH or VV |
| Start of season—day of the year | sos_doy |
| End of season—day of the year | eos_doy |
| Length of season—days | sos_doy |
| correlation_winter | |
| slope_winter |
| Ascending (Orbit 73) | Descending (Orbit 22) | ||
|---|---|---|---|
| HPAR | |||
| Cosine 1 | VH | HPAR-C1_2018_VH_A073 | HPAR-C1_2018_VH_D022 |
| VV | HPAR-C1_2018_VV_A073 | HPAR-C1_2018_VV_D022 | |
| Cosine 2 | VH | HPAR-C2_2018_VH_A073 | HPAR-C2_2018_VH_D022 |
| VV | HPAR-C2_2018_VV_A073 | HPAR-C2_2018_VV_D022 | |
| Cosine 3 | VH | HPAR-C3_2018_VH_A073 | HPAR-C3_2018_VH_D022 |
| VV | HPAR-C3_2018_VV_A073 | HPAR-C3_2018_VV_D022 | |
| Sine 1 | VH | HPAR-S1_2018_VH_A073 | HPAR-S1_2018_VH_D022 |
| VV | HPAR-S1_2018_VV_A073 | HPAR-S1_2018_VV_D022 | |
| Sine 2 | VH | HPAR-S2_2018_VH_A073 | HPAR-S2_2018_VH_D022 |
| VV | HPAR-S2_2018_VV_A073 | HPAR-S2_2018_VV_D022 | |
| Sine 3 | VH | HPAR-S3_2018_VH_A073 | HPAR-S3_2018_VH_D022 |
| VV | HPAR-S3_2018_VV_A073 | HPAR-S3_2018_VV_D022 | |
| HPAR temporal average | |||
| VH | HPAR-M0_2018_VH_A073 | HPAR-M0_2018_VH_D022 | |
| VV | HPAR-M0_2018_VV_A073 | HPAR-M0_2018_VV_D022 | |
| HPAR model error | |||
| VH | HPAR-STD_2018_VH_A073 | HPAR-STD_2018_VH_D022 | |
| VV | HPAR-STD_2018_VV_A073 | HPAR-STD_2018_VV_D022 |
| Test Case | Acronym | Features | Comment |
|---|---|---|---|
| 1 | S1 | 43 | multitemporal S1 parameters |
| 2 | S2 (MULTI) | 574 | multitemporal image data of S2 |
| 3 | S1 + S2 (MULTI) | 617 | multitemporal S1 parameters + multitemporal image data of S2 |
| 4 | S2 (MONO) | 41 | monotemporal image data of S2 |
| 5 | S1 + S2 (MONO) | 84 | multitemporal S1 parameters + monotemporal image data of S2 |
| 6 | S2 (MAS) | 123 | image data of Most Accurate S2 Scene of each growing season |
| 7 | S1 + S2 (MAS) | 166 | multitemporal S1 parameters + image data of Most Accurate S2 Scene of each growing season |
| 8 | S2 (LAS) | 123 | image data of Least Accurate S2 Scene of each growing season |
| 9 | S1 + S2 (LAS) | 166 | multitemporal S1 parameters + image data of Least Accurate S2 Scene of each growing season |
| Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS | AG | FE | QU | PR | CP | AC | PA | PN | PS | LD | PM | UA | F1-Score | |||
| Sentinel-1 − Test case 1 | Classification | FS | 236 | 33 | 36 | 86 | 16 | 39 | 34 | 0 | 10 | 4 | 14 | 0 | 46.5% | 0.584 |
| AG | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 1 | NA | NA | ||
| FE | 3 | 2 | 6 | 2 | 1 | 1 | 4 | 0 | 0 | 0 | 0 | 0 | 31.6% | 0.113 | ||
| QU | 47 | 14 | 40 | 140 | 2 | 18 | 13 | 0 | 4 | 1 | 0 | 0 | 50.2% | 0.550 | ||
| PR | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 50.0% | 0.074 | ||
| CP | 0 | 0 | 2 | 0 | 0 | 4 | 0 | 1 | 0 | 0 | 0 | 1 | 50.0% | 0.101 | ||
| AC | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | NA | NA | ||
| PA | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 119 | 2 | 7 | 0 | 24 | 76.8% | 0.821 | ||
| PN | 7 | 2 | 2 | 1 | 0 | 5 | 2 | 1 | 114 | 19 | 1 | 5 | 71.7% | 0.755 | ||
| PS | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 5 | 11 | 44 | 1 | 5 | 65.7% | 0.603 | ||
| LD | 5 | 1 | 1 | 0 | 4 | 1 | 2 | 1 | 0 | 2 | 31 | 1 | 63.3% | 0.626 | ||
| PM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 8 | 2 | 2 | 3 | 19 | 55.9% | 0.422 | ||
| ∑ Reference | 300 | 52 | 87 | 230 | 25 | 71 | 55 | 135 | 143 | 79 | 50 | 56 | ||||
| PA | 78.7% | 0.0% | 6.9% | 60.9% | 4.0% | 5.6% | 0.0% | 88.1% | 79.7% | 55.7% | 62.0% | 33.9% | ||||
| OA 55.7% | Kappa 0.469 | |||||||||||||||
| Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS | AG | FE | QU | PR | CP | AC | PA | PN | PS | LD | PM | UA | F1-Score | |||
| Sentinel-2 − Test case 2 | Classification | FS | 272 | 4 | 10 | 26 | 5 | 22 | 15 | 0 | 2 | 0 | 6 | 0 | 75.1% | 0.822 |
| AG | 1 | 42 | 3 | 1 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 85.7% | 0.832 | ||
| FE | 6 | 2 | 64 | 5 | 2 | 4 | 3 | 0 | 2 | 0 | 2 | 0 | 71.1% | 0.723 | ||
| QU | 13 | 1 | 7 | 195 | 5 | 4 | 4 | 0 | 0 | 0 | 1 | 0 | 84.8% | 0.848 | ||
| PR | 0 | 0 | 0 | 0 | 12 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100.0% | 0.649 | ||
| CP | 3 | 2 | 1 | 1 | 0 | 40 | 1 | 0 | 0 | 0 | 0 | 0 | 83.3% | 0.672 | ||
| AC | 5 | 1 | 0 | 1 | 0 | 0 | 30 | 0 | 0 | 0 | 0 | 0 | 81.1% | 0.652 | ||
| PA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 127 | 1 | 4 | 1 | 3 | 93.4% | 0.937 | ||
| PN | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 133 | 1 | 1 | 1 | 94.3% | 0.937 | ||
| PS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 3 | 72 | 3 | 5 | 83.7% | 0.873 | ||
| LD | 0 | 0 | 2 | 1 | 0 | 0 | 0 | 0 | 2 | 2 | 36 | 2 | 80.0% | 0.758 | ||
| PM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 45 | 95.7% | 0.874 | ||
| ∑ Reference | 300 | 52 | 87 | 230 | 25 | 71 | 55 | 135 | 143 | 79 | 50 | 56 | ||||
| PA | 90.7% | 80.8% | 73.6% | 84.8% | 48.0% | 56.3% | 54.5% | 94.1% | 93.0% | 91.1% | 72.0% | 80.4% | ||||
| OA 83.2% | Kappa 0.806 | |||||||||||||||
| Reference | ||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| FS | AG | FE | QU | PR | CP | AC | PA | PN | PS | LD | PM | UA | F1-Score | |||
| Sentinel-1 + Sentinel-2 − Test case 3 | Classification | FS | 271 | 2 | 9 | 28 | 8 | 19 | 16 | 0 | 1 | 0 | 3 | 0 | 75.9% | 0.825 |
| AG | 0 | 43 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 95.6% | 0.887 | ||
| FE | 6 | 2 | 64 | 6 | 0 | 3 | 4 | 0 | 1 | 0 | 2 | 0 | 72.7% | 0.731 | ||
| QU | 16 | 1 | 9 | 193 | 6 | 4 | 3 | 0 | 0 | 0 | 0 | 0 | 83.2% | 0.835 | ||
| PR | 0 | 0 | 0 | 0 | 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 100% | 0.571 | ||
| CP | 2 | 3 | 3 | 2 | 0 | 44 | 1 | 0 | 1 | 0 | 0 | 0 | 78.6% | 0.693 | ||
| AC | 4 | 1 | 0 | 0 | 1 | 0 | 29 | 0 | 0 | 0 | 0 | 0 | 82.9% | 0.644 | ||
| PA | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 129 | 1 | 4 | 1 | 5 | 92.1% | 0.938 | ||
| PN | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 135 | 2 | 2 | 0 | 93.1% | 0.938 | ||
| PS | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 3 | 70 | 2 | 3 | 87.5% | 0.881 | ||
| LD | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 1 | 3 | 40 | 2 | 83.3% | 0.816 | ||
| PM | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 46 | 97.9% | 0.893 | ||
| ∑ Reference | 300 | 52 | 87 | 230 | 25 | 71 | 55 | 135 | 143 | 79 | 50 | 56 | ||||
| PA | 90.3% | 82.7% | 73.6% | 83.9% | 40.0% | 62.0% | 52.7% | 95.6% | 94.4% | 88.6% | 80.0% | 82.1% | ||||
| OA 83.7% | Kappa 0.811 | |||||||||||||||
| Satellite | No. of Species | Species Names | OA | Reference |
|---|---|---|---|---|
| S1 | 3 | Quercus spp., Fagus sylvatica, Picea abies | 72% | [16] |
| S1 | 4 | Quercus robur, Betula spp., Picea abies, Pinus sylvestris | 66% | [17] |
| S1 | 12 | Fagus sylvatica, Alnus glutinosa, Fraxinus excelsior, Quercus spp., Prunus spp., Carpinus betulus, Acer spp., Picea abies, Pinus nigra, Pinus sylvestris, Larix decidua, Pseudotsuga menziesii | 58% | Table 6 |
| S2 | 4 | Fagus sylvatica, Quercus spp., other broadleaf trees, coniferous trees | 88% | [36] |
| S2 | 4 | Sabina przewalskii, Picea crassifolia, Betula spp., Populus spp. | 90% | [37] |
| S2 | 5 | Larix spp., Pinus spp., Pinus mugo, Abies alba/Picea abies, broadleaf trees | 84% | [38] |
| S2 | 5 | Picea abies, Pinus silvestris, Larix × marschlinsii, Betula sp., Quercus robur | 88% | [39] |
| S2 | 7 | Picea sp., Pinus sp., Larix sp., Abies sp., Fagus sp., Quercus sp., other broadleaf trees | 66% | [3] |
| S2 | 7 | Acacia mearnsii, Eucalyptus dunnii, Eucalyptus grandiis, Eucalyptus mix, Pinus tecunumanii, Pinus elliotii, Pinus taedea | 84% | [40] |
| S2 | 8 | Fagus sylvatica, Quercus spp., Alnus spp., Betula pendula, Picea abies, Pinus sylvestris, Abies alba, Larix decidua | 82% | [8] |
| S2 | 9 | Fagus sylvatica, Betula pendula, Carpinus betulus, Abies alba, Acer pseudoplatanus, Larix decidua, European larch, Alnus incana, Pinus sylvestris, Picea abies | 92% | [41] |
| S2 | 11 | Alnus spp., Acer pseudoplatanus, Fagus sylvatica, Betula pendula, Carpinus betulus, Quercus spp., Picea abies, Pinus sylvestris, Larix decidua, Pseudotsuga menziesii | 87% | [42] |
| S2 | 12 | Fagus sylvatica, Alnus glutinosa, Fraxinus excelsior, Quercus spp., Prunus spp., Carpinus betulus, Acer spp., Picea abies, Pinus nigra, Pinus sylvestris, Larix decidua, Pseudotsuga menziesii | 83% | Table 7 |
| S2 | 12 | Fagus sylvatica, Alnus glutinosa, Fraxinus excelsior, Quercus spp., Prunus spp., Carpinus betulus, Acer spp., Picea abies, Pinus nigra, Pinus sylvestris, Larix decidua, Pseudotsuga menziesii | 90% | [6] |
| S2 | 12 | Betula pendula, Quercus robur/pubescens/petraea, Quercus rubra, Populus spp., Fraxinus excelsior, Robinia pseudoacacia, Salix spp., Eucalyptus spp., Pinus nigra subsp. laricio, Pinus pinaster, Pinus nigra, Abies alba, Pseudotsuga menziesii, Cupressus spp. | 0.90 * | [43] |
| S2 | 17 | Fagus sylvatica, Alnus spp., Quercus petraea/robur, Quercus rubra, Betula pendula, Robinia pseudoacacia, Tilia cordata, Acer pseudoplatanus, Fraxinus excelsior, Populus spp., Carpinus betulus, Picea abies, Larix spp., Pseudotsuga menziesii, Pinus sylvestris, Pinus strobus, Pinus nigra | 96% ** | [44] |
| S1 + LS | 4 | Shorea siamensis, Shorea obtuse, Dipterocarpus tuberculatus, semi-evergreen/evergreen | 79% | [18] |
| S1 + S2 | 6 | Fagus sylvatica, Quercus spp., other broadleaves, Picea sp., Pinus sp., other conifers | 63% | [7] |
| S1 + S2 | 6 | Quercus mongolia, Betula spp., Populus spp., Armeniaca sibirica Larix spp., Pinus tabulaeformis | 78% | [45] |
| S1 + S2 | 7 | Acacia mearnsii, Eucalyptus dunnii, Eucalyptus grandiis, Eucalyptus mix, Pinus tecunumanii, Pinus elliotii, Pinus taedea | 88% | [40] |
| S1 + S2 | 12 | Fagus sylvatica, Alnus glutinosa, Fraxinus excelsior, Quercus spp., Prunus spp., Carpinus betulus, Acer spp., Picea abies, Pinus nigra, Pinus sylvestris, Larix decidua, Pseudotsuga menziesii | 84% | Table 8 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Lechner, M.; Dostálová, A.; Hollaus, M.; Atzberger, C.; Immitzer, M. Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve. Remote Sens. 2022, 14, 2687. https://doi.org/10.3390/rs14112687
Lechner M, Dostálová A, Hollaus M, Atzberger C, Immitzer M. Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve. Remote Sensing. 2022; 14(11):2687. https://doi.org/10.3390/rs14112687
Chicago/Turabian StyleLechner, Michael, Alena Dostálová, Markus Hollaus, Clement Atzberger, and Markus Immitzer. 2022. "Combination of Sentinel-1 and Sentinel-2 Data for Tree Species Classification in a Central European Biosphere Reserve" Remote Sensing 14, no. 11: 2687. https://doi.org/10.3390/rs14112687