1. Introduction
Root, butt, and stem rot (RBSR) is a group of fungal diseases that is one of the most significant forest health issues in Europe and throughout the northern hemisphere. RBSR is primarily characterized by the decay of tree roots and the lower tree bole, which can result in reduced tree growth and ultimately lead to tree mortality. In Norway, over 80% of RBSR is caused by fungi from the genera
Heterobasidion and
Armillaria [
1]. Most of the losses occur in forests of Norway spruce (
Picea abies L. Karst). Two species in the
Heterobasidion genus exist in Norway:
Heterobasidion parviporum (Niemelä & Korhonen 1998) and
H. annosum s.s.
H. parviporum is more common in Norway and primarily infects spruce trees, although pines are also somewhat susceptible, while
H. annosum s.s. occurs more often in pine than in spruce. Both species spread via spores and mycelia. Spores can land on tree wounds caused by logging or storm damage, which allows the fungus to spread to healthy stands of trees. Once a tree is infected, the fungus can spread through the root network to adjacent trees (although not directly through the soil) [
2].
Armillaria species account for a smaller but significant portion of RBSR infection in Norway. Compared to
Heterobasidion,
Armillaria tend to cause more significant hollowing of the tree bole but are generally limited to a maximum height of 2 m. In comparison,
Heterobasidion may extend as high as 7 m within the stem. Other fungal species which may cause RBSR in Norway include
Stereum sanguinolentum (Alb. & Schwein.) Fr.,
Phellinus chrysoloma (Fr.) Donk, and
Climacocystis borealis (Fr.) Kotl. & Pouzar [
3]. These species are generally believed to account for a small proportion of RBSR cases in Norway. However,
Phellinus chrysoloma can account for a large proportion of infection in high-elevation spruce forests.
In Norway, RBSR is responsible for an estimated 10 million euros in losses for the forest products industry every year, while Europe-wide losses have been estimated at over 800 million euros annually [
1,
2]. Some losses occur due to the reduction in wood quality in infected trees: logs that could otherwise be used as sawtimber may become pulpwood or biomass feedstock instead, reducing the price forest owners receive for the wood. In contrast, other losses occur due to reduced volume growth and tree death. While
H. parviporum, the most common RBSR pathogen in Norway, is rarely fatal (and only after several decades of infection),
H. annosum s.s. and
H. irregulare both cause tree mortality. The latter is also an invasive pathogen in Europe and other places around the world, with the potential to cause severe ecological damage. There is significant interest in monitoring the presence of these fungi to facilitate control methods, both for ecological and financial reasons. Unfortunately, detecting RBSR infection in the field can be difficult or impossible until the very late stages of infection, and the cost of fieldwork can make effective monitoring programs cost-prohibitive for large areas [
4].
One approach for reducing the cost of monitoring programs is the use of remotely sensed data. Remote sensing platforms such as satellites, fixed-wing aircraft, and UAVs facilitate the collection of wall-to-wall data throughout an area of interest, something which is either impractical or impossible with field surveys. These platforms allow for the use of cameras that detect infrared light, making possible the detection of changes in vegetation properties that are invisible to the naked eye. Changes in cellular structure due to stress are usually apparent in the near-infrared (NIR) portion of the spectrum [
5], while changes in foliar water content are shown in the shortwave infrared (SWIR) [
6]. Consequently, remote sensing can replace expensive sampling methods such as tree coring or felling, which may be necessary to detect pathogens in the field.
Previous research has examined the use of remotely sensed data for more broadly studying a variety of plant pathogens and health issues. Much of the work has focused on the use of passive spectral cameras, both multi-spectral and hyperspectral. Multi-spectral sensors have been more widely utilized due to their lower cost, while hyperspectral sensors offer the advantages of collecting information across a wider portion of the electromagnetic spectrum and of providing finer spectral resolution. Some studies have also incorporated airborne laser scanning (ALS) data to provide information about changes to crown density and other structural properties of trees. Leckie et al. used a multi-spectral sensor with 2.5 m resolution to detect jack pine budworm in Ontario, Canada [
7]. They found that classification models using just three bands (red, NIR, and SWIR bands) could provide classification accuracies greater than 80%. Meng et al., used hyperspectral imagery and ALS data to detect defoliation in a mixed-pine oak forest due to gypsy moth infestation [
8]. Their analysis used regression models to predict the amount of defoliation, with the best models delivering R
2 values of 0.81. From the hyperspectral imagery, red-edge and near-infrared bands proved to be the most useful for detecting defoliation.
A few studies have specifically examined spectral information’s use for detecting root and stem rot fungi in woody plants. Examples include Leckie et al., who classified the severity of
Phellinus weirii infection in Douglas fir with accuracies as high as 80%, and LeLong et al., who detected the presence of
Ganoderma in oil palm [
7,
9]. This work shows that detecting root rot diseases with remote sensing is possible. An important caveat is that both of these diseases cause significant changes in tree foliage, which are visible to the naked eye, which is not generally true of RBSR in Norway spruce. Kankaanhuhta et al. attempted to detect the presence of RBSR in forests in Finland [
10]. They had some success with detecting
Heterobasidion in pine forests, but not in spruce, a disparity which is likely due to the greater effect on foliage color in pine trees. Thus, current field-based studies of RBSR detection cannot settle the question of whether pre-visual detection is possible.
Calamita et al. used a laboratory-based hyperspectral camera to classify grape leaves based on the presence of
Armillaria in the grapevine of origin [
11]. They found significant differences around 705 nm (the red edge) and 550 nm. They were able to identify both visibly diseased and infected but visually asymptomatic plants with an accuracy of 90% and 75%, respectively. This indicates that
Armillaria does affect the spectral properties of at least some woody plants. However, it leaves open the question of whether such differences will be noticeable in a field setting. Allen et al. classified the presence of RBSR in spruce trees at a site in Norway using hyperspectral imagery with two different classifiers [
12]. Results from this study indicated that detection of RBSR in spruce trees is possible, although classification accuracies were modest (64%).
UAV-based sensors offer several advantages over sensors mounted on conventional fixed-wing aircraft. UAVs can be deployed more rapidly and at a lower cost for very small areas (although costs are higher for large areas). UAVs also allow for more rapid deployment in response to favorable weather conditions. The low flying height of UAVs allows for higher spatial resolution than airborne systems. High spatial resolution UAV imagery has been found to allow for the detection of finer scale variation in plant health than is possible with satellite-based or airborne sensors [
13]. Overall, UAVs are highly suitable for gathering remotely sensed data on small tracts of land and provide valuable information for forest owners to aid in management decisions.
Numerous studies have examined using UAV-based sensors to monitor forest pathogens and other health issues. Otsu et al. detected pine processionary moth defoliation in Spain using a combination of Landsat 8 and multi-spectral UAV imagery [
14]. Zhang et al. predicted the extent of defoliation on
Pinus tabulaeformis Carrière due to the Chinese pine caterpillar using data from a UAV-based hyperspectral camera [
15]. Lin et al. used hyperspectral imagery and ALS data to predict damage from pine shoot beetle in Yunnan pine [
16]. Their study found that hyperspectral imagery-based estimates of chlorophyll levels and ALS intensity metrics were the most significant predictors of pine shoot beetle infestation. Näsi et al. used a hyperspectral camera with a spectral range of 500–900 nm to classify Norway spruce trees based on bark beetle infection status (healthy, infested, dead) with an overall accuracy of 76% [
17]. Subsequent work showed that UAV imagery delivered greater classification accuracies than satellite imagery, likely due to the finer spatial resolution of the UAV imagery [
18]. Honkavaara et al. used multitemporal hyperspectral and multi-spectral imagery to attempt to detect RBSR and European spruce beetle attacks at a site in Finland. Their work focused specifically on detecting beetle infestation before any visible changes in foliage color had occurred, rather than beetle infestation with visible foliar symptoms. The overall accuracy obtained in their study was around 45%, although their study used a relatively small sample size, which could have limited the accuracy of their models. While RBSR-infected trees were included in their classification, detecting RBSR was not their study’s primary goal [
19].
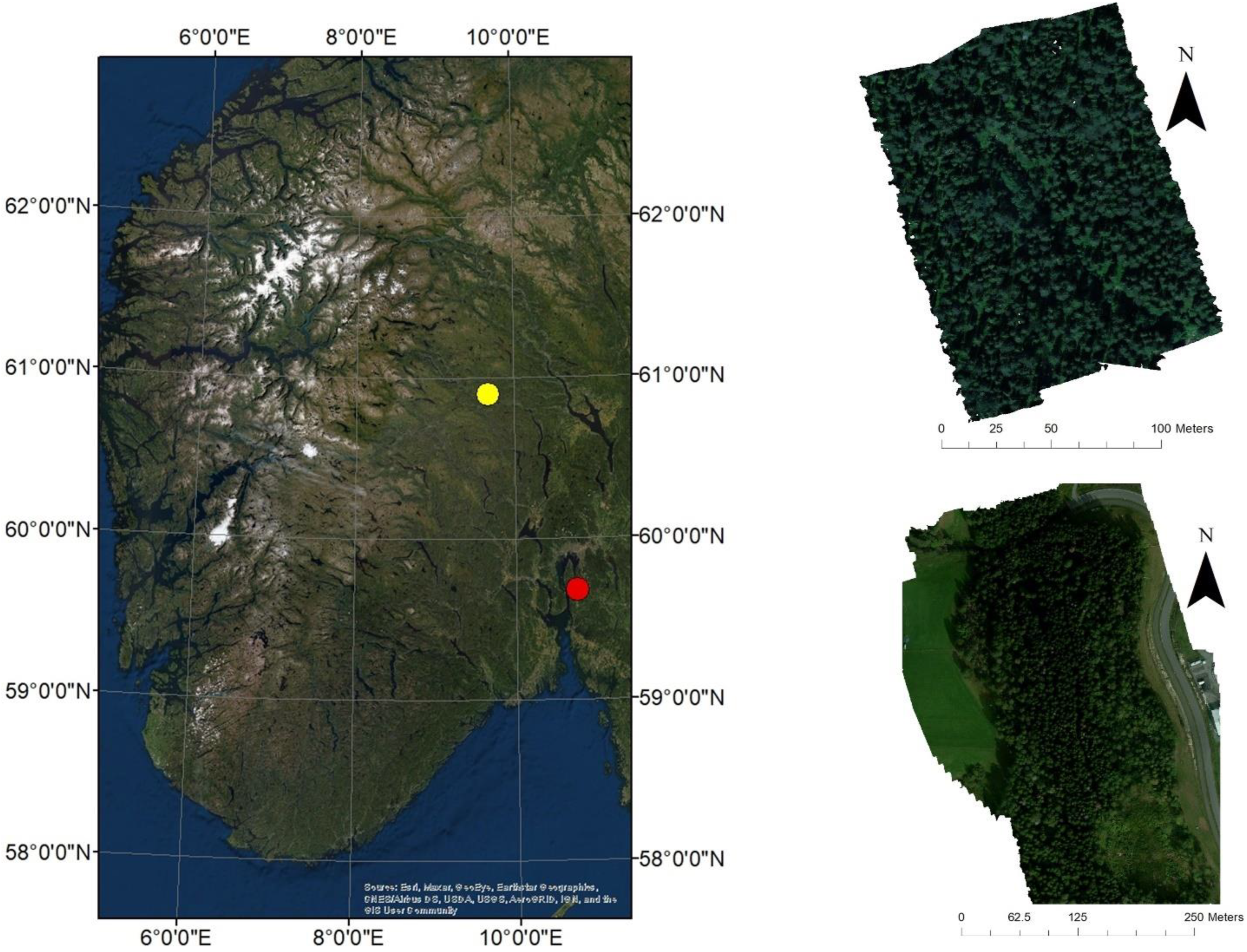
The primary objective of this study was to assess the potential of remotely sensed data from UAV-based cameras to facilitate the detection of RBSR in Norway Spruce by producing classification models. Unlike most previous studies of UAV applications in forest pathology, this study focused solely on pre-visual detection of infection since RBSR rarely produces visible changes to foliage in Norwegian conditions. Two cameras were used, allowing for a comparison between two sensor types. With the use of two sensors, it was hoped that the effect of different spectral regions or numbers of spectral bands on the accuracy of classification could be assessed. Since the cameras used in this study covered different wavelength regions, resampling was also performed on one of the images to enable such comparisons. Based on the spectral data collected from the imagery, trees were classified according to rot infection status. The second major objective of the study was to determine which vegetation indices were statistically different between infected and healthy trees. Such a comparison could also shed light on the physiological changes induced in spruce trees by RBSR infection.
4. Discussion
The results of this paper indicate that detection of RBSR in Picea abies is possible with UAV-based hyperspectral sensors. However, accuracies and kappa values were relatively modest. Classification accuracies were greater in the Etnedal study area, where a 490-band hyperspectral sensor was used, than in the Nordskogen study area, where a 29-band hyperspectral sensor was used. Likely explanations include the greater number of bands and the wider spectral range of the sensor used in the Etnedal study area. The sensor used in Etnedal contained enough bands with small enough bandwidths to calculate spectral derivatives, while the camera used in Nordskogen did not. A second explanation for these results is that the 29-band sensor used in this study covered a narrower portion of the spectrum than the 490-band sensor (450–895 nm vs. 400–2500 nm). Consequently, the 29-band camera lacked data from much of the near-infrared spectrum and the entire shortwave infrared spectrum. Given the importance of these bands for detecting changes to leaf cellular structure and water content, the difference in spectral coverage remains a plausible explanation for the difference in classification accuracy between the two sensors. Future research should aim to control these differences more carefully by using sensors covering identical portions of the electromagnetic spectrum, ideally in the same study area.
An important limitation of this study is the fact that the imagery was collected from two separate study areas with different field data collection procedures. Classification models using resampled imagery from Etnedal provided accuracies that were greater than the Nordskogen imagery but lesser than the original Etnedal imagery. Furthermore, only models including spectral bands and derivatives from the original Etnedal imagery provided greater accuracies than models from the resampled imagery. Models using spectral indices alone offered identical performance.
Given the difference in classification accuracy between the resampled imagery and the original 29-band imagery, site effects or field data collection discrepancies may have played a role in the accuracy differences observed between the study areas. The smaller size of the Etnedal study area could have also led to a greater spatial correlation between the trees in the training and testing samples, which could produce higher accuracies at the Etnedal site. Thus, while this study suggests that hyperspectral imagery with hundreds of bands may be more effective for RBSR detection than multi-spectral imagery or hyperspectral imagery with only 20–30 bands, this is not necessarily a universal rule. It may be worthwhile to test multi-spectral sensors in the future, provided the sensors cover the near-infrared and shortwave infrared portions of the spectrum. Our analysis of the effectiveness of vegetation indices for the classification of RBSR offers clues to potential physiological changes caused by RBSR infection. In the Etnedal study area, the Moisture Stress Index (MSI), Normalized Difference Infrared Index (NDII), and Enhanced Vegetation Index (EVI) were found to be the most important indices for detecting RBSR with SVM classification when only a single vegetation index was used. Both the MSI and NDII are strongly correlated with leaf moisture content, suggesting that RBSR infection could compromise water transport within infected trees. EVI is a greenness index that is strongly influenced by canopy structural variables such as leaf area. This suggests that RBSR is potentially altering the canopy of infected trees, possibly through lower foliage density. Such changes to canopy structure would be consistent with previous studies, which have found changes in crown density due to RBSR infection [
45,
46]. If so, these changes in canopy density could potentially be detected through the use of ALS data, and this should be tested in future research. ALS offers a greater potential to detect changes in canopy structure than spectral data and is suitable for both UAV platforms (for small areas) and airborne platforms for large-scale studies.
In the Nordskogen study area, MSI and NDII were not calculated due to the lack of necessary infrared bands. The most effective vegetation indices for detecting RBSR in this study area were the Normalized Difference Vegetation Index (NDVI) and the Red-Edge Normalized Difference Vegetation Index. These indices are influenced by leaf area and chlorophyll content, among other variables. This suggests that both foliar pigment content and total leaf area could be altered by RBSR infection, although additional research would be needed to assess the extent to which RBSR alters leaf pigments.
Statistical significance tests showed that most vegetation indices exhibited statistically significant differences between infected and healthy trees in the Etnedal site. These included indices of vegetation greenness such as NDVI, indices of water content such as NDII and Normalized Difference Water Index (NDWI), as well as measures of leaf pigments such as Anthocyanin Reflectance Index 2 (ARI2) and Carotenoid Reflectance Index 2 (CRI2). Interestingly, while ARI2 and CRI2 were statistically different between infected and healthy trees, ARI1 and CRI1 were not. One possible explanation is that ARI 2 and CRI2 are superior for detecting high values of anthocyanins and carotenoids, respectively [
38,
39]. A similar phenomenon occurred with leaf water content: NDII and NDWI showed statistical differences, while the Water Band Index (WBI) did not. Future remote sensing-based studies of plant health should therefore consider including two or three vegetation indices for each plant functional trait, rather than just one, as one vegetation index can detect a change in a specific plant pigment when another index does not. By contrast, vegetation indices in the Nordskogen site were generally not statistically different between the rot and rot-free classes. This could be due to the sensor used or confounding site factors.
Even though the accuracies obtained thus far are modest, there is still potential value in detecting RBSR with UAV-based sensors. In Norway, where RBSR fungi are native and rarely cause tree death or severe ecosystem disruption, knowing which specific trees are infected is likely less important than in a context where RBSR fungi are invasive (such as H. irregulare in Italy), and accurate detection of infected trees is of paramount importance to allow for severe control measures. Estimating the percentage of trees in a stand infested with RBSR could help forest owners optimize the timing of harvests to minimize losses from rot damage. While UAVs are expensive for use over large areas, their flexibility and low start-up cost make them potentially attractive for small forest owners. To determine the applicability of these methods for practical forestry, it will be necessary to test external classification models, i.e., models developed at one study site and applied to other study sites. If models developed at one site can be applied to others, then UAV-based imagery could offer a way to detect RBSR without fieldwork. In cases where RBSR is likely to cause tree death and/or severe ecological damage, greater classification accuracies than those obtained in this study may be necessary for the classifications to be useful. Classification accuracies will likely be higher in these contexts since rapid tree death caused by H. irregulare would be expected to provide a stronger spectral signature than the slow tree decline caused by RBSR species in Norway. Future research should test this hypothesis by evaluating the effectiveness of hyperspectral and multi-spectral imagery for detecting other RBSR species.