Prediction of Strawberry Dry Biomass from UAV Multispectral Imagery Using Multiple Machine Learning Methods
Abstract
:1. Introduction
2. Materials and Methods
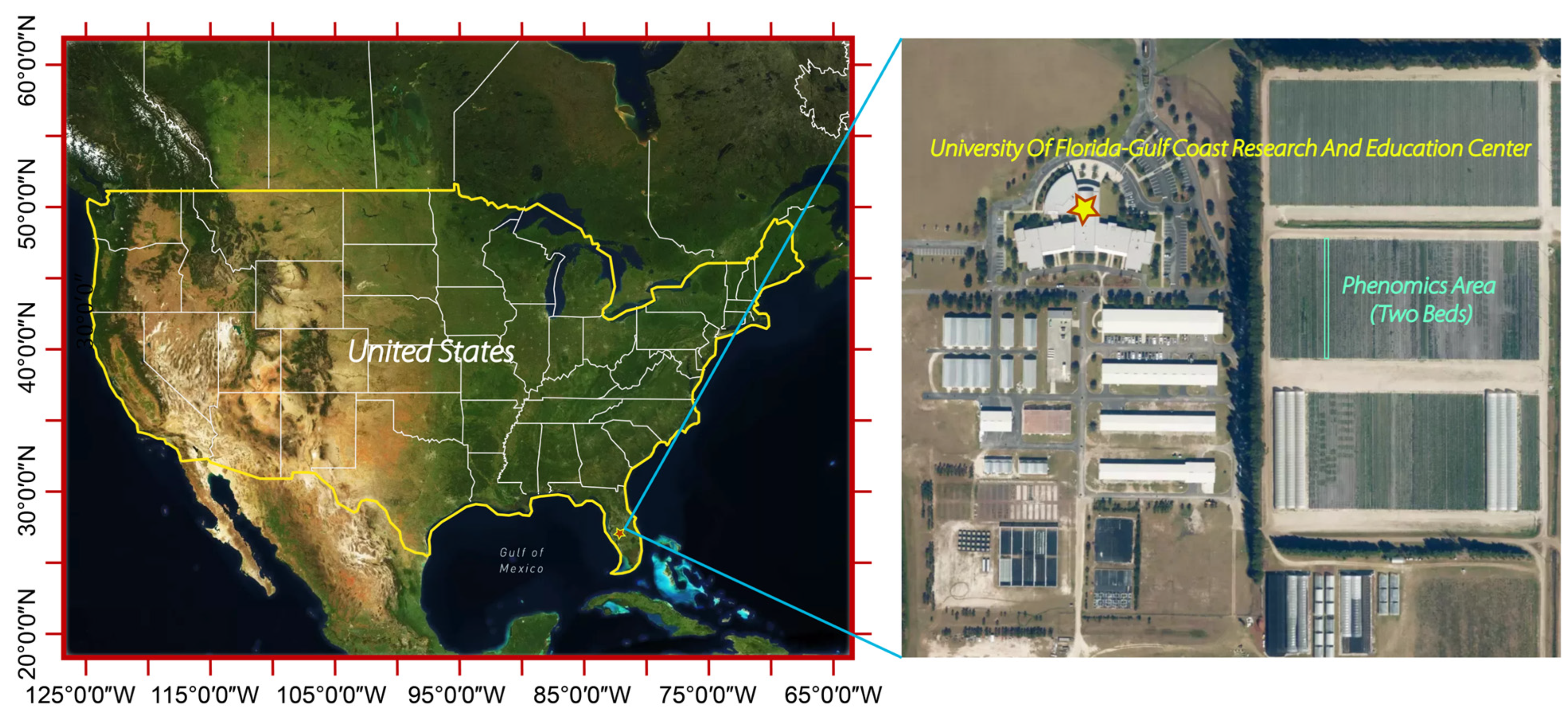
2.1. Study Site and Plant Materials
2.2. UAV Image Acquisition and Ground Control Points (GCP)
2.3. Ground-Based Image Collection
2.4. Experimental Workflow
2.4.1. UAV Image Preprocessing Using Agisoft Metashape Software
2.4.2. Canopy Delineation and Structural Parameter Extraction

2.4.3. Selection of Vegetation Indices
2.4.4. Biomass Modeling
- (1)
- Multiple linear regression (MLR) is a statistical method that uses several independent variables to predict the outcome of one dependent variable. The regression equation is designed to establish a linear relationship between the response variable with each independent variable and uses the least squares method to calculate the model parameters by minimizing the sum of squared errors [59].
- (2)
- Random forest (RF) is a supervised machine learning algorithm that uses an ensemble learning (bagging) strategy to solve classification and regression problems. Bagging, also referred to as bootstrap aggregation, is a sampling technique that generates a fixed number of subset training samples from the original dataset. The idea of random forest is to build multiple decision trees by selecting a random number of samples and features (bagging) during the training process and output the average prediction of all regression trees.
- (3)
- Support vector machine (SVM) is also a popular machine learning algorithm, widely used in pattern recognition, classification and prediction. There are two types of SVM, support vector classification (SVC) [60] for classification and support vector regression (SVR) [61]. The objective of SVM is to construct a hyperplane in a high-dimensional space that distinctly classifies the data points for classification or contains the largest number of points for regression tasks. The most important feature of SVM is a kernel function that maps the data from the original finite-dimensional space into a higher-dimensional space. In this work, we used SVR and selected the radial basis function (RBF) as the kernel.
- (4)
- Multivariate adaptive regression spline (MARS) is a non-parametric algorithm for nonlinear problems, introduced by Friedman [62]. It constructs a flexible prediction model by applying piecewise linear regressions. This means that various regression slopes are determined for each predictor’s interval. It consists of two steps: forward selection and backward deletion. MARS begins with a model with only an intercept term and iteratively adds basis functions in pairs to the model until a threshold of the residual error or number of terms is reached. Typically, the obtained model in this process (forward selection) has too many knots with high complexity, which leads to overfitting. Then, the knots that do not contribute significantly to the model performance are removed, which is also known as “pruning” (backward selection).
- (5)
- The eXtreme Gradient Boosting (XGBoost) algorithm is a scalable end-to-end tree boosting method introduced by Chen and Guestrin [63]. It uses a gradient boosting framework and performs a second-order Taylor expansion to optimize the objective function. A regularization term was also added to the objective function to control the model complexity and prevent model overfitting. Different from the RF model, which trains each tree independently, XGBoost grows each tree on the residuals of the previous tree. In addition, another advantage of XGBoost is its scalability in all scenarios, so it can solve the problems of sparse data.
- (6)
- Artificial neural networks (ANN) have become a hot research topic in the field of artificial intelligence. They simulate the working mechanism of neurons in the human brain and consist of one input layer, one output layer and one or more hidden layers. Each layer includes neurons, and each neuron has an activation function for introducing nonlinearity into relationships. A connection between two neurons represents the weighted value of the signal passing through that connection. ANN contains two phases: forward propagation and backward propagation. Forward propagation is the process of sequentially computing and storing intermediate variables (including outputs) from the input layer to the output layer. Backpropagation is a method of updating the weights in the model and requires an optimization function and a loss function.
2.4.5. Model Validation
3. Results
3.1. Comparison of Ground-Based and UAV Imagery
3.2. Modeling and Validation of Biomass
3.2.1. Variable Importance Evaluation
3.2.2. Performance of Prediction Methods in Strawberry Dry Biomass Prediction
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Hofius, D.; Börnke, F.A. Photosynthesis, carbohydrate metabolism and source–sink relations. In Potato Biology and Biotechnology; Elsevier: Amsterdam, The Netherlands, 2007; pp. 257–285. [Google Scholar]
- Yoo, C.G.; Pu, Y.; Ragauskas, A.J. Measuring Biomass-Derived Products in Biological Conversion and Metabolic Process. In Metabolic Pathway Engineering; Springer: Berlin/Heidelberg, Germany, 2020; pp. 113–124. [Google Scholar]
- Johansen, K.; Morton, M.; Malbeteau, Y.; Aragon Solorio, B.J.L.; Almashharawi, S.; Ziliani, M.; Angel, Y.; Fiene, G.; Negrão, S.; Mousa, M. Predicting biomass and yield at harvest of salt-stressed tomato plants using UAV imagery. Int. Arch. Photogramm. Remote Sens. Spatial Inf. Sci. 2019, XLII-2/W13, 407–411. [Google Scholar] [CrossRef]
- Bendig, J.; Yu, K.; Aasen, H.; Bolten, A.; Bennertz, S.; Broscheit, J.; Gnyp, M.L.; Bareth, G. Combining UAV-based plant height from crop surface models, visible, and near infrared vegetation indices for biomass monitoring in barley. Int. J. Appl. Earth Obs. Geoinf. 2015, 39, 79–87. [Google Scholar] [CrossRef]
- Hensgen, F.; Bühle, L.; Wachendorf, M. The effect of harvest, mulching and low-dose fertilization of liquid digestate on above ground biomass yield and diversity of lower mountain semi-natural grasslands. Agric. Ecosyst. Environ. 2016, 216, 283–292. [Google Scholar] [CrossRef]
- Yuan, M.; Burjel, J.; Isermann, J.; Goeser, N.; Pittelkow, C. Unmanned aerial vehicle–based assessment of cover crop biomass and nitrogen uptake variability. J. Soil Water Conserv. 2019, 74, 350–359. [Google Scholar] [CrossRef]
- Zheng, C.; Abd-Elrahman, A.; Whitaker, V. Remote sensing and machine learning in crop phenotyping and management, with an emphasis on applications in strawberry farming. Remote Sens. 2021, 13, 531. [Google Scholar] [CrossRef]
- Yang, K.-W.; Chapman, S.; Carpenter, N.; Hammer, G.; McLean, G.; Zheng, B.; Chen, Y.; Delp, E.; Masjedi, A.; Crawford, M. Integrating crop growth models with remote sensing for predicting biomass yield of sorghum. in silico Plants 2021, 3, diab001. [Google Scholar] [CrossRef]
- Catchpole, W.; Wheeler, C. Estimating plant biomass: A review of techniques. Aust. J. Ecol. 1992, 17, 121–131. [Google Scholar] [CrossRef]
- Wang, T.; Liu, Y.; Wang, M.; Fan, Q.; Tian, H.; Qiao, X.; Li, Y. Applications of UAS in crop biomass monitoring: A review. Front. Plant Sci. 2021, 12, 616689. [Google Scholar] [CrossRef]
- Chen, D.; Shi, R.; Pape, J.-M.; Neumann, K.; Arend, D.; Graner, A.; Chen, M.; Klukas, C. Predicting plant biomass accumulation from image-derived parameters. GigaScience 2018, 7, giy001. [Google Scholar] [CrossRef]
- Yang, S.; Feng, Q.; Liang, T.; Liu, B.; Zhang, W.; Xie, H. Modeling grassland above-ground biomass based on artificial neural network and remote sensing in the Three-River Headwaters Region. Remote Sens. Environ. 2018, 204, 448–455. [Google Scholar] [CrossRef]
- Chao, Z.; Liu, N.; Zhang, P.; Ying, T.; Song, K. Estimation methods developing with remote sensing information for energy crop biomass: A comparative review. Biomass Bioenergy 2019, 122, 414–425. [Google Scholar] [CrossRef]
- Réjou-Méchain, M.; Barbier, N.; Couteron, P.; Ploton, P.; Vincent, G.; Herold, M.; Mermoz, S.; Saatchi, S.; Chave, J.; De Boissieu, F. Upscaling forest biomass from field to satellite measurements: Sources of errors and ways to reduce them. Surv. Geophys. 2019, 40, 881–911. [Google Scholar] [CrossRef]
- Rodríguez-Veiga, P.; Quegan, S.; Carreiras, J.; Persson, H.J.; Fransson, J.E.; Hoscilo, A.; Ziółkowski, D.; Stereńczak, K.; Lohberger, S.; Stängel, M. Forest biomass retrieval approaches from earth observation in different biomes. Int. J. Appl. Earth Obs. Geoinf. 2019, 77, 53–68. [Google Scholar] [CrossRef]
- Santoro, M.; Cartus, O.; Carvalhais, N.; Rozendaal, D.; Avitabile, V.; Araza, A.; De Bruin, S.; Herold, M.; Quegan, S.; Rodríguez-Veiga, P. The global forest above-ground biomass pool for 2010 estimated from high-resolution satellite observations. Earth Syst. Sci. Data 2021, 13, 3927–3950. [Google Scholar] [CrossRef]
- Ahmad, A.; Gilani, H.; Ahmad, S.R. Forest aboveground biomass estimation and mapping through high-resolution optical satellite imagery—A literature review. Forests 2021, 12, 914. [Google Scholar] [CrossRef]
- Naik, P.; Dalponte, M.; Bruzzone, L. Prediction of Forest Aboveground Biomass Using Multitemporal Multispectral Remote Sensing Data. Remote Sens. 2021, 13, 1282. [Google Scholar] [CrossRef]
- Zheng, H.; Cheng, T.; Zhou, M.; Li, D.; Yao, X.; Tian, Y.; Cao, W.; Zhu, Y. Improved estimation of rice aboveground biomass combining textural and spectral analysis of UAV imagery. Precis. Agric. 2019, 20, 611–629. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Tian, Q.; Feng, H.; Xu, K.; Zhou, C. Estimate of winter-wheat above-ground biomass based on UAV ultrahigh-ground-resolution image textures and vegetation indices. ISPRS J. Photogramm. Remote Sens. 2019, 150, 226–244. [Google Scholar] [CrossRef]
- Toda, Y.; Kaga, A.; Kajiya-Kanegae, H.; Hattori, T.; Yamaoka, S.; Okamoto, M.; Iwata, H. Genomic prediction modeling of soybean biomass using UAV-based remote sensing and longitudinal model parameters. Plant Genome 2021, 14, e20157. [Google Scholar] [CrossRef]
- Geng, L.; Che, T.; Ma, M.; Tan, J.; Wang, H. Corn biomass estimation by integrating remote sensing and long-term observation data based on machine learning techniques. Remote Sens. 2021, 13, 2352. [Google Scholar] [CrossRef]
- Dong, T.; Liu, J.; Qian, B.; He, L.; Liu, J.; Wang, R.; Jing, Q.; Champagne, C.; McNairn, H.; Powers, J. Estimating crop biomass using leaf area index derived from Landsat 8 and Sentinel-2 data. ISPRS J. Photogramm. Remote Sens. 2020, 168, 236–250. [Google Scholar] [CrossRef]
- Stagakis, S.; Markos, N.; Sykioti, O.; Kyparissis, A. Monitoring canopy biophysical and biochemical parameters in ecosystem scale using satellite hyperspectral imagery: An application on a Phlomis fruticosa Mediterranean ecosystem using multiangular CHRIS/PROBA observations. Remote Sens. Environ. 2010, 114, 977–994. [Google Scholar] [CrossRef]
- Yang, G.; Liu, J.; Zhao, C.; Li, Z.; Huang, Y.; Yu, H.; Xu, B.; Yang, X.; Zhu, D.; Zhang, X. Unmanned aerial vehicle remote sensing for field-based crop phenotyping: Current status and perspectives. Front. Plant Sci. 2017, 8, 1111. [Google Scholar] [CrossRef] [PubMed]
- Tsouros, D.C.; Bibi, S.; Sarigiannidis, P.G. A review on UAV-based applications for precision agriculture. Information 2019, 10, 349. [Google Scholar] [CrossRef]
- Xie, C.; Yang, C. A review on plant high-throughput phenotyping traits using UAV-based sensors. Comput. Electron. Agric. 2020, 178, 105731. [Google Scholar] [CrossRef]
- Shi, Y.; Thomasson, J.A.; Murray, S.C.; Pugh, N.A.; Rooney, W.L.; Shafian, S.; Rajan, N.; Rouze, G.; Morgan, C.L.; Neely, H.L. Unmanned aerial vehicles for high-throughput phenotyping and agronomic research. PLoS ONE 2016, 11, e0159781. [Google Scholar] [CrossRef]
- Toth, C.; Jóźków, G. Remote sensing platforms and sensors: A survey. ISPRS J. Photogramm. Remote Sens. 2016, 115, 22–36. [Google Scholar] [CrossRef]
- Westoby, M.J.; Brasington, J.; Glasser, N.F.; Hambrey, M.J.; Reynolds, J.M. ‘Structure-from-Motion’photogrammetry: A low-cost, effective tool for geoscience applications. Geomorphology 2012, 179, 300–314. [Google Scholar] [CrossRef]
- De Souza, C.H.W.; Lamparelli, R.A.C.; Rocha, J.V.; Magalhães, P.S.G. Height estimation of sugarcane using an unmanned aerial system (UAS) based on structure from motion (SfM) point clouds. Int. J. Remote Sens. 2017, 38, 2218–2230. [Google Scholar] [CrossRef]
- Fraser, B.T.; Congalton, R.G. Issues in Unmanned Aerial Systems (UAS) data collection of complex forest environments. Remote Sens. 2018, 10, 908. [Google Scholar] [CrossRef] [Green Version]
- Fu, Y.; Yang, G.; Song, X.; Li, Z.; Xu, X.; Feng, H.; Zhao, C. Improved estimation of winter wheat aboveground biomass using multiscale textures extracted from UAV-based digital images and hyperspectral feature analysis. Remote Sens. 2021, 13, 581. [Google Scholar] [CrossRef]
- Johansen, K.; Morton, M.J.; Malbeteau, Y.; Aragon, B.; Al-Mashharawi, S.; Ziliani, M.G.; Angel, Y.; Fiene, G.; Negrão, S.; Mousa, M.A. Predicting biomass and yield in a tomato phenotyping experiment using UAV imagery and random forest. Front. Artif. Intell. 2020, 3, 28. [Google Scholar] [CrossRef] [PubMed]
- Wang, F.; Yang, M.; Ma, L.; Zhang, T.; Qin, W.; Li, W.; Zhang, Y.; Sun, Z.; Wang, Z.; Li, F. Estimation of Above-Ground Biomass of Winter Wheat Based on Consumer-Grade Multi-Spectral UAV. Remote Sens. 2022, 14, 1251. [Google Scholar] [CrossRef]
- Che, S.; Du, G.; Wang, N.; He, K.; Mo, Z.; Sun, B.; Chen, Y.; Cao, Y.; Wang, J.; Mao, Y. Biomass estimation of cultivated red algae Pyropia using unmanned aerial platform based multispectral imaging. Plant Methods 2021, 17, 1–13. [Google Scholar] [CrossRef] [PubMed]
- Mutanga, O.; Skidmore, A.K. Narrow band vegetation indices overcome the saturation problem in biomass estimation. Int. J. Remote Sens. 2004, 25, 3999–4014. [Google Scholar] [CrossRef]
- Tesfaye, A.A.; Awoke, B.G. Evaluation of the saturation property of vegetation indices derived from sentinel-2 in mixed crop-forest ecosystem. Spat. Inf. Res 2021, 29, 109–121. [Google Scholar] [CrossRef]
- Guan, Z.; Abd-Elrahman, A.; Fan, Z.; Whitaker, V.M.; Wilkinson, B. Modeling strawberry biomass and leaf area using object-based analysis of high-resolution images. ISPRS J. Photogramm. Remote Sens. 2020, 163, 171–186. [Google Scholar] [CrossRef]
- Abd-Elrahman, A.; Guan, Z.; Dalid, C.; Whitaker, V.; Britt, K.; Wilkinson, B.; Gonzalez, A. Automated canopy delineation and size metrics extraction for strawberry dry weight modeling using raster analysis of high-resolution imagery. Remote Sens. 2020, 12, 3632. [Google Scholar] [CrossRef]
- Abd-Elrahman, A.; Pande-Chhetri, R.; Vallad, G. Design and development of a multi-purpose low-cost hyperspectral imaging system. Remote Sens. 2011, 3, 570–586. [Google Scholar] [CrossRef]
- Abd-Elrahman, A.; Sassi, N.; Wilkinson, B.; Dewitt, B. Georeferencing of mobile ground-based hyperspectral digital single-lens reflex imagery. J. Appl. Remote Sens. 2016, 10, 014002. [Google Scholar] [CrossRef] [Green Version]
- Carlson, T.N.; Ripley, D.A. On the relation between NDVI, fractional vegetation cover, and leaf area index. Remote Sens. Environ. 1997, 62, 241–252. [Google Scholar] [CrossRef]
- Roujean, J.-L.; Breon, F.-M. Estimating PAR absorbed by vegetation from bidirectional reflectance measurements. Remote Sens. Environ. 1995, 51, 375–384. [Google Scholar] [CrossRef]
- Banerjee, B.P.; Sharma, V.; Spangenberg, G.; Kant, S. Machine learning regression analysis for estimation of crop emergence using multispectral UAV imagery. Remote Sens. 2021, 13, 2918. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N. Remote sensing of chlorophyll concentration in higher plant leaves. Adv. Space Res. 1998, 22, 689–692. [Google Scholar] [CrossRef]
- Strong, C.J.; Burnside, N.G.; Llewellyn, D. The potential of small-Unmanned Aircraft Systems for the rapid detection of threatened unimproved grassland communities using an Enhanced Normalized Difference Vegetation Index. PLoS ONE 2017, 12, e0186193. [Google Scholar] [CrossRef]
- Motohka, T.; Nasahara, K.N.; Oguma, H.; Tsuchida, S. Applicability of green-red vegetation index for remote sensing of vegetation phenology. Remote Sens. 2010, 2, 2369–2387. [Google Scholar] [CrossRef]
- Liu, X.; Wang, L. Feasibility of using consumer-grade unmanned aerial vehicles to estimate leaf area index in mangrove forest. Remote Sens. Lett. 2018, 9, 1040–1049. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Pattey, E.; Zarco-Tejada, P.J.; Strachan, I.B. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Wang, F.-M.; Huang, J.-F.; Tang, Y.-L.; Wang, X.-Z. New vegetation index and its application in estimating leaf area index of rice. Rice Sci. 2007, 14, 195–203. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Gitelson, A.A. Wide dynamic range vegetation index for remote quantification of biophysical characteristics of vegetation. J. Plant Physiol. 2004, 161, 165–173. [Google Scholar] [CrossRef] [PubMed]
- Jordan, C.F. Derivation of leaf-area index from quality of light on the forest floor. Ecology 1969, 50, 663–666. [Google Scholar] [CrossRef]
- Santos, A.; Lacerda, L.; Gobbo, S.; Tofannin, A.; Silva, R.; Vellidis, G. Using remote sensing to map in-field variability of peanut maturity. Precis. Agric. 2019, 19, 91–101. [Google Scholar]
- Chen, J.M. Evaluation of vegetation indices and a modified simple ratio for boreal applications. Can. J. Remote Sens. 1996, 22, 229–242. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Gritz, Y.; Merzlyak, M.N. Relationships between leaf chlorophyll content and spectral reflectance and algorithms for non-destructive chlorophyll assessment in higher plant leaves. J. Plant Physiol. 2003, 160, 271–282. [Google Scholar] [CrossRef]
- Thompson, C.N.; Guo, W.; Sharma, B.; Ritchie, G.L. Using normalized difference red edge index to assess maturity in cotton. Crop Sci. 2019, 59, 2167–2177. [Google Scholar] [CrossRef]
- Uyanık, G.K.; Güler, N. A study on multiple linear regression analysis. Procedia-Soc. Behav. Sci. 2013, 106, 234–240. [Google Scholar] [CrossRef]
- Boser, B.E.; Guyon, I.M.; Vapnik, V.N. A training algorithm for optimal margin classifiers. In Proceedings of the Fifth Annual Workshop on Computational Learning Theory, Pittsburgh, PA, USA, 27–29 July 1992; pp. 144–152. [Google Scholar]
- Drucker, H.; Burges, C.J.; Kaufman, L.; Smola, A.; Vapnik, V. Support vector regression machines. Adv. Neural Inf. Processing Syst. 1996, 9, 155–161. [Google Scholar]
- Friedman, J.H. Multivariate adaptive regression splines. Ann. Stat. 1991, 19, 1–67. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. Xgboost: A scalable tree boosting system. In Proceedings of the 22nd Acm Sigkdd International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Yue, J.; Feng, H.; Yang, G.; Li, Z. A comparison of regression techniques for estimation of above-ground winter wheat biomass using near-surface spectroscopy. Remote Sens. 2018, 10, 66. [Google Scholar] [CrossRef]
- Xue, J.; Su, B. Significant remote sensing vegetation indices: A review of developments and applications. J. Sens. 2017, 2017, 1353691. [Google Scholar] [CrossRef]
- Lee, H.; Wang, J.; Leblon, B. Using linear regression, random forests, and support vector machine with unmanned aerial vehicle multispectral images to predict canopy nitrogen weight in corn. Remote Sens. 2020, 12, 2071. [Google Scholar] [CrossRef]
- Maimaitijiang, M.; Sagan, V.; Sidike, P.; Hartling, S.; Esposito, F.; Fritschi, F.B. Soybean yield prediction from UAV using multimodal data fusion and deep learning. Remote Sens. Environ. 2020, 237, 111599. [Google Scholar] [CrossRef]
- Santos, A.F.; Lacerda, L.N.; Rossi, C.; Moreno, L.d.A.; Oliveira, M.F.; Pilon, C.; Silva, R.P.; Vellidis, G. Using UAV and Multispectral Images to Estimate Peanut Maturity Variability on Irrigated and Rainfed Fields Applying Linear Models and Artificial Neural Networks. Remote Sens. 2021, 14, 93. [Google Scholar] [CrossRef]
- Wu, C.; Niu, Z.; Tang, Q.; Huang, W. Estimating chlorophyll content from hyperspectral vegetation indices: Modeling and validation. Agric. For. Meteorol. 2008, 148, 1230–1241. [Google Scholar] [CrossRef]
- Xie, Q.; Dash, J.; Huang, W.; Peng, D.; Qin, Q.; Mortimer, H.; Casa, R.; Pignatti, S.; Laneve, G.; Pascucci, S. Vegetation indices combining the red and red-edge spectral information for leaf area index retrieval. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2018, 11, 1482–1493. [Google Scholar] [CrossRef]
- Yue, J.; Yang, G.; Feng, H. Comparative of remote sensing estimation models of winter wheat biomass based on random forest algorithm. Trans. Chin. Soc. Agric. Eng. 2016, 32, 175–182. [Google Scholar]
- Yuan, H.; Yang, G.; Li, C.; Wang, Y.; Liu, J.; Yu, H.; Feng, H.; Xu, B.; Zhao, X.; Yang, X. Retrieving soybean leaf area index from unmanned aerial vehicle hyperspectral remote sensing: Analysis of RF, ANN, and SVM regression models. Remote Sens. 2017, 9, 309. [Google Scholar] [CrossRef]
- Guan, Z.; Abd-Elrahman, A.; Whitaker, V.; Agehara, S.; Wilkinson, B.; Gastellu-Etchegorry, J.P.; Dewitt, B. Radiative transfer image simulation using L-system modeled strawberry canopies. Remote Sens. 2022, 14, 548. [Google Scholar] [CrossRef]








| Code | Genotype | Dry Biomass | |||
|---|---|---|---|---|---|
| Mean | Max | Min | Std | ||
| A | 15.25–6 | 20.45 | 37 | 2.70 | 10.32 |
| B | 15.74–11 | 16.40 | 38 | 1.60 | 9.72 |
| C | 16.30–128 | 17.46 | 32 | 3.30 | 9.27 |
| D | Brilliance | 24.15 | 42 | 1.80 | 12.49 |
| E | 13.42–5 | 16.97 | 42 | 1.80 | 10.29 |
| F | 16.33–8 | 31.80 | 66 | 6.40 | 15.52 |
| G | 17.14–155 | 28.82 | 61 | 6.40 | 14.16 |
| H | 17.17–127 | 27.04 | 59 | 5.30 | 13.88 |
| I | Beauty | 19.64 | 43 | 2.70 | 11.73 |
| J | Elyana | 36.98 | 80 | 9.50 | 17.22 |
| K | Festival | 43.20 | 109 | 6.90 | 26.70 |
| L | 18.19–294 | 30.12 | 68 | 3.60 | 17.17 |
| M | Florida127 | 47.25 | 123 | 6.70 | 31.48 |
| N | Fronteras | 35.70 | 89 | 6.80 | 20.32 |
| O | Radiance | 28.21 | 72 | 2.10 | 18.22 |
| P | Treasure | 41.90 | 80 | 3.90 | 23.72 |
| Q | Winterstar | 27.26 | 60 | 1.50 | 18.17 |
| VIs | Equation | Reference |
|---|---|---|
| Normalized Difference Vegetation Index (NDVI) | [43] | |
| Renormalized Difference Vegetation Index (RDVI) | [44] | |
| Blue Normalized Difference Vegetation Index (BNDVI) | [45] | |
| Green Normalized Difference Vegetation Index (GNDVI) | [46] | |
| NIR–Green Difference Vegetation Index (GDVI) | [47] | |
| Green–Red Vegetation Index (GRVI) | [48] | |
| Green Leaf Index (GLI) | [49] | |
| Modified Triangular Vegetation Index 1 (MTVI-1) | [50] | |
| Modified Triangular Vegetation Index 2 (MTVI-2) | [50] | |
| Pan Normalized Difference Vegetation Index (PNDVI) | [51] | |
| Enhanced Vegetation Index (EVI) | [52] | |
| Wide Dynamic Range Vegetation Index (WDRVI) | [53] | |
| Simple Ratio Index (SR) | [54] | |
| Red-Edge Simple Ratio Index (SRRedEdge) | [55] | |
| Modified Simple Ratio (MSR) | [56] | |
| Modified Simple Ratio Red-Edge (MSRRedEdge) | [56] | |
| Chlorophyll Index Red and Red-Edge (CIred&RE) | [57] | |
| Chlorophyll Index Green (CIgreen) | [57] | |
| Normalized Difference Red-Edge Index (NDRE) | [58] | |
| Normalized Difference Red-Edge/Red Index (NDRE-R) | [45] |
| Method | RF | SVM | ||||
| Parameters | Best_Mtry | Best_Ntree | Cost | Sigma | ||
| Min value | 1 | 100 | 1 | 0 | ||
| Max value | 20 | 600 | 10 | 1 | ||
| Method | MARS | ANN | ||||
| Parameters | nprune | degree | hidden layer 1 | hidden layer 2 | ||
| Min value | 10 | 1 | 1 | 1 | ||
| Max value | 100 | 5 | 10 | 10 | ||
| Method | XGBoost | |||||
| Parameters | eta | max_depth | min_child_weight | subsample | colsample_bytree | optimal_trees |
| Min value | 0 | 1 | 1 | 0.5 | 0.5 | 0 |
| Max value | 0.3 | 10 | 10 | 1 | 1 | 100 |
| Model | R2 | RMSE (g) | MAE (g) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Geometric | Geometric and VImean | Geometric and VIsum | Geometric | Geometric and VImean | Geometric and VIsum | Geometric | Geometric and VImean | Geometric and VIsum | |
| MLR | 0.78 | 0.80 | 0.85 | 9.37 | 8.85 | 7.54 | 6.66 | 6.40 | 5.33 |
| RF | 0.80 | 0.81 | 0.85 | 8.89 | 8.72 | 7.73 | 6.30 | 5.95 | 5.37 |
| SVM | 0.78 | 0.80 | 0.85 | 9.58 | 8.92 | 7.67 | 6.58 | 6.09 | 5.26 |
| XGBoost | 0.77 | 0.82 | 0.83 | 9.41 | 8.67 | 8.09 | 6.70 | 5.85 | 5.47 |
| MARS | 0.79 | 0.81 | 0.86 | 9.27 | 8.80 | 7.35 | 6.53 | 6.14 | 5.25 |
| ANN | 0.89 | 0.90 | 0.93 | 8.98 | 8.61 | 7.16 | 6.29 | 5.93 | 5.06 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Zheng, C.; Abd-Elrahman, A.; Whitaker, V.; Dalid, C. Prediction of Strawberry Dry Biomass from UAV Multispectral Imagery Using Multiple Machine Learning Methods. Remote Sens. 2022, 14, 4511. https://doi.org/10.3390/rs14184511
Zheng C, Abd-Elrahman A, Whitaker V, Dalid C. Prediction of Strawberry Dry Biomass from UAV Multispectral Imagery Using Multiple Machine Learning Methods. Remote Sensing. 2022; 14(18):4511. https://doi.org/10.3390/rs14184511
Chicago/Turabian StyleZheng, Caiwang, Amr Abd-Elrahman, Vance Whitaker, and Cheryl Dalid. 2022. "Prediction of Strawberry Dry Biomass from UAV Multispectral Imagery Using Multiple Machine Learning Methods" Remote Sensing 14, no. 18: 4511. https://doi.org/10.3390/rs14184511
APA StyleZheng, C., Abd-Elrahman, A., Whitaker, V., & Dalid, C. (2022). Prediction of Strawberry Dry Biomass from UAV Multispectral Imagery Using Multiple Machine Learning Methods. Remote Sensing, 14(18), 4511. https://doi.org/10.3390/rs14184511






