Unlocking the Potential of Deep Learning for Migratory Waterbirds Monitoring Using Surveillance Video
Abstract
:1. Introduction
2. Materials and Methods
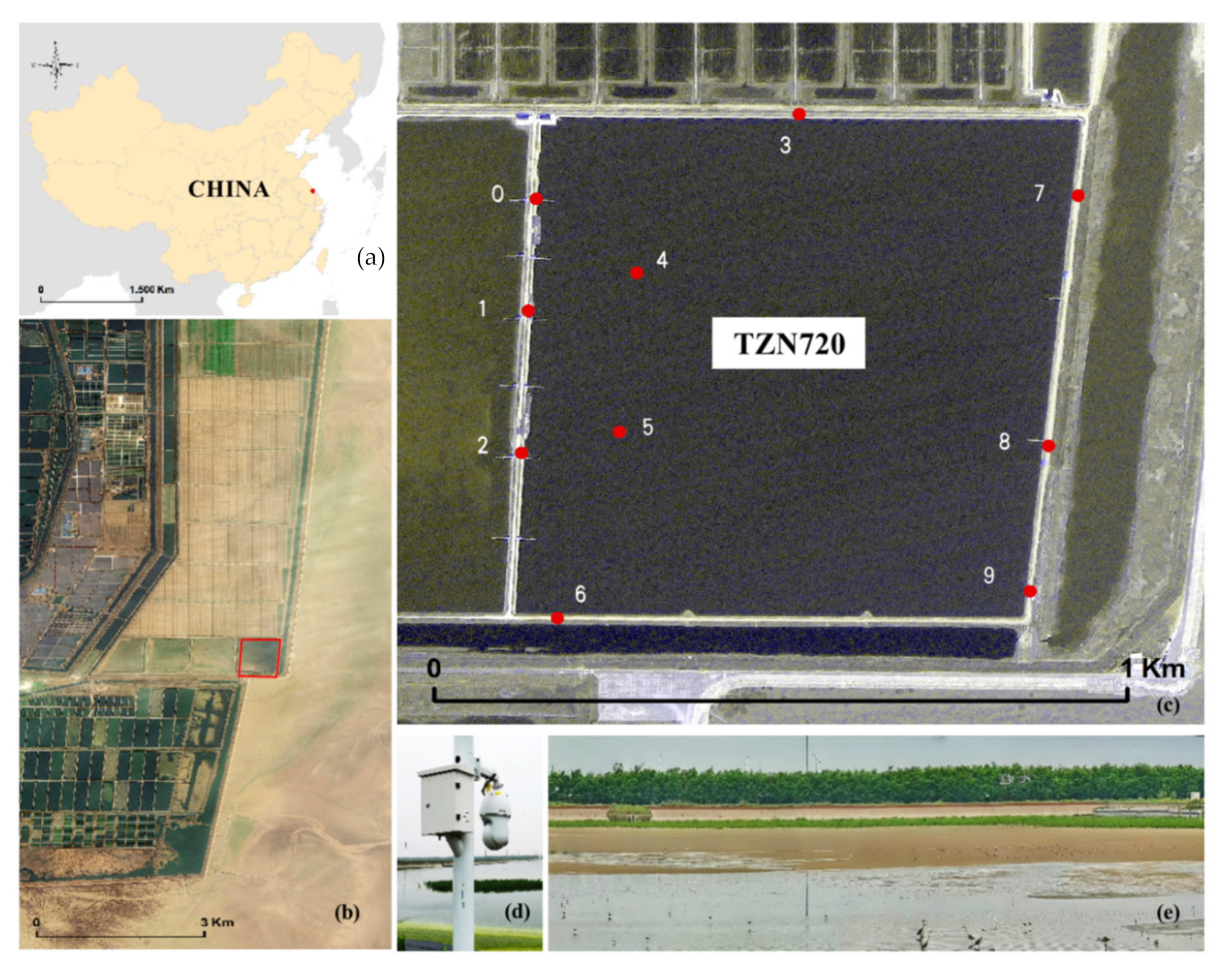
2.1. Study Site
2.2. Build a Bird Population Counting Dataset
2.2.1. Dataset Collection
2.2.2. Data Filtering
- A.
- Image resolution: Because the image features are not apparent, the image mosaic is incomplete, and some data are lost, so the images with a resolution lower than 4k in the dataset are removed.
- B.
- Image sharpness: Since the images collected by the camera in the rotation process are affected by motion blur, it is necessary to remove the samples with unclear photos in the data set.
- C.
- Data statistical characteristics: To ensure the rationality of data set distribution and the effectiveness of neural network training, it is necessary to reasonably allocate the number of samples with different number scales in the data set. We removed the pictures with less than 10 targets in the dataset and screened the images with a larger number of scales (ranging from 50 to 20,000) to ensure a reasonable distribution of the dataset.
2.2.3. Data Annotation
- A.
- Determining the minimum unit of image clipping: The input size of the density estimation module designed in this method is fixed at 1024 × 768, which can ensure the computational efficiency of the algorithm and retain the valuable feature information of the image as much as possible.
- B.
- Resizing the original image: The size of the original image is not regular. The length range is 4k~30k, and the width is 1k~1.2k. It cannot be cut into multiple 1024 × 768 images. Therefore, it is necessary to resize the original image and round it to a multiple of 1024 × 768.
- C.
- Clipping the resized image: After three steps of clipping preprocessing, the original dataset is clipped into many small images of 1024 × 768, which improves the labeling efficiency, and the labeled label information can be mapped to the original data through the corresponding relationship between file name and image size.
2.3. Build a Panorama Bird Population Counting Network
2.3.1. Image Clipping Module
2.3.2. Depth Density Estimation Network
2.4. Model Training
2.5. Performance Evaluation
2.6. Motivating Application: Exploring Daily Movement of Waterbirds by Using High-Frequency Population Counting
3. Results
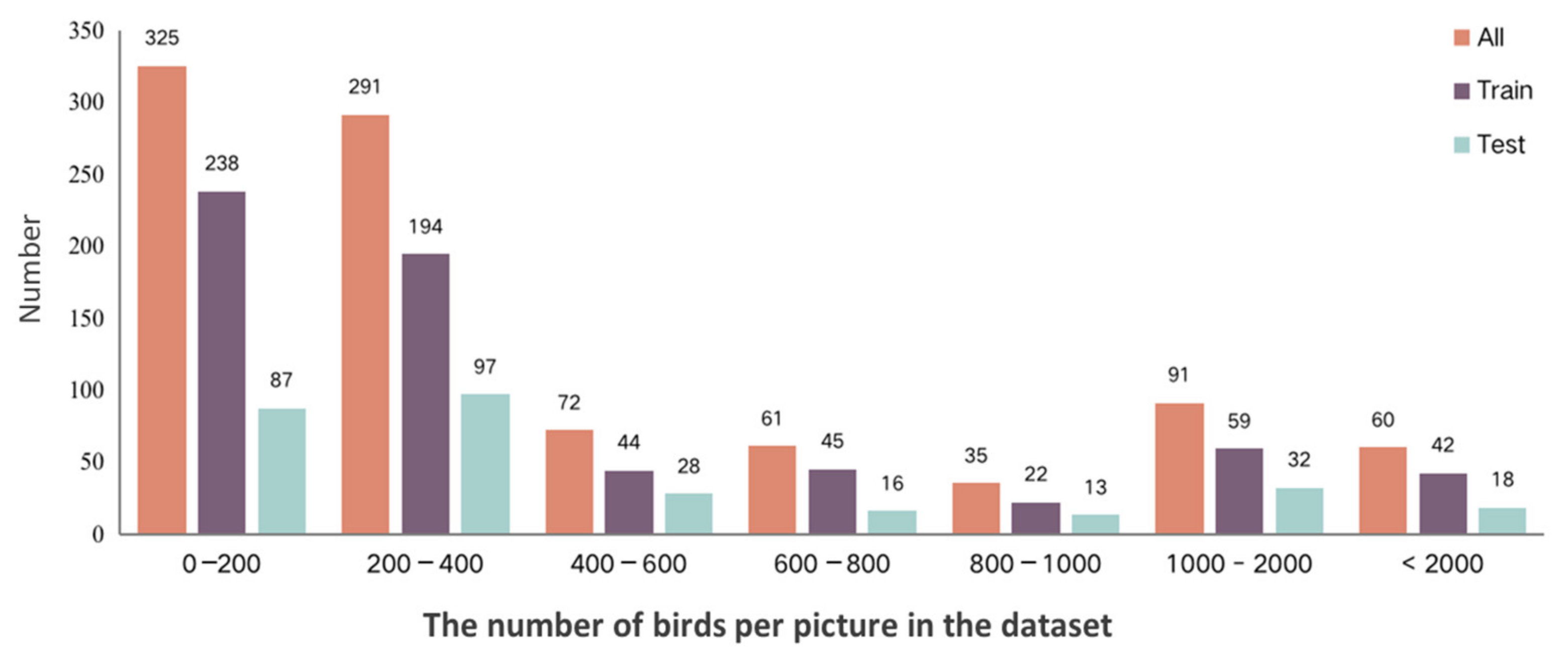
3.1. The Panoramic Bird Population Counting Dataset
3.2. Model Performance
3.3. Comparison with Other Deep Learning Algorithms
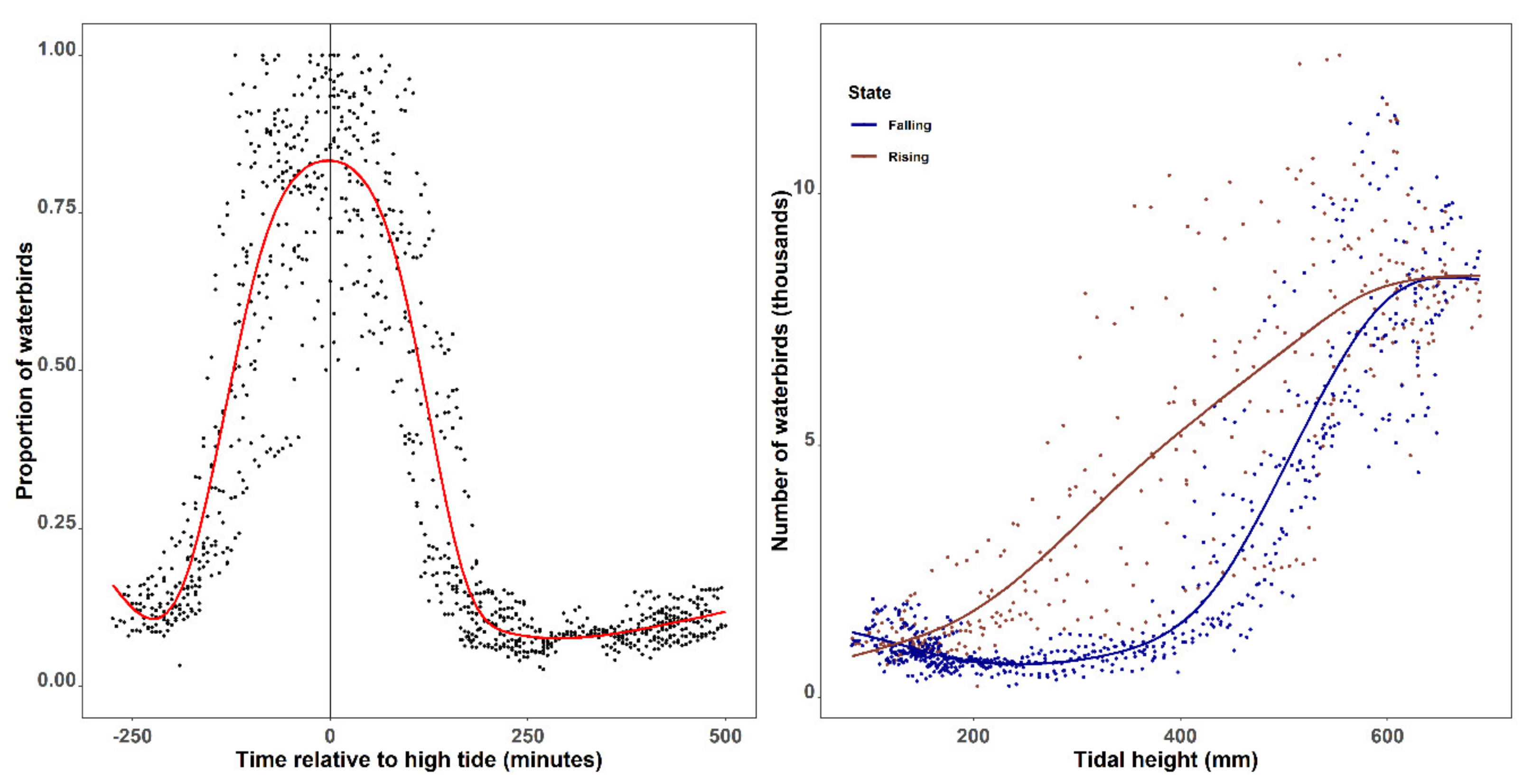
3.4. Tidal-Driven Waterbird Movement in Tiaozini
4. Discussion
4.1. Overall Accuracy and Innovation
4.2. Limitations and Prospects
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Conflicts of Interest
References
- Boitani, L.; Cowling, R.M.; Dublin, H.T.; Mace, G.M.; Parrish, J.; Possingham, H.P. Change the IUCN Protected Area Categories to Reflect Biodiversity Outcomes. PLoS Biol. 2008, 6, e66. [Google Scholar] [CrossRef] [Green Version]
- Myers, N.; Mittermeier, R.A.; Mittermeier, C.G.; Da Fonseca, G.A.B.; Kent, J. Biodiversity hotspots for conservation priorities. Nature 2000, 403, 853–858. [Google Scholar] [CrossRef]
- Albouy, C.; Delattre, V.L.; Mérigot, B.; Meynard, C.N.; Leprieur, F. Multifaceted biodiversity hotspots of marine mammals for conservation priorities. Divers. Distrib. 2017, 23, 615–626. [Google Scholar] [CrossRef] [Green Version]
- Nummelin, M.; Urho, U. International environmental conventions on biodiversity. In Oxford Research Encyclopedia of Environmental Science; Oxford University Press: Oxford, UK, 2018. [Google Scholar] [CrossRef]
- Kremen, C. Assessing the Indicator Properties of Species Assemblages for Natural Areas Monitoring. Ecol. Appl. 1992, 2, 203–217. [Google Scholar] [CrossRef] [Green Version]
- Edney, A.J.; Wood, M.J. Applications of digital imaging and analysis in seabird monitoring and research. Ibis 2021, 163, 317–337. [Google Scholar] [CrossRef]
- Sutherland, W.J.; Newton, I.; Green, R. Bird Ecology and Conservation: A Handbook of Techniques; OUP Oxford: Oxford, UK, 2004. [Google Scholar]
- Witmer, G.W. Wildlife population monitoring: Some practical considerations. Wildl. Res. 2005, 32, 259–263. [Google Scholar] [CrossRef]
- Lahoz-Monfort, J.J.; Magrath, M.J.L. A Comprehensive Overview of Technologies for Species and Habitat Monitoring and Conservation. BioScience 2021, 71, 1038–1062. [Google Scholar] [CrossRef] [PubMed]
- Kellenberger, B.; Veen, T.; Folmer, E.; Tuia, D. 21,000 birds in 4.5 h: Efficient large-scale seabird detection with machine learning. Remote Sens. Ecol. Conserv. 2021, 7, 445–460. [Google Scholar] [CrossRef]
- Lyons, M.B.; Brandis, K.J.; Murray, N.J.; Wilshire, J.H.; McCann, J.A.; Kingsford, R.T.; Callaghan, C.T. Monitoring large and complex wildlife aggregations with drones. Methods Ecol. Evol. 2019, 10, 1024–1035. [Google Scholar] [CrossRef] [Green Version]
- Zhao, P.; Liu, S.; Zhou, Y.; Lynch, T.; Lu, W.; Zhang, T.; Yang, H. Estimating animal population size with very high-resolution satellite imagery. Conserv. Biol. 2021, 35, 316–324. [Google Scholar] [CrossRef]
- Christin, S.; Hervet, É.; Lecomte, N. Applications for deep learning in ecology. Methods Ecol Evol. 2019, 10, 1632–1644. [Google Scholar] [CrossRef]
- Jarić, I.; Correia, R.A.; Brook, B.W.; Buettel, J.C.; Courchamp, F.; Di Minin, E.; Firth, J.A.; Gaston, K.J.; Jepson, P.; Kalinkat, G.; et al. iEcology: Harnessing large online resources to generate ecological insights. Trends Ecol. Evol. 2020, 35, 630–639. [Google Scholar] [CrossRef] [PubMed]
- Lopez-Vazquez, V.; Lopez-Guede, J.M.; Marini, S.; Fanelli, E.; Johnsen, E.; Aguzzi, J. Video image enhancement and machine learning pipeline for underwater animal detection and classification at cabled observatories. Sensors 2020, 20, 726. [Google Scholar] [CrossRef] [Green Version]
- Stewart, P.D.; Ellwood, S.A.; Macdonald, D.W. Remote video-surveillance of wildlife—An introduction from experience with the European badger Meles meles. Mammal Rev. 1997, 27, 185–204. [Google Scholar] [CrossRef]
- Elharrouss, O.; Almaadeed, N.; Al-Maadeed, S. A review of video surveillance systems. J. Vis. Commun. Image Represent 2021, 77, 103116. [Google Scholar] [CrossRef]
- Rasool, M.A.; Zhang, X.; Hassan, M.A.; Hussain, T.; Lu, C.; Zeng, Q.; Peng, B.; Wen, L.; Lei, G. Construct social-behavioral association network to study management impact on waterbirds community ecology using digital video recording cameras. Ecol. Evol. 2021, 11, 2321–2335. [Google Scholar] [CrossRef] [PubMed]
- Emogor, C.A.; Ingram, D.J.; Coad, L.; Worthington, T.A.; Dunn, A.; Imong, I.; Balmford, A. The scale of Nigeria’s involvement in the trans-national illegal pangolin trade: Temporal and spatial patterns and the effectiveness of wildlife trade regulations. Biol. Conserv. 2021, 264, 109365. [Google Scholar] [CrossRef]
- Edrén, S.M.C.; Teilmann, J.; Dietz, R. Effect from the Construction of Nysted Offshore Wind Farm on Seals in Rødsand Seal Sanctuary Based on Remote Video Monitoring; Ministry of the Environment: Copenhagen, Denmark, 2004; p. 21. [Google Scholar]
- Su, J.; Kurtek, S.; Klassen, E. Statistical analysis of trajectories on Riemannian manifolds: Bird migration, hurricane tracking and video surveillance. Ann. Appl. Stat. 2014, 8, 530–552. [Google Scholar] [CrossRef] [Green Version]
- Nassauer, A.; Legewie, N.M. Analyzing 21st Century Video Data on Situational Dynamics—Issues and Challenges in Video Data Analysis. Soc. Sci. 2019, 8, 100. [Google Scholar] [CrossRef] [Green Version]
- Pöysä, H.; Kotilainen, J.; Väänänen, V.M.; Kunnasranta, M. Estimating production in ducks: A comparison between ground surveys and unmanned aircraft surveys. Eur. J. Wildl. Res. 2018, 64, 1–4. [Google Scholar] [CrossRef] [Green Version]
- Olden, J.D.; Lawler, J.J.; Poff, N.L. Machine learning methods without tears: A primer for ecologists. Q. Rev. Biol. 2016, 83, 171–193. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Shao, Q.Q.; Guo, X.J.; Li, Y.Z.; Wang, Y.C.; Wang, D.L.; Liu, J.Y.; Yang, F. Using UAV remote sensing to analyze the population and distribution of large wild herbivores. J. Remote Sens. 2018, 22, 497–507. [Google Scholar] [CrossRef]
- Goodwin, M.; Halvorsen, K.T.; Jiao, L.; Knausgård, K.M.; Martin, A.H.; Moyano, M.; Oomen, R.A.; Rasmussen, J.H.; Sørdalen, T.K.; Thorbjørnsen, S.H. Unlocking the potential of deep learning for marine ecology: Overview, applications, and outlook. arXiv 2021, arXiv:2109.14737. [Google Scholar] [CrossRef]
- Lamba, A.; Cassey, P.; Segaran, R.R.; Koh, L.P. Deep learning for environmental conservation. Curr. Biol. 2019, 29, R977–R982. [Google Scholar] [CrossRef]
- Weinstein, B.G. A computer vision for animal ecology. J. Anim. Ecol. 2018, 87, 533–545. [Google Scholar] [CrossRef]
- Wäldchen, J.; Mäder, P. Machine learning for image based species identification. Methods Ecol. Evol. 2018, 9, 2216–2225. [Google Scholar] [CrossRef]
- Egnor, S.R.; Branson, K. Computational analysis of behavior. Annu. Rev. Neurosci. 2016, 39, 217–236. [Google Scholar] [CrossRef]
- Gunasekaran, H.; Ramalakshmi, K.; Rex Macedo Arokiaraj, A.; Deepa Kanmani, S.; Venkatesan, C.; Suresh Gnana Dhas, C. Analysis of DNA Sequence Classification Using CNN and Hybrid Models. Comput. Math. Methods Med. 2021, 2021, 1835056. [Google Scholar] [CrossRef]
- Lei, J.; Jia, Y.; Zuo, A.; Zeng, Q.; Shi, L.; Zhou, Y.; Zhang, H.; Lu, C.; Lei, G.; Wen, L. Bird satellite tracking revealed critical protection gaps in East Asian–Australasian Flyway. Int. J. Environ. Res. Public Health 2019, 16, 1147. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Runge, C.A.; Watson, J.E.; Butchart, S.H.; Hanson, J.O.; Possingham, H.P.; Fuller, R.A. Protected areas and global conservation of migratory birds. Science 2015, 350, 1255–1258. [Google Scholar] [CrossRef] [Green Version]
- Yong, D.L.; Jain, A.; Liu, Y.; Iqbal, M.; Choi, C.Y.; Crockford, N.J.; Millington, S.; Provencher, J. Challenges and opportunities for transboundary conservation of migratory birds in the East Asian-Australasian flyway. Conserv. Biol. 2018, 32, 740–743. [Google Scholar] [CrossRef]
- Amano, T.; Székely, T.; Koyama, K.; Amano, H.; Sutherland, W.J. A framework for monitoring the status of populations: An example from wader populations in the East Asian–Australasian flyway. Biol. Conserv. 2010, 143, 2238–2247. [Google Scholar] [CrossRef]
- Runge, C.A.; Martin, T.G.; Possingham, H.P. Conserving mobile species. Front. Ecol. Environ. 2014, 12, 395–402. [Google Scholar] [CrossRef] [Green Version]
- Peele, A.M.; Marra, P.M.; Sillett, T.S.; Sherry, T.W. Combining survey methods to estimate abundance and transience of migratory birds among tropical nonbreeding habitats. Auk Ornithol. Adv. 2015, 132, 926–937. [Google Scholar] [CrossRef]
- Sindagi, V.A.; Patel, V.M. A survey of recent advances in cnn-based single image crowd counting and density estimation. Pattern Recognit. Lett. 2018, 107, 3–16. [Google Scholar] [CrossRef] [Green Version]
- Akçay, H.G.; Kabasakal, B.; Aksu, D.; Demir, N.; Öz, M.; Erdoğan, A. Automated Bird Counting with Deep Learning for Regional Bird Distribution Mapping. Animals 2020, 10, 1207. [Google Scholar] [CrossRef]
- Liu, Y.; Wang, S. A quantitative detection algorithm based on improved faster R-CNN for marine benthos. Ecol. Inform. 2021, 61, 101228. [Google Scholar] [CrossRef]
- Peng, J.; Wang, D.; Liao, X.; Shao, Q.; Sun, Z.; Yue, H.; Ye, H. Wild animal survey using UAS imagery and deep learning: Modified Faster R-CNN for kiang detection in Tibetan Plateau. ISPRS J. Photogramm. Remote Sens. 2020, 169, 364–376. [Google Scholar] [CrossRef]
- Wang, C.; Wang, G.; Dai, L.; Liu, H.; Li, Y.; Zhou, Y.; Chen, H.; Dong, B.; Lv, S.; Zhao, Y. Diverse usage of shorebirds habitats and spatial management in Yancheng coastal wetlands. Ecol. Indic. 2020, 117, 106583. [Google Scholar] [CrossRef]
- Peng, H.-B.; Anderson, G.Q.A.; Chang, Q.; Choi, C.-Y.; Chowdhury, S.U.; Clark, N.A.; Zöckler, C. The intertidal wetlands of southern Jiangsu Province, China—Globally important for Spoon-billed Sandpipers and other threatened shorebirds, but facing multiple serious threats. Bird Conserv. Int. 2017, 27, 305–322. [Google Scholar] [CrossRef] [Green Version]
- Peng, H.-B.; Choi, C.-Y.; Zhang, L.; Gan, X.; Liu, W.-L.; Li, J.; Ma, Z.-J. Distribution and conservation status of the Spoon-billed Sandpipers in China. Chin. J. Zool. 2017, 52, 158–166. [Google Scholar]
- Jackson, M.V.; Carrasco, L.R.; Choi, C.; Li, J.; Ma, Z.; Melville, D.S.; Mu, T.; Peng, H.; Woodworth, B.K.; Yang, Z.; et al. Multiple habitat use by declining migratory birds necessitates joined-up conservation. Ecol. Evol. 2019, 9, 2505–2515. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Harris, C.R.; Millman, K.J.; van der Walt, S.J.; Gommers, R.; Virtanen, P.; Cournapeau, D.; Wieser, E.; Taylor, J.; Berg, S.; Smith, N.J.; et al. Array programming with NumPy. Nature 2020, 585, 357–362. [Google Scholar] [CrossRef]
- Culjak, I.; Abram, D.; Pribanic, T.; Dzapo, H.; Cifrek, M. A brief introduction to OpenCV. IEEE Int. Conv. MIPRO 2012, 35, 1725–1730. [Google Scholar]
- Myers, J.P.; Myers, L.P. Waterbirds of coastal Buenos Aires Province, Argentina. Ibis 1979, 121, 186–200. [Google Scholar] [CrossRef]
- He, K.; Zhang, X.; Ren, S.; Sun, J. Deep residual learning for image recognition. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition, Honolulu, HI, USA, 26 June–1 July 2016; pp. 770–778. [Google Scholar]
- Simonyan, K.; Zisserman, A. Very deep convolutional networks for large-scale image recognition. arXiv 2014, arXiv:1409.1556. [Google Scholar]
- Krizhevsky, A.; Sutskever, I.; Hinton, G.E. Imagenet classification with deep convolutional neural networks. Adv. Neural Inf. Processing Syst. 2012, 25, 1097–1105. [Google Scholar] [CrossRef]
- Wang, Y.; Song, R.; Wei, X.S.; Zhang, L. An adversarial domain adaptation network for cross-domain fine-grained recognition. In Proceedings of the IEEE/CVF Winter Conference on Applications of Computer Vision (WACV 2020), Snowmass Village, CO, USA, 2–5 May 2020; pp. 1228–1236. [Google Scholar]
- Zhang, Y.; Zhou, D.; Chen, S.; Gao, S.; Ma, Y. single-image crowd counting via multi-column convolutional neural network. In Proceedings of the 2016 IEEE Conference on Computer Vision and Pattern Recognition (2016 CVPR), Las Vegas, NV, USA, 26 June–1 July 2016; pp. 589–597. [Google Scholar]
- Zeng, L.; Xu, X.; Cai, B.; Qiu, S.; Zhang, T. Multi-scale convolutional neural networks for crowd counting. In Proceedings of the 2017 IEEE International Conference on Image Processing (2017 ICIP), Beijing, China, 24–28 September 2017; pp. 465–469. [Google Scholar]
- Liu, W.; Salzmann, M.; Fua, P. Context-aware crowd counting. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition (2019 CVPR), Long Beach, CA, USA, 15–21 June 2019; pp. 5099–5108. [Google Scholar]
- Wei, B.; Yuan, Y.; Wang, Q. MSPNET: Multi-supervised parallel network for crowd counting. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (2020 ICASSP), Barcelona, Spain, 4–7 May 2020; pp. 2418–2422. [Google Scholar]
- Cao, X.; Wang, Z.; Zhao, Y.; Su, F. Scale aggregation network for accurate and efficient crowd counting. In Proceedings of the European Conference on Computer Vision (2018 ECCV), Munich, Germany, 8–14 August 2018; pp. 734–750. [Google Scholar]
- Li, Y.; Zhang, X.; Chen, D. Csrnet: Dilated convolutional neural networks for understanding the highly congested scenes. In Proceedings of the IEEE Conference on Computer Vision and Pattern Recognition (2018 CVPR), Salt Lake City, UT, USA, 19–21 June 2018; pp. 1091–1100. [Google Scholar]
- Granadeiro, J.P.; Dias, M.P.; Martins, R.C.; Palmeirim, J.M. Variation in numbers and behaviour of waders during the tidal cycle: Implications for the use of estuarine sediment flats. Acta Oecol. 2016, 29, 293–300. [Google Scholar] [CrossRef]
- Connors, P.G.; Myers, J.P.; Connors, C.S.; Pitelka, F.A. Interhabitat movements by Sanderlings in relation to foraging profitability and the tidal cycle. Auk 1981, 98, 49–64. [Google Scholar] [CrossRef]
- The China Maritime Safety Administration. Available online: https://www.cnss.com.cn/tide/ (accessed on 15 October 2021).
- Wilke, C.O. cowplot: Streamlined Plot Theme and Plot Annotations for ‘ggplot2’; R Package Version 0.9, 4; R Project: Vienna, Austria, 2019. [Google Scholar]
- Wickham, H. ggplot2. Comput. Stat. 2011, 3, 180–185. [Google Scholar] [CrossRef]
- Neuwirth, E.; Neuwirth, M.E. Package ‘RColorBrewer’; CRAN 2011-06-17 08: 34: 00. Apache License 2.0; R Project: Vienna, Austria, 2018. [Google Scholar]
- Chabot, D.; Francis, C.M. Computer-automated bird detection and counts in high-resolution aerial images: A review. J. Field Ornithol. 2016, 87, 343–359. [Google Scholar] [CrossRef]
- Hou, J.; He, Y.; Yang, H.; Connor, T.; Gao, J.; Wang, Y.; Zhou, S. Identification of animal individuals using deep learning: A case study of giant panda. Biol. Conserv. 2020, 242, 108–414. [Google Scholar] [CrossRef]
- Hodgson, J.C.; Mott, R.; Baylis, S.M.; Pham, T.T.; Wotherspoon, S.; Kilpatrick, A.D.; Raja Segaran, R.; Reid, I.; Terauds, A.; Koh, L. Drones count wildlife more accurately and precisely than humans. Methods Ecol. Evol. 2018, 9, 1160–1167. [Google Scholar] [CrossRef] [Green Version]
- McClure, E.C.; Sievers, M.; Brown, C.J.; Buelow, C.A.; Ditria, E.M.; Hayes, M.A.; Pearson, R.M.; Tulloch, V.J.; Unsworth, R.K. Connolly, R.M. Artificial intelligence meets citizen science to supercharge ecological monitoring. Patterns 2020, 1, 100109. [Google Scholar] [CrossRef] [PubMed]
- Isola, P.; Zhu, J.Y.; Zhou, T.; Efros, A.A. Image-to-image translation with conditional adversarial networks. In Proceedings of the IEEE conference on computer vision and pattern recognition (2017 CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 1125–1134. [Google Scholar]
- Dujon, A.M.; Schofield, G. Importance of machine learning for enhancing ecological studies using information-rich imagery. Endangered Species Research. 2019, 39, 91–104. [Google Scholar] [CrossRef] [Green Version]
- Xu, M.X. An overview of image recognition technology based on deep learning. Comput. Prod. Circ. 2019, 1, 213. [Google Scholar]
- Sindagi, V.A.; Patel, V.M. Generating high-quality crowd density maps using contextual pyramid cnns. In Proceedings of the IEEE International Conference on Computer Vision (ICCV 2017), Venice, Italy, 22–29 October 2017; pp. 1879–1888. [Google Scholar]
- Shen, Z.; Xu, Y.; Ni, B.; Wang, M.; Hu, J.; Yang, X. Crowd counting via adversarial cross-scale consistency pursuit. In Proceedings of the IEEE conference on computer vision and pattern recognition (2018 CVPR), Salt Lake City, UT, USA, 19–21 June 2018; pp. 5245–5254. [Google Scholar]
- Huang, S.; Li, X.; Cheng, Z.Q.; Zhang, Z.; Hauptmann, A. Stacked pooling for boosting scale invariance of crowd counting. In Proceedings of the ICASSP 2020-2020 IEEE International Conference on Acoustics, Speech and Signal Processing (ICASSP 2020), Barcelona, Spain, 4–7 May 2020; pp. 2578–2582. [Google Scholar]
- Babu Sam, D.; Surya, S.; Venkatesh Babu, R. Switching convolutional neural network for crowd counting. In Proceedings of the IEEE conference on computer vision and pattern recognition(2017 CVPR), Honolulu, HI, USA, 21–26 July 2017; pp. 5744–5752. [Google Scholar]
- Kahl, S.; Wood, C.M.; Eibl, M.; Klinck, H. BirdNET: A deep learning solution for avian diversity monitoring. Ecol. Inform. 2021, 61, 101236. [Google Scholar] [CrossRef]
- Ketkar, N.; Santana, E. Deep learning with Python; Apress: Berkeley, CA, USA, 2017; ISBN 978-1-4842-2766-4. [Google Scholar]
- Burton, N.H.K.; Musgrove, A.J.; Rehfisch, M.M. Tidal variation in numbers of waterbirds: How frequently should birds be counted to detect change and do low tide counts provide a realistic average? Bird Study 2004, 51, 48–57. [Google Scholar] [CrossRef]
- Borowiec, M.L.; Frandsen, P.; Dikow, R.; McKeeken, A.; Valentini, G.; White, A.E. Deep learning as a tool for ecology and evolution. EcoEvoRxiv 2021, 1–30. [Google Scholar] [CrossRef]
- Pimm, S.L.; Pimm, S.L. The Balance of Nature? Ecological Issues in the Conservation of Species and Communities; University of Chicago Press: Chicago, IL, USA, 1991. [Google Scholar]





| Density Map | Train | Test | All |
|---|---|---|---|
| 1024 × 768 | 1495.29 | 2252.23 | 3747.52 |
| 512 × 384 | 1495.32 | 2252.24 | 3747.56 |
| 256 × 192 | 1515.80 | 2264.97 | 3780.77 |
| 128 × 96 | 9226.60 | 6256.52 | 15,483.12 |
| 64 × 48 | 53,659.82 | 28,304.39 | 81,964.21 |
| 32 × 24 | 126,574.40 | 65,526.26 | 192,100.66 |
| Method | MAE | RMSE | Error Rate | Error Rate * | FPS |
|---|---|---|---|---|---|
| MCNN [53] | 253.89 | 751.34 | 29.72% | 21.99% | 9.57 |
| MSCNN [54] | 330.26 | 817.51 | 38.66% | 19.31% | 1.91 |
| ECAN [55] | 322.49 | 804.82 | 37.75% | 14.35% | 1.35 |
| MSPNet [56] | 257.30 | 747.76 | 30.12% | 16.25% | 2.41 |
| SANet [57] | 207.16 | 789.65 | 24.25% | 15.43% | 2.45 |
| CSRNet [58] | 191.02 | 720.80 | 22.36% | 15.56% | 3.87 |
| ResNet34 [49] | 120.86 | 599.74 | 14.14% | -- | 2.12 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Wu, E.; Wang, H.; Lu, H.; Zhu, W.; Jia, Y.; Wen, L.; Choi, C.-Y.; Guo, H.; Li, B.; Sun, L.; et al. Unlocking the Potential of Deep Learning for Migratory Waterbirds Monitoring Using Surveillance Video. Remote Sens. 2022, 14, 514. https://doi.org/10.3390/rs14030514
Wu E, Wang H, Lu H, Zhu W, Jia Y, Wen L, Choi C-Y, Guo H, Li B, Sun L, et al. Unlocking the Potential of Deep Learning for Migratory Waterbirds Monitoring Using Surveillance Video. Remote Sensing. 2022; 14(3):514. https://doi.org/10.3390/rs14030514
Chicago/Turabian StyleWu, Entao, Hongchang Wang, Huaxiang Lu, Wenqi Zhu, Yifei Jia, Li Wen, Chi-Yeung Choi, Huimin Guo, Bin Li, Lili Sun, and et al. 2022. "Unlocking the Potential of Deep Learning for Migratory Waterbirds Monitoring Using Surveillance Video" Remote Sensing 14, no. 3: 514. https://doi.org/10.3390/rs14030514
APA StyleWu, E., Wang, H., Lu, H., Zhu, W., Jia, Y., Wen, L., Choi, C.-Y., Guo, H., Li, B., Sun, L., Lei, G., Lei, J., & Jian, H. (2022). Unlocking the Potential of Deep Learning for Migratory Waterbirds Monitoring Using Surveillance Video. Remote Sensing, 14(3), 514. https://doi.org/10.3390/rs14030514








