A Machine Learning Framework for the Classification of Natura 2000 Habitat Types at Large Spatial Scales Using MODIS Surface Reflectance Data
Abstract
:1. Introduction
2. Materials and Methods
2.1. Data Pre-Processing
2.1.1. Reflectance Data, Cloud Mask, and Land Cover Mask
2.1.2. Regions of Interest
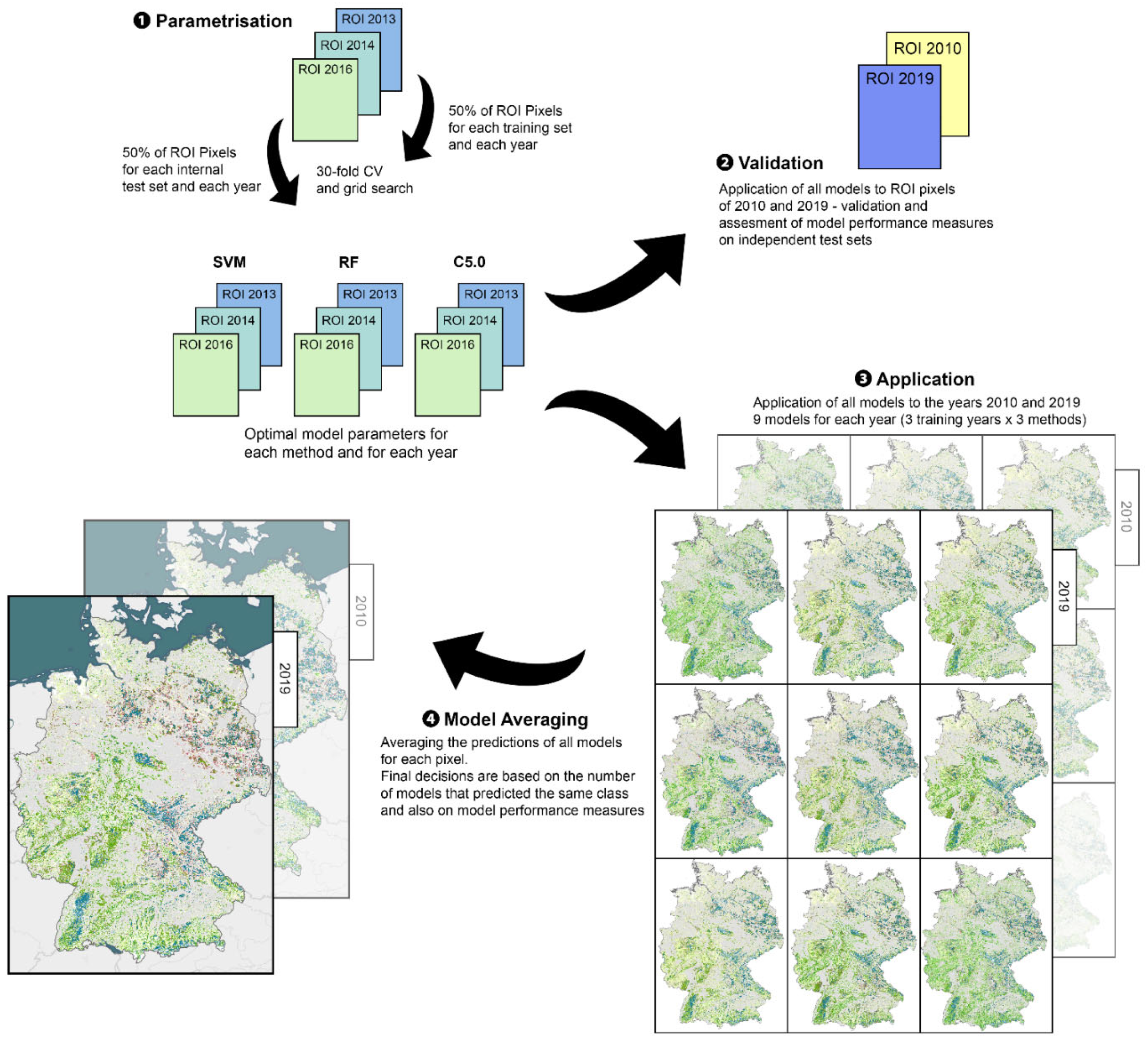
2.2. Machine Learning Framework
| Name | Spatial Resolution | Dates of Acquisition | Spectral Bands | Reference |
|---|---|---|---|---|
| Surface Reflectance Band 1 (620–670 nm) | ||||
| Years: 2010, 2013, 2014, 2016, 2019 | Surface Reflectance Band 2 (841–876 nm) | |||
| MODIS/Terra Surface Reflectance 8-Day L3 | Recording dates *: | Surface Reflectance Band 3 (459–479 nm) | ||
| Global 500 m SIN Grid (MOD09A1 Version 006) | 500 Meters | 15/04, 23/04, 02/06 | Surface Reflectance Band 4 (545–565 nm) | [27] |
| (Multispectral Reflectance Data) | 26/06, 04/07, 20/07 | Surface Reflectance Band 5 (1230–1250 nm) | ||
| 21/08, 29/08, 06/09 | Surface Reflectance Band 6 (1628–1652 nm) | |||
| Surface Reflectance Band 7 (2105–2155 nm) | ||||
| Corine Land Cover (CLC) 2018, Version 2020_20u | 100 Meters | 2018 | – | |
| (Landuse Data) | [34] |
2.2.1. Parametrisation and Validation
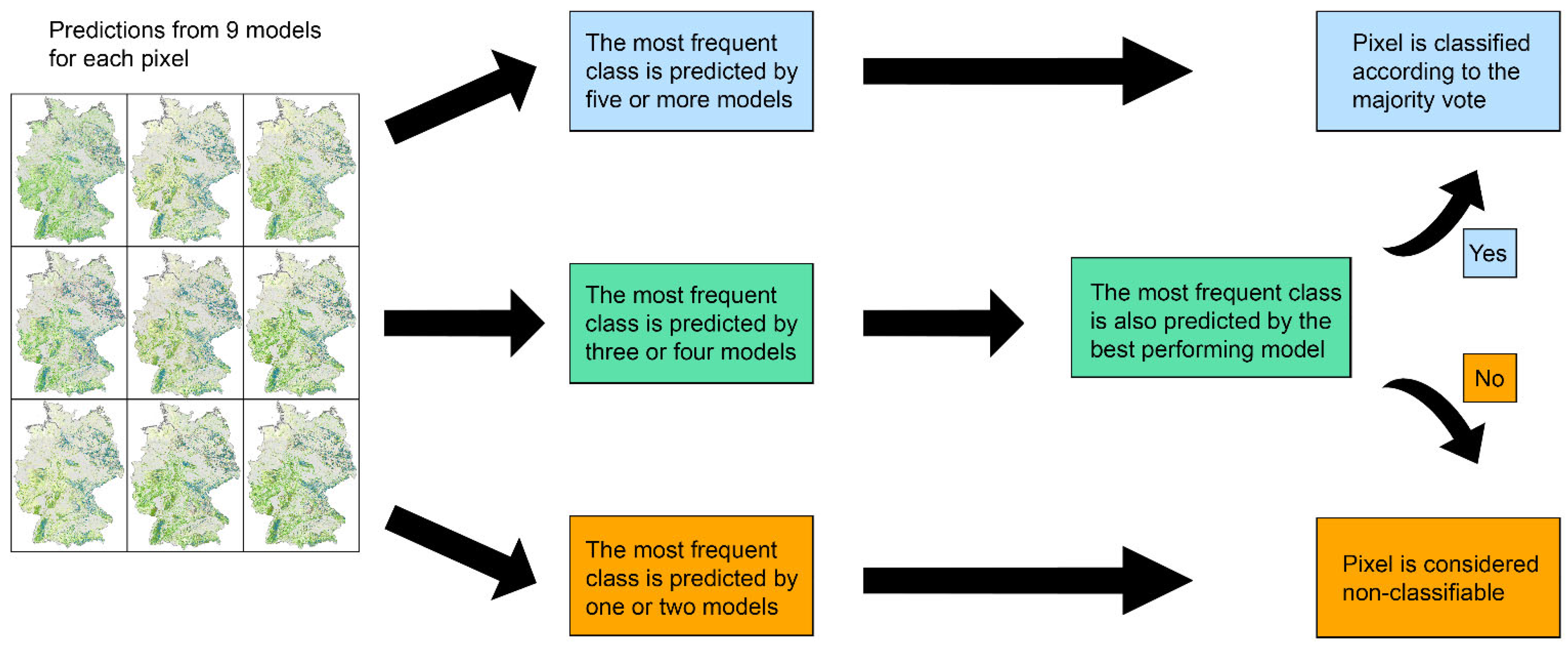
2.2.2. Distance Measure and Model Averaging
3. Results
3.1. Model Validation
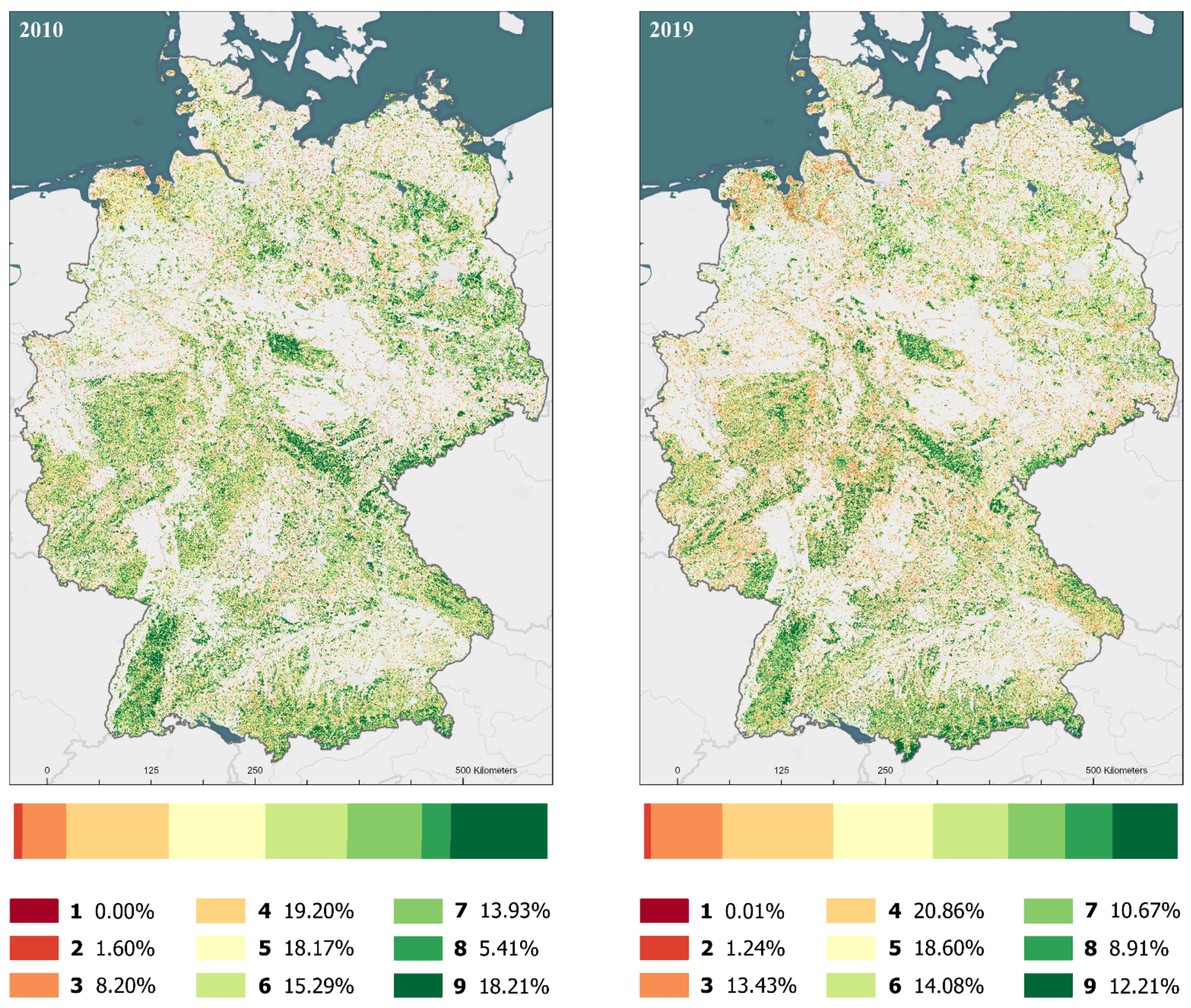
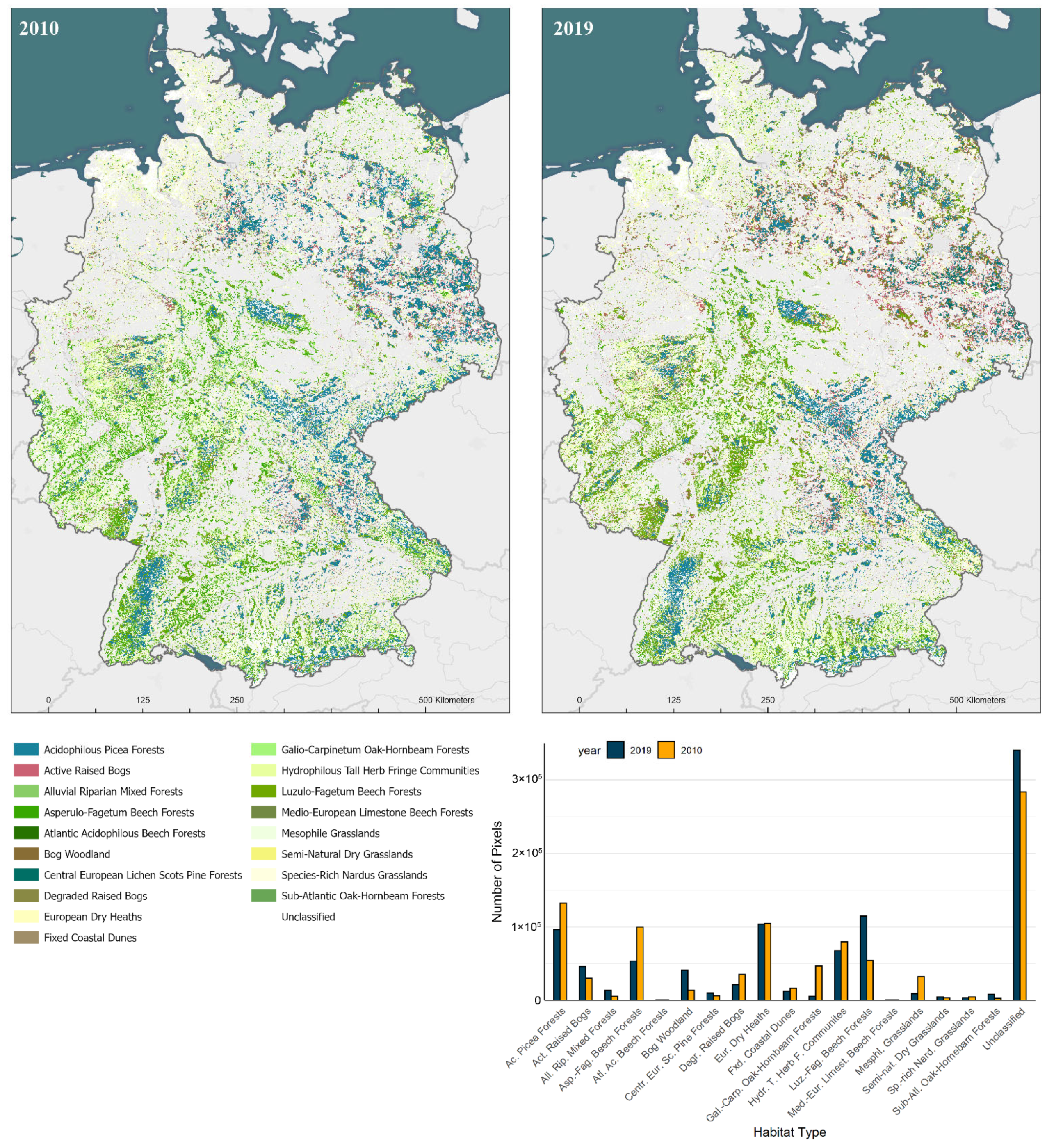
3.2. Habitat Classification
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
Appendix A
| Corine Land Use Types | Status | Corine Land Use Types | Status |
|---|---|---|---|
| Artificial surfaces | Forests and semi natural areas | ||
| 111–Continuous urban fabric | – | 311–Broad–leaved forest | included |
| 112–Discontinuous urban fabric | – | 312–Coniferous forest | included |
| 121–Industrial or commercial units | – | 313–Mixed forest | included |
| 122–Road and rail networks and associated land | – | 321–Natural grasslands | included |
| 123–Port areas | – | 322–Moors and heathland | included |
| 124–Airports | – | 323–Sclerophyllous vegetation | included |
| 131–Mineral extraction sites | – | 324–Transitional woodland–shrub | included |
| 132–Dump sites | – | 331–Beaches dunes sands | included |
| 133–Construction sites | – | 332–Bare rocks | – |
| 141–Green urban areas | – | 333–Sparsely vegetated areas | included |
| 142–Sport and leisure facilities | – | 334–Burnt areas | – |
| 335–Glaciers and perpetual snow | – | ||
| Agricultural Areas | |||
| 211–Non–irrigated arable land | – | Wetlands | |
| 212–Permanently irrigated land | – | 411–Inland marshes | – |
| 213–Rice fields | – | 412–Peat bogs | included |
| 221–Vineyards | – | 421–Salt marshes | – |
| 222–Fruit trees and berry plantations | – | 422–Salines | – |
| 223–Olive groves | – | 423–Intertidal flats | – |
| 231–Pastures | included | ||
| 241–Annual crops associated with permanent crops | – | Water bodies | |
| 242–Complex cultivation patterns | – | 511–Water courses | – |
| 243–Land principally occupied by agriculture with significant areas of natural vegetation | included | 512–Water bodies | – |
| 244–Agro–forestry areas | included | 521–Coastal lagoons | – |
| 522–Estuaries | – | ||
| 523–Sea and ocean | – |
Appendix B
| Habitat Type | Abbreviation | Habitat Code |
|---|---|---|
| Asperulo-Fagetum beech forests | Asp.-Fag. Beech Forests | 9130 |
| European Dry Heaths | Eur. Dry Heaths | 4030 |
| Active raised bogs | Act. Raised Bogs | 7110 |
| Luzulo-Fagetum beech forests | Luz.-Fag. Beech Forests | 9110 |
| Species-rich Nardus grasslands, on silicious substrates in (sub)mountain areas | Sp.-rich Nard. Grassl. | 6230 |
| Sub-Atlantic oak-hornbeam forests and Tilio-Acerion Forests | Sub-Atl. Oak- Hornebam Forests | 9160 + 9180 |
| Hydrophilous tall herb fringe communities | Hydr. T. Herb F. Communities | 6430 |
| Acidophilous Picea forests of the montane to alpine levels (Vaccinio-Piceetea) | Ac. Picea Forests | 9410 |
| Degraded raised bogs still capable of natural regeneration | Degr. Raised Bogs | 7120 + 7140 |
| Galio-Carpinetum oak-hornbeam forests | Gal.-Carp. Oak-Hornbeam Forests | 9170 |
| Alluvial Forests and Riparian mixed forests | All. Rip. Mixed Forests | 91F0 + 91E0 |
| Medio-European limestone beechforests of the Cephalanthero-Fagion | Med.-Eur. Limest. Beech Forests | 9150 |
| Atlantic acidophilous beech forests with Ilex and Taxus in the shrublayer | Atl. Ac. Beech Forests | 9120 |
| Central European lichen Scots pine forests | Centr. Eur. Sc. Pine Forests | 91T0 |
| Bog woodland | Bog Woodland | 91D0 |
| Semi-natural dry grasslands or Molinia Meadows on calcareous substrates | Semi-nat. Dry Grassl. | 6210 + 6410 |
| Fixed coastal dunes with herbaceous vegetation (‘grey dunes’) | Fxd. Coastal Dunes | 2130 |
| Mesophile Grasslands (Lowland and Mountain Hay Meadows) | Mesphl. Grassl. | 6510 + 6520 |
Appendix C
| 2010 Overall Accuracy: 0.942 Kappa: 0.9353 | Reference | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Fxd. Coastal Dunes | Eur. Dry Heaths | Semi-nat. Dry Grassl. | Sp.-rich Nard. Grassl. | Hydr. T. Herb F. Communities | Mesphl. Grassl. | Act. Raised Bogs | Degr. Raised Bogs | Luz.-Fag. Beech Forests | Bog Woodland | Centr. Eur. Sc. Pine Forests | All. Rip. Mixed Forests | Atl. Ac. Beech Forests | Asp.-Fag. Beech Forests | Med.-Eur. Limest. Beech Forests | Sub-Atl. Oak- Hb. Forests | Gal.-Carp. Oak-Hb. Forests | Ac. Picea Forests |
| Fxd. Coastal Dunes | 76 | 1 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eur. Dry Heaths | 0 | 343 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Semi-nat. Dry Grassl. | 0 | 3 | 79 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 |
| Sp.-rich Nard. Grassl. | 0 | 0 | 0 | 295 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hydr. T. Herb F. Communities | 0 | 0 | 0 | 0 | 206 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 1 | 0 | 0 |
| Mesphl. Grassl. | 0 | 2 | 0 | 1 | 6 | 74 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Act. Raised Bogs | 0 | 0 | 0 | 0 | 0 | 0 | 386 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Degr. Raised Bogs | 1 | 7 | 0 | 0 | 0 | 0 | 17 | 138 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Luz.-Fag. Beech Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 327 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 |
| Bog Woodland | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 87 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Centr. Eur. Sc. Pine Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 86 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| All. Rip. Mixed Forests | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 8 | 0 | 0 | 114 | 0 | 3 | 0 | 12 | 0 | 0 |
| Atl. Ac. Beech Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 9 | 0 | 0 | 0 | 100 | 1 | 0 | 0 | 0 | 0 |
| Asp.-Fag. Beech Forests | 0 | 1 | 0 | 0 | 3 | 0 | 0 | 0 | 66 | 0 | 0 | 1 | 0 | 796 | 0 | 2 | 1 | 0 |
| Med.-Eur. Limest. Beech Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 101 | 0 | 0 | 0 |
| Sub-Atl. Oak- Hb. Forests | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 5 | 0 | 171 | 0 | 0 |
| Gal.-Carp. Oak-Hb. Forests | 0 | 5 | 0 | 0 | 1 | 0 | 0 | 0 | 42 | 0 | 0 | 0 | 0 | 13 | 0 | 2 | 106 | 0 |
| Ac. Picea Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 183 |
| 2019 Overall Accuracy: 0.801 Kappa: 0.7736 | Reference | |||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Prediction | Fxd. Coastal Dunes | Eur. Dry Heaths | Semi-nat. Dry Grassl. | Sp.-rich Nard. Grassl. | Hydr. T. Herb F. Communities | Mesphl. Grassl. | Act. Raised Bogs | Degr. Raised Bogs | Luz.-Fag. Beech Forests | Bog Woodland | Centr. Eur. Sc. Pine Forests | All. Rip. Mixed Forests | Atl. Ac. Beech Forests | Asp.-Fag. Beech Forests | Med.-Eur. Limest. Beech Forests | Sub-Atl. Oak- Hb. Forests | Gal.-Carp. Oak-Hb. Forests | Ac. Picea Forests |
| Fxd. Coastal Dunes | 56 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Eur. Dry Heaths | 1 | 317 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Semi-nat. Dry Grassl. | 0 | 10 | 50 | 0 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Sp.-rich Nard. Grassl. | 0 | 16 | 6 | 328 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Hydr. T. Herb F. Communities | 0 | 0 | 0 | 0 | 180 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 0 |
| Mesphl. Grassl. | 0 | 1 | 0 | 0 | 0 | 45 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Act. Raised Bogs | 0 | 3 | 0 | 0 | 0 | 0 | 373 | 15 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Degr. Raised Bogs | 0 | 31 | 0 | 0 | 0 | 0 | 24 | 93 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 1 | 0 |
| Luz.-Fag. Beech Forests | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 327 | 0 | 0 | 1 | 0 | 52 | 0 | 1 | 0 | 0 |
| Bog Woodland | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 2 | 58 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 |
| Centr. Eur. Sc. Pine Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 72 | 0 | 0 | 0 | 0 | 0 | 0 | 1 |
| All. Rip. Mixed Forests | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 1 | 11 | 0 | 0 | 62 | 0 | 27 | 0 | 5 | 0 | 0 |
| Atl. Ac. Beech Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 43 | 48 | 0 | 0 | 0 | 0 |
| Asp.-Fag. Beech Forests | 0 | 0 | 0 | 0 | 2 | 0 | 0 | 0 | 221 | 0 | 0 | 30 | 1 | 901 | 0 | 65 | 16 | 0 |
| Med.-Eur. Limest. Beech Forests | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 23 | 63 | 0 | 0 | 0 |
| Sub-Atl. Oak- Hb. Forests | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 84 | 0 | 24 | 0 | 176 | 0 | 0 |
| Gal.-Carp. Oak-Hb. Forests | 0 | 8 | 0 | 0 | 0 | 0 | 0 | 4 | 32 | 0 | 0 | 3 | 0 | 31 | 0 | 5 | 70 | 0 |
| Ac. Picea Forests | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 167 |
Appendix D
| 2013 | 2014 | 2016 | |||||||
|---|---|---|---|---|---|---|---|---|---|
| Training Year | Method | Best Parameters | Overall Accuracy | Kappa | Overall Accuracy | Kappa | Overall Accuracy | Kappa | |
| SVM | γ: 0.09 | C: 9 | - | - | 0.772 | 0.706 | 0.776 | 0.782 | |
| 2013 | RF | nodesize: 4 | ntrees: 2500 | - | - | 0.755 | 0.709 | 0.792 | 0.762 |
| C5.0 | Trials: 50 | MinCases: 5 | - | - | 0.726 | 0.659 | 0.798 | 0.776 | |
| SVM | γ: 0.05 | C: 10 | 0.721 | 0.745 | - | - | 0.731 | 0.751 | |
| 2014 | RF | nodesize: 3 | ntrees: 2000 | 0.738 | 0.707 | - | - | 0.733 | 0.718 |
| C5.0 | Trials: 30 | MinCases: 3 | 0.658 | 0.673 | - | - | 0.681 | 0.675 | |
| SVM | γ: 0.09 | C: 15 | 0.83 | 0.836 | 0.83 | 0.757 | - | - | |
| 2016 | RF | nodesize: 5 | ntrees: 750 | 0.807 | 0.807 | 0.831 | 0.779 | - | - |
| C5.0 | Trials: 50 | MinCases: 8 | 0.762 | 0.753 | 0.75 | 0.705 | - | - | |
References
- Randin, C.F.; Ashcroft, M.B.; Bolliger, J.; Cavender-Bares, J.; Coops, N.C.; Dullinger, S.; Dirnböck, T.; Eckert, S.; Ellis, E.; Fernández, N.; et al. Monitoring biodiversity in the Anthropocene using remote sensing in species distribution models. Remote Sens. Environ. 2020, 239, 111626. [Google Scholar] [CrossRef]
- Bittner, T.; Jaeschke, A.; Reineking, B.; Beierkuhnlein, C. Comparing modelling approaches at two levels of biological organisation—Climate change impacts on selected Natura 2000 habitats. J. Veg. Sci. 2011, 22, 699–710. [Google Scholar] [CrossRef]
- Mahecha, M.D.; Gans, F.; Sippel, S.; Donges, J.F.; Kaminski, T.; Metzger, S.; Migliavacca, M.; Papale, D.; Rammig, A.; Zscheischler, J. Detecting impacts of extreme events with ecological in situ monitoring networks. Biogeosciences 2017, 14, 4255–4277. [Google Scholar] [CrossRef] [Green Version]
- Pressey, R.L.; Cabeza, M.; Watts, M.E.; Cowling, R.M.; Wilson, K.A. Conservation planning in a changing world. Trends Ecol. Evol. 2007, 22, 583–592. [Google Scholar] [CrossRef] [PubMed]
- Álvarez-Martínez, J.M.; Jiménez-Alfaro, B.; Barquín, J.; Ondiviela, B.; Recio, M.; Silió-Calzada, A.; Juanes, J.A. Modelling the area of occupancy of habitat types with remote sensing. Methods Ecol. Evol. 2018, 9, 580–593. [Google Scholar] [CrossRef]
- Vanden Borre, J.; Paelinckx, D.; Mücher, C.A.; Kooistra, L.; Haest, B.; de Blust, G.; Schmidt, A.M. Integrating remote sensing in Natura 2000 habitat monitoring: Prospects on the way forward. J. Nat. Conserv. 2011, 19, 116–125. [Google Scholar] [CrossRef]
- Roelofsen, H.D.; Kooistra, L.; van Bodegom, P.M.; Verrelst, J.; Krol, J.; Witte, J.-P.M. Mapping a priori defined plant associations using remotely sensed vegetation characteristics. Remote Sens. Environ. 2014, 140, 639–651. [Google Scholar] [CrossRef]
- Nagendra, H.; Lucas, R.; Honrado, J.P.; Jongman, R.H.G.; Tarantino, C.; Adamo, M.; Mairota, P. Remote sensing for conservation monitoring: Assessing protected areas, habitat extent, habitat condition, species diversity, and threats. Ecol. Indic. 2013, 33, 45–59. [Google Scholar] [CrossRef]
- Keshtkar, H.; Voigt, W.; Alizadeh, E. Land-cover classification and analysis of change using machine-learning classifiers and multi-temporal remote sensing imagery. Arab. J. Geosci. 2017, 10, 154. [Google Scholar] [CrossRef]
- Feilhauer, H.; Dahlke, C.; Doktor, D.; Lausch, A.; Schmidtlein, S.; Schulz, G.; Stenzel, S. Mapping the local variability of Natura 2000 habitats with remote sensing. Appl. Veg. Sci. 2014, 17, 765–779. [Google Scholar] [CrossRef]
- Kopéc, D.; Michalska-Hejduk, D.; Sſawik, ſ.; Berezowski, T.; Borowski, M.; Rosadziſski, S.; Chormaſski, J. Application of multisensoral remote sensing data in the mapping of alkaline fens Natura 2000 habitat. Ecol. Indic. 2016, 70, 196–208. [Google Scholar] [CrossRef]
- Vanden Borre, J.; Spanhove, T.; Haest, B. Towards a Mature Age of Remote Sensing for Natura 2000 Habitat Conservation: Poor Method Transferability as a Prime Obstacle. In The Roles of Remote Sensing in Nature Conservation: A Practical Guide and Case Studies; Lucas, R., Hurford, C., Díaz-Delgado, R., Eds.; Springer: Cham, Switzerland, 2017; pp. 11–37. ISBN 978-3-319-64332-8. [Google Scholar]
- Lengyel, S.; Déri, E.; Varga, Z.; Horváth, R.; Tóthmérész, B.; Henry, P.-Y.; Kobler, A.; Kutnar, L.; Babij, V.; Seliškar, A.; et al. Habitat monitoring in Europe: A description of current practices. Biodivers. Conserv 2008, 17, 3327–3339. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Vanden Borre, J.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status—New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Council Directive 92/43/EEC of 21 May 1992 on the Conservation of Natural Habitats and of Wild Fauna and Flora. Off. J. Eur. Union 1992, OJ L 206, 7–50.
- EEA. Natura 2000 Data—The European Network of Protected Sites; European Environmental Agency: Copenhagen, Denmark, 2020. [Google Scholar]
- Harley, M.; de Soye, Y.; Dickson, B.; Tucker, G.; Keder, G. Biodiversity and climate change in relation to the Natura 2000 network. Adv. Sci. Res. 2009, 3, 35–37. [Google Scholar] [CrossRef]
- Steinacker, C.; Beierkuhnlein, C.; Jaeschke, A. Assessing the exposure of forest habitat types to projected climate change-Implications for Bavarian protected areas. Ecol. Evol. 2019, 9, 14417–14429. [Google Scholar] [CrossRef]
- O’Keeffe, J.; Marcinkowski, P.; Utratna, M.; Piniewski, M.; Kardel, I.; Kundzewicz, Z.; Okruszko, T. Modelling Climate Change’s Impact on the Hydrology of Natura 2000 Wetland Habitats in the Vistula and Odra River Basins in Poland. Water 2019, 11, 2191. [Google Scholar] [CrossRef] [Green Version]
- Sommerfeld, A.; Rammer, W.; Heurich, M.; Hilmers, T.; Müller, J.; Seidl, R. Do bark beetle outbreaks amplify or dampen future bark beetle disturbances in Central Europe? J. Ecol. 2021, 109, 737–749. [Google Scholar] [CrossRef]
- Bonn, A.; Reed, M.S.; Evans, C.D.; Joosten, H.; Bain, C.; Farmer, J.; Emmer, I.; Couwenberg, J.; Moxey, A.; Artz, R.; et al. Investing in nature: Developing ecosystem service markets for peatland restoration. Ecosyst. Serv. 2014, 9, 54–65. [Google Scholar] [CrossRef] [Green Version]
- Haest, B.; Vanden Borre, J.; Spanhove, T.; Thoonen, G.; Delalieux, S.; Kooistra, L.; Mücher, C.; Paelinckx, D.; Scheunders, P.; Kempeneers, P. Habitat Mapping and Quality Assessment of NATURA 2000 Heathland Using Airborne Imaging Spectroscopy. Remote Sens. 2017, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocińska, A.; Sławik, Ł. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef] [Green Version]
- Demarchi, L.; Kania, A.; Ciężkowski, W.; Piórkowski, H.; Oświecimska-Piasko, Z.; Chormański, J. Recursive Feature Elimination and Random Forest Classification of Natura 2000 Grasslands in Lowland River Valleys of Poland Based on Airborne Hyperspectral and LiDAR Data Fusion. Remote Sens. 2020, 12, 1842. [Google Scholar] [CrossRef]
- Eigenbrod, F.; Gonzalez, P.; Dash, J.; Steyl, I. Vulnerability of ecosystems to climate change moderated by habitat intactness. Glob. Chang. Biol. 2015, 21, 275–286. [Google Scholar] [CrossRef] [PubMed]
- Pacifici, M.; Foden, W.B.; Visconti, P.; Watson, J.E.M.; Butchart, S.H.M.; Kovacs, K.M.; Scheffers, B.R.; Hole, D.G.; Martin, T.G.; Akçakaya, H.R.; et al. Assessing species vulnerability to climate change. Nat. Clim. Chang. 2015, 5, 215–224. [Google Scholar] [CrossRef]
- Vermote, E. MOD09A1 MODIS/Terra Surface Reflectance 8-Day L3 Global 500m SIN Grid V006; 2015, distributed by NASA EOSDIS Land Processes DAAC. Available online: https://lpdaac.usgs.gov/products/mod09a1v006/ (accessed on 31 March 2021).
- Busetto, L.; Ranghetti, L. MODIStsp: An R package for automatic preprocessing of MODIS Land Products time series. Comput. Geosci. 2016, 97, 40–48. [Google Scholar] [CrossRef] [Green Version]
- EEA. Corine Land Cover (CLC) 2018, Version 2020_20u1. In © European Union Copernicus Land Monitoring Service; European Environmental Agency: Copenhagen, Denmark, 2018; Available online: https://land.copernicus.eu/pan-european/corine-land-cover/clc2018 (accessed on 31 March 2021).
- Metzger, M.J.; Bunce, R.G.H.; Jongman, R.H.G.; Mücher, C.A.; Watkins, J.W. A climatic stratification of the environment of Europe. Glob. Ecol. Biogeogr. 2005, 14, 549–563. [Google Scholar] [CrossRef]
- R Core Team. R: A Language and Environment for Statistical Computing; R Foundation for Statistical Computing: Vienna, Austria, 2019. [Google Scholar]
- Esri Inc. ArcGIS Pro (Version 2.7); Esri Inc.: Redlands, CA, USA, 2021. [Google Scholar]
- Adam, E.; Mutanga, O.; Odindi, J.; Abdel-Rahman, E.M. Land-use/cover classification in a heterogeneous coastal landscape using RapidEye imagery: Evaluating the performance of random forest and support vector machines classifiers. Int. J. Remote Sens. 2014, 35, 3440–3458. [Google Scholar] [CrossRef]
- Sun, Z.; Leinenkugel, P.; Guo, H.; Huang, C.; Kuenzer, C. Extracting distribution and expansion of rubber plantations from Landsat imagery using the C5.0 decision tree method. J. Appl. Remote Sens. 2017, 11, 26011. [Google Scholar] [CrossRef] [Green Version]
- Berhane, T.M.; Lane, C.R.; Wu, Q.; Autrey, B.C.; Anenkhonov, O.A.; Chepinoga, V.V.; Liu, H. Decision-Tree, Rule-Based, and Random Forest Classification of High-Resolution Multispectral Imagery for Wetland Mapping and Inventory. Remote Sens. 2018, 10, 580. [Google Scholar] [CrossRef] [Green Version]
- Golkarian, A.; Naghibi, S.A.; Kalantar, B.; Pradhan, B. Groundwater potential mapping using C5.0, random forest, and multivariate adaptive regression spline models in GIS. Environ. Monit. Assess. 2018, 190, 149. [Google Scholar] [CrossRef]
- Liaw, A.; Wiener, M. Classification and regression by randomForest. R News 2002, 2, 18–22. [Google Scholar]
- Kuhn, M.; Quinlan, R. C50: C5.0 Decision Trees and Rule-Based Models. R Package Version 0.1.3.1. 2020. Available online: https://cran.r-project.org/web/packages/C50/index.html (accessed on 31 March 2021).
- Meyer, D.; Dimitriadou, E.; Hornik, K.; Weingessel, A.; Leisch, F. e1071: Misc Functions of the Department of Statistics; Probability Theory Group (Formerly: E1071), TU Wien: Vienna, Austria, 2019. [Google Scholar]
- Pal, M. Random forest classifier for remote sensing classification. Int. J. Remote Sens. 2005, 26, 217–222. [Google Scholar] [CrossRef]
- Melgani, F.; Bruzzone, L. Classification of hyperspectral remote sensing images with support vector machines. IEEE Trans. Geosci. Remote Sens. 2004, 42, 1778–1790. [Google Scholar] [CrossRef] [Green Version]
- Mountrakis, G.; Im, J.; Ogole, C. Support vector machines in remote sensing: A review. ISPRS J. Photogramm. Remote Sens. 2011, 66, 247–259. [Google Scholar] [CrossRef]
- Hastie, T.; Tibshirani, R.; Friedman, J.H. The Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2017; ISBN 978-0-387-84858-7. [Google Scholar]
- Duro, D.C.; Franklin, S.E.; Dubé, M.G. A comparison of pixel-based and object-based image analysis with selected machine learning algorithms for the classification of agricultural landscapes using SPOT-5 HRG imagery. Remote Sens. Environ. 2012, 118, 259–272. [Google Scholar] [CrossRef]
- Preidl, S.; Lange, M.; Doktor, D. Introducing APiC for regionalised land cover mapping on the national scale using Sentinel-2A imagery. Remote Sens. Environ. 2020, 240, 111673. [Google Scholar] [CrossRef]
- Sperle, T.; Bruelheide, H. Climate change aggravates bog species extinctions in the Black Forest (Germany). Divers. Distrib. 2021, 27, 282–295. [Google Scholar] [CrossRef]
- Janssen, M. European Red List of Habitats—Part 2 Terrestrial and Freshwater Habitats; Publications Office of the European Union: Luxembourg, 2016. [Google Scholar] [CrossRef]

| 2010 | 2019 | ||||||
|---|---|---|---|---|---|---|---|
| Training Year | Method | Best Parameters | Overall Accuracy | Kappa | Overall Accuracy | Kappa | |
| SVM | γ: 0.09 | C: 9 | 0.778 | 0.784 | 0.721 | 0.738 | |
| 2013 | RF | nodesize: 4 | ntrees: 2500 | 0.801 | 0.769 | 0.724 | 0.702 |
| C5.0 | Trials: 50 | MinCases: 5 | 0.805 | 0.781 | 0.655 | 0.674 | |
| SVM | γ: 0.05 | C: 10 | 0.74 | 0.757 | 0.721 | 0.758 | |
| 2014 | RF | nodesize: 3 | ntrees: 2000 | 0.745 | 0.731 | 0.736 | 0.733 |
| C5.0 | Trials: 30 | MinCases: 3 | 0.689 | 0.678 | 0.675 | 0.668 | |
| SVM | γ: 0.09 | C: 15 | 0.932 | 0.935 | 0.78 | 0.793 | |
| 2016 | RF | nodesize: 5 | ntrees: 750 | 0.94 | 0.934 | 0.78 | 0.747 |
| C5.0 | Trials: 50 | MinCases: 8 | 0.932 | 0.916 | 0.716 | 0.672 | |
| Habitat Type | User’s Accuracy | Producer’s Accuracy | F1-Score | |||||||
|---|---|---|---|---|---|---|---|---|---|---|
| 2010 | CI | 2019 | CI | 2010 | CI | 2019 | CI | 2010 | 2019 | |
| European Dry Heaths | 1.000 | 0.989–1.000 | 0.984 | 0.964–0.995 | 0.945 | 0.916–0.966 | 0.805 | 0.762–0.843 | 0.972 | 0.885 |
| Active raised bogs | 1.000 | 0.990–1.000 | 0.954 | 0.928–0.972 | 0.958 | 0.933–0.975 | 0.935 | 0.906–0.957 | 0.978 | 0.944 |
| Degraded raised bogs still capable of natural regeneration | 0.847 | 0.782–0.898 | 0.620 | 0.537–0.698 | 0.986 | 0.949–0.998 | 0.802 | 0.717–0.870 | 0.911 | 0.699 |
| Alluvial Forests and Riparian mixed forests | 0.826 | 0.752–0.885 | 0.574 | 0.475–0.669 | 0.983 | 0.939–0.998 | 0.344 | 0.275–0.419 | 0.898 | 0.431 |
| Luzulo-Fagetum beech forests | 0.997 | 0.983–1.000 | 0.856 | 0.817–0.890 | 0.723 | 0.680–0.764 | 0.551 | 0.509–0.591 | 0.838 | 0.670 |
| Sub-Atlantic oak-hornbeam forests and Tilio-Acerion Forests | 0.961 | 0.921–0.984 | 0.615 | 0.556–0.672 | 0.905 | 0.854–0.943 | 0.696 | 0.635–0.752 | 0.932 | 0.653 |
| Semi-natural dry grasslands or Molinia Meadows on calcareous substrates | 0.940 | 0.867–0.980 | 0.806 | 0.686–0.896 | 1.000 | 0.954–1.000 | 0.893 | 0.781–0.960 | 0.969 | 0.847 |
| Bog woodland | 1.000 | 0.958–1.000 | 0.892 | 0.791–0.956 | 0.989 | 0.938–1.000 | 1.000 | 0.938–1.000 | 0.994 | 0.943 |
| Mesophile Grasslands (Lowland and Mountain Hay Meadows) | 0.892 | 0.804–0.949 | 0.978 | 0.885–0.999 | 1.000 | 0.951–1.000 | 1.000 | 0.921–1.000 | 0.943 | 0.989 |
| Fixed coastal dunes with herbaceous vegetation (‘grey dunes’) | 0.962 | 0.893–0.992 | 0.933 | 0.838–0.982 | 0.987 | 0.930–1.000 | 0.982 | 0.906–1.000 | 0.974 | 0.957 |
| Asperulo-Fagetum beech forests | 0.915 | 0.894–0.933 | 0.729 | 0.703–0.754 | 0.967 | 0.953–0.978 | 0.813 | 0.789–0.836 | 0.940 | 0.769 |
| Galio-Carpinetum oak-hornbeam forests | 0.627 | 0.550–0.700 | 0.458 | 0.377–0.540 | 0.991 | 0.949–1.000 | 0.805 | 0.706–0.882 | 0.768 | 0.583 |
| Central European lichen Scots pine forests | 0.989 | 0.938–1.000 | 0.986 | 0.926–1.000 | 1.000 | 0.958–1.000 | 0.947 | 0.871–0.985 | 0.994 | 0.966 |
| Atlantic acidophilous beech forests with Ilex and Taxus in the shrublayer | 0.909 | 0.839–0.956 | 0.473 | 0.367–0.580 | 1.000 | 0.964–1.000 | 0.977 | 0.880–0.999 | 0.952 | 0.637 |
| Acidophilous Picea forests of the montane to alpine levels (Vaccinio-Piceetea) | 0.995 | 0.970–1.000 | 0.977 | 0.941–0.994 | 0.995 | 0.970–1.000 | 0.971 | 0.933–0.990 | 0.995 | 0.974 |
| Medio–European limestone beechforests of the Cephalanthero-Fagion | 0.981 | 0.932–0.998 | 0.708 | 0.602–0.799 | 1.000 | 0.964–1.000 | 1.000 | 0.943–1.000 | 0.990 | 0.829 |
| Species-rich Nardus grasslands on silicious substrates in (sub)mountain areas | 1.000 | 0.988–1.000 | 0.937 | 0.906–0.960 | 0.993 | 0.976–0.999 | 0.997 | 0.983–1.000 | 0.997 | 0.966 |
| Hydrophilous tall herb fringe communities | 0.986 | 0.959–0.997 | 0.989 | 0.961–0.999 | 0.949 | 0.911–0.974 | 0.968 | 0.931–0.988 | 0.967 | 0.978 |
| Habitat Type | Number of Pixels | Area (km2) | Change (%) | ||
|---|---|---|---|---|---|
| 2019 | 2010 | 2019 | 2010 | ||
| European Dry Heaths | 103,741 | 104,298 | 25,935 | 26,075 | −0.53 |
| Active raised bogs | 45,864 | 29,904 | 11,466 | 7476 | 53.37 |
| Degraded raised bogs still capable of natural regeneration | 21,085 | 34,972 | 5271 | 8743 | −39.71 |
| Alluvial Forests and Riparian mixed forests | 13,374 | 5298 | 3344 | 1325 | 152.43 |
| Luzulo-Fagetum beech forests | 114,230 | 53,938 | 28,558 | 13,485 | 111.78 |
| Sub-Atlantic oak-hornbeam forests and Tilio-Acerion Forests | 8017 | 2243 | 2004 | 561 | 257.42 |
| Semi-natural dry grasslands and Molinia Meadows on calcareous substrates | 4600 | 2910 | 1150 | 728 | 58.08 |
| Bog woodland | 40,777 | 13,455 | 10,194 | 3364 | 203.06 |
| Mesophile Grasslands (Lowland and Mountain Hay Meadows) | 8973 | 32,197 | 2243 | 8049 | −72.13 |
| Fixed coastal dunes with herbaceous vegetation (‘grey dunes’) | 11,884 | 16,123 | 2971 | 4031 | −26.29 |
| Asperulo-Fagetum beech forests | 53,166 | 99,378 | 13,292 | 24,845 | −46.50 |
| Galio-Carpinetum oak-hornbeam forests | 5175 | 46,654 | 1294 | 11,664 | −88.91 |
| Central European lichen Scots pine forests | 9682 | 6096 | 2421 | 1524 | 58.83 |
| Atlantic acidophilous beech forests with Ilex and Taxus in the shrublayer | 150 | 285 | 38 | 71 | −47.37 |
| Acidophilous Picea forests of the montane to alpine levels (Vaccinio-Piceetea) | 96,253 | 132,286 | 24,063 | 33,072 | −27.24 |
| Medio-European limestone beechforests of the Cephalanthero-Fagion | 156 | 386 | 39 | 97 | −59.59 |
| Species-rich Nardus grasslands, on silicious substrates in (sub)mountain areas | 2878 | 4579 | 720 | 1145 | −37.15 |
| Hydrophilous tall herb fringe communities | 67,282 | 79,284 | 16,821 | 19,821 | −15.14 |
| Unclassified | 340,712 | 283,713 | 85,178 | 70,928 | 20.09 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Sittaro, F.; Hutengs, C.; Semella, S.; Vohland, M. A Machine Learning Framework for the Classification of Natura 2000 Habitat Types at Large Spatial Scales Using MODIS Surface Reflectance Data. Remote Sens. 2022, 14, 823. https://doi.org/10.3390/rs14040823
Sittaro F, Hutengs C, Semella S, Vohland M. A Machine Learning Framework for the Classification of Natura 2000 Habitat Types at Large Spatial Scales Using MODIS Surface Reflectance Data. Remote Sensing. 2022; 14(4):823. https://doi.org/10.3390/rs14040823
Chicago/Turabian StyleSittaro, Fabian, Christopher Hutengs, Sebastian Semella, and Michael Vohland. 2022. "A Machine Learning Framework for the Classification of Natura 2000 Habitat Types at Large Spatial Scales Using MODIS Surface Reflectance Data" Remote Sensing 14, no. 4: 823. https://doi.org/10.3390/rs14040823
APA StyleSittaro, F., Hutengs, C., Semella, S., & Vohland, M. (2022). A Machine Learning Framework for the Classification of Natura 2000 Habitat Types at Large Spatial Scales Using MODIS Surface Reflectance Data. Remote Sensing, 14(4), 823. https://doi.org/10.3390/rs14040823







