Functional Analysis for Habitat Mapping in a Special Area of Conservation Using Sentinel-2 Time-Series Data
Abstract
:1. Introduction
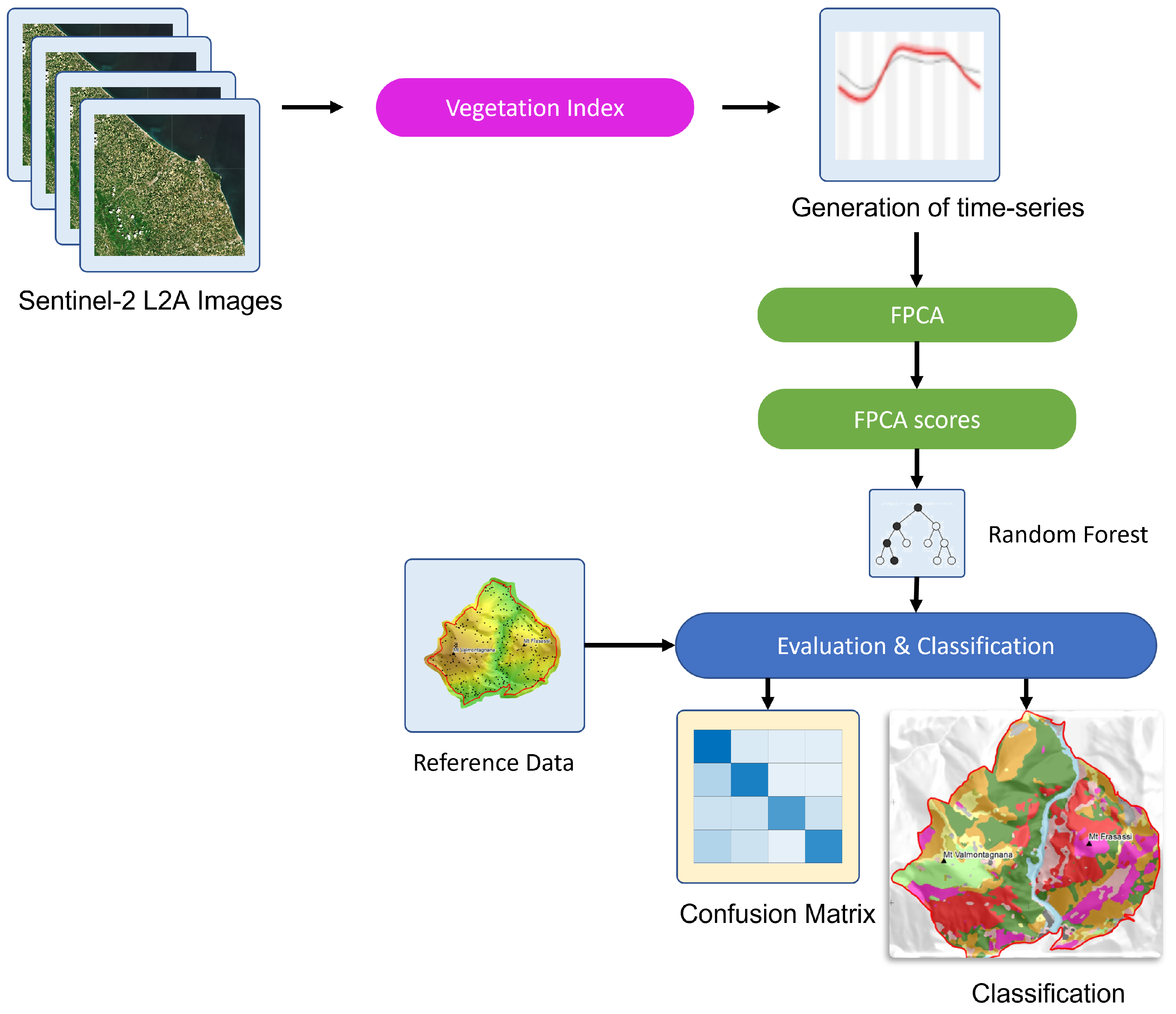
2. Materials and Methods
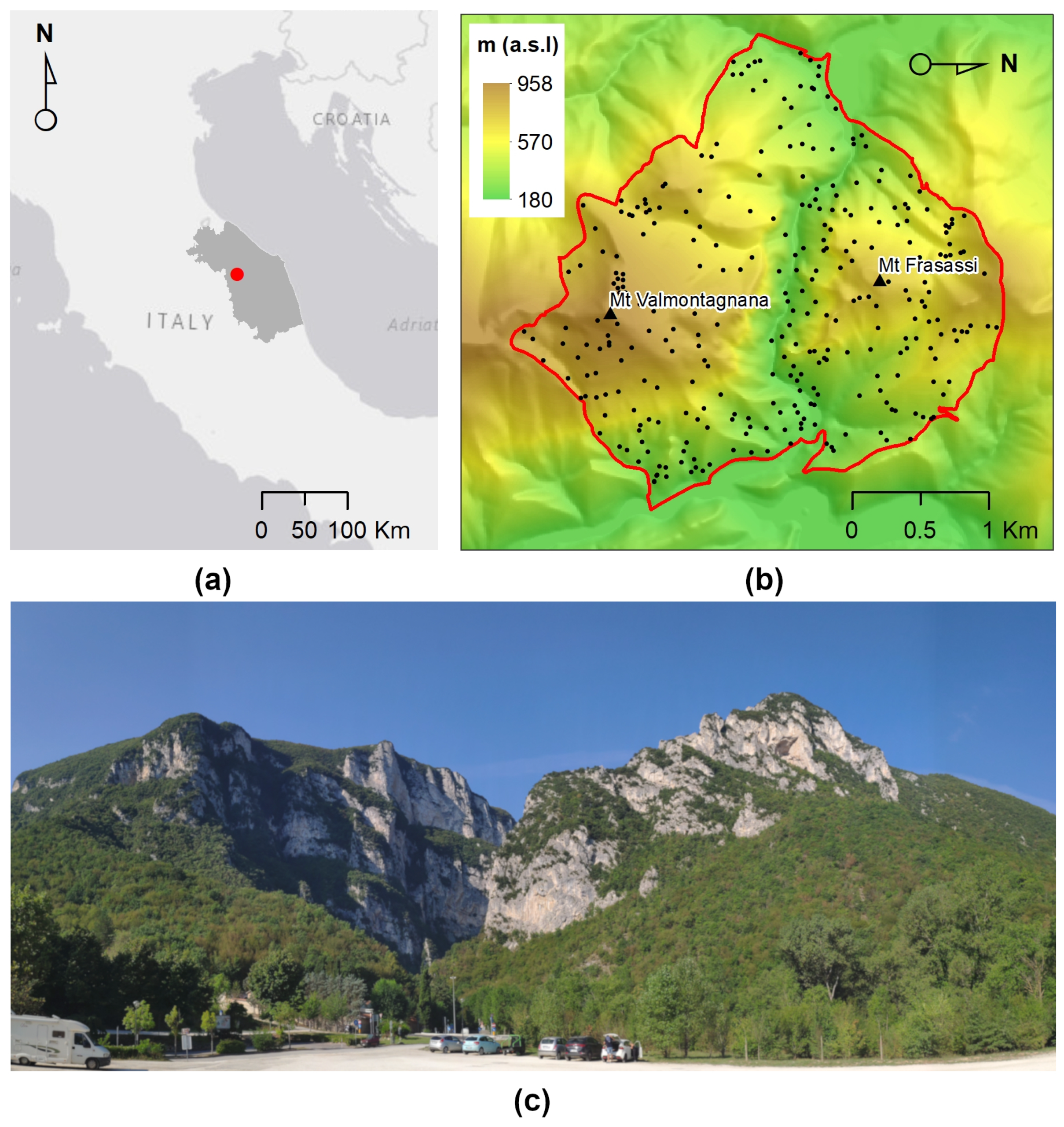
2.1. Study Site
2.2. Target Classes and Reference Data
2.3. Topographic and Lithological Factors
2.4. Remote Sensing Time-Series
Functional Principal Component Analysis (FPCA)
2.5. Supervised Mapping Using Random Forest
- pixel-based FPCA scores, representing the main seasonal (intra-annual) variations of the time-series;
- pixel-based PCA scores, representing the topographic and lithological features.
2.6. Classification and Mapping Accuracy Assessment
3. Results
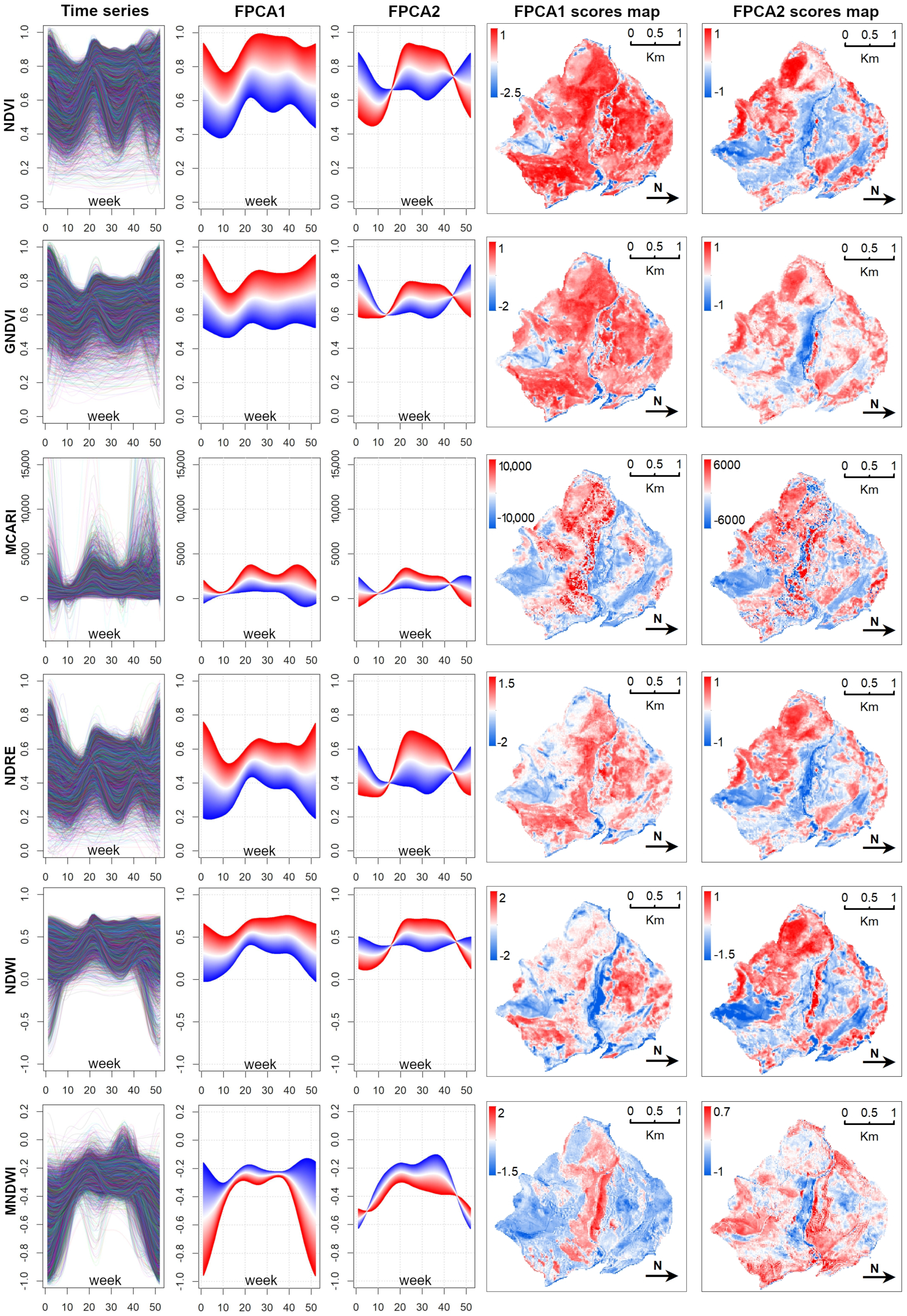
3.1. Main Seasonal Variations of Time-Series
3.2. Plant Community Modeling Using the Main Topographic–Lithological and Phenological Predictors
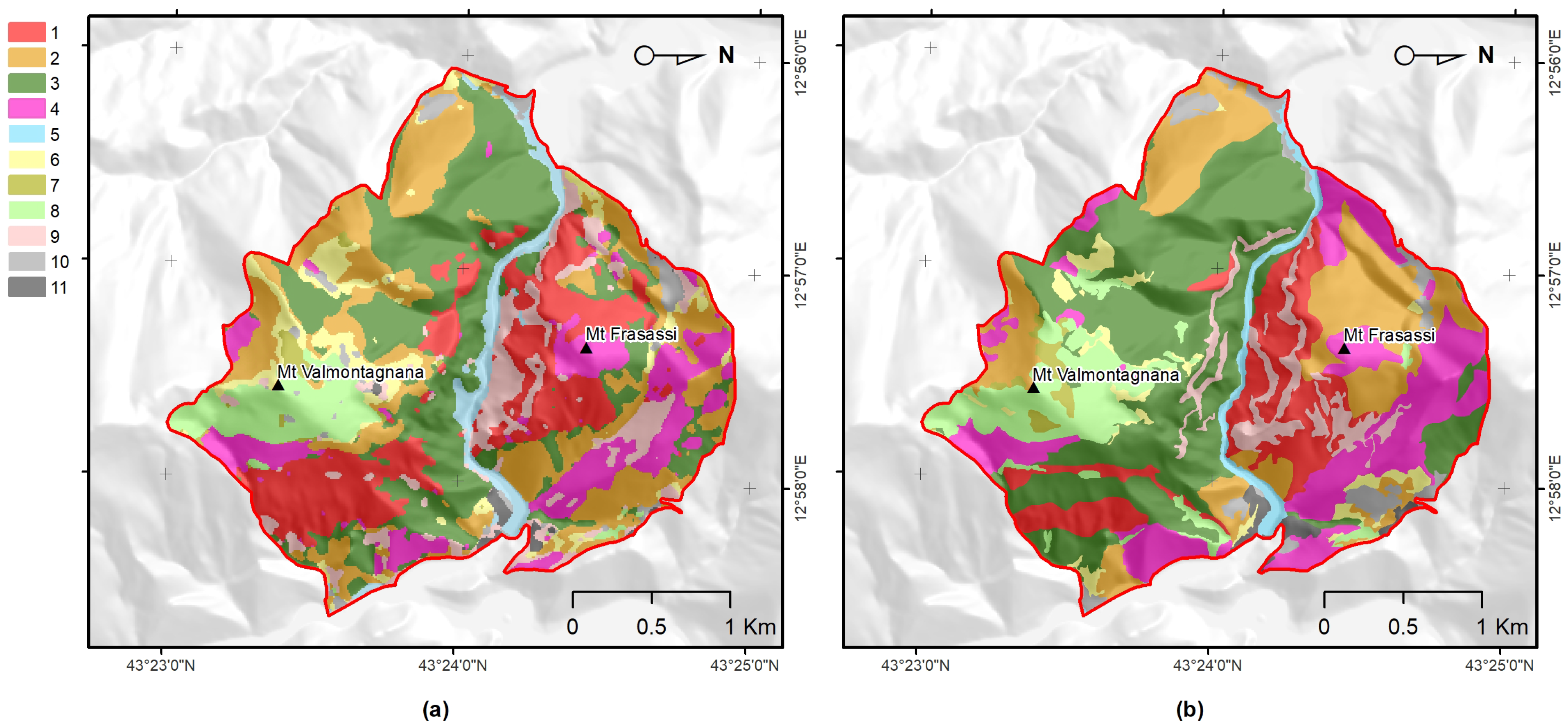
3.3. Comparison of Obtained Results with Ancillary Data
4. Discussion
- 1.
- Main results. Our results confirmed that remotely sensed data can be used to (automatically) map plant associations and habitats, particularly due to their multi-spectral seasonal profiles (phenological behaviors). The main seasonal multi-spectral variations were effective predictors for the production of accurate maps as previously discussed in [16,37].In particular, the main seasonal variations extracted from Sentinel-2 time-series data using FPCA, according to the methodology proposed in Pesaresi et al. [37], proved to be an effective tool for mapping several plant associations (of different structure and specific composition) for an entire SAC. We used supervised random forest classification, similarly to Zhu and Liu [29,73,74,75], and revealed that the main spectral (phenological) seasonal variations contributed to the OA (here, 85.58%) of the produced map, much more strongly than the lithological and topographic features (see Table A1).
- 2.
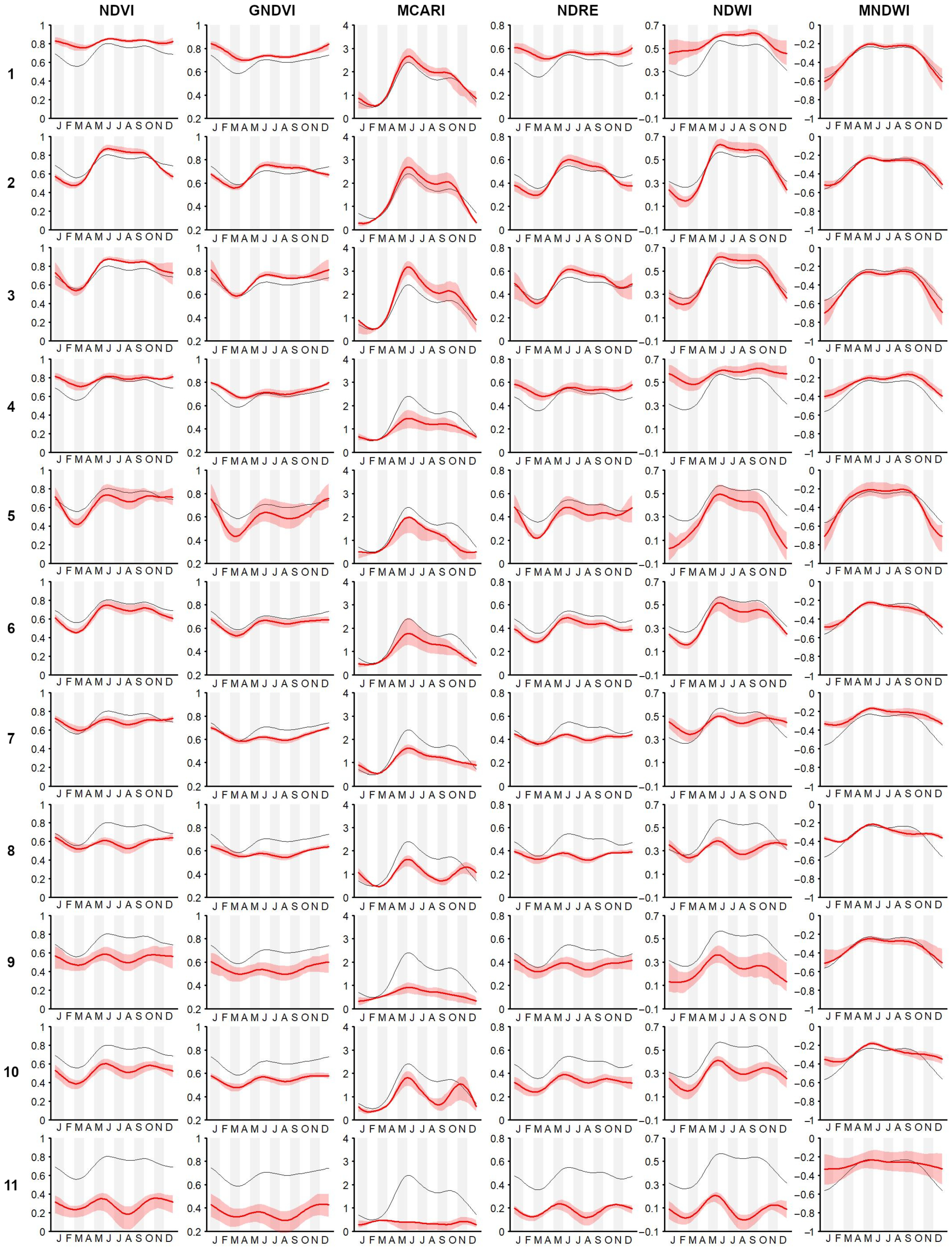
- Simultaneous use of multiple time-series We confirmed the importance of using the popular NDVI time-series [76]. It was demonstrated (as in other related works, e.g., [8,24,75]) that the integration of multi-spectral (vegetation index) time-series can improve the performance of vegetation and habitat mapping, as seasonal patterns manifest differently in different spectral bands and vegetation indices [28] (Figure A3).For example, holm-oak wood and Pinus sp. plantations had similar seasonal temporal profiles, in terms of the NDVI, while presenting clearly different MCARI profiles (Figure A3). The supervised RF models constructed using the time-series individually revealed that, for the study area, seasonal variations of the NDVI time-series were the most explanatory (OA 73.13% alone), while RF model constructed using the time-series jointly revealed some complementarity between the time-series, where each appeared to be useful in classifying different plant associations.The integration of all six considered spectral vegetation indices supplied the RF with crucial variations characterizing and discriminating the different plant associations. With the combined model, an improvement of 9% in the OA over the best single time-series was obtained. When we also considered the topographic and lithological features, an additional 3.4% improvement was achieved (Table A1).
- 3.
- Mapping accuracy The obtained results, in terms of the OA (85.58%), demonstrated a meaningful gain of performance when compared with the existing map [72]. Habitat (92/43/EEC) and plant association maps (using the Braun–Blanquet approach) rarely reach an overall accuracy greater than 80% [17]. In most cases, the number of classes is up to five [11,14,37,77,78,79,80], while an increase in the number of classes typically has a negative impact on the overall performance, reaching values close to 75%, as in [9,16,80,81,82,83,84].From a methodological point of view, it is necessary to define an acceptable level of accuracy of a map generated using remotely sensed data [85,86], taking into consideration the related use-cases and scenarios. Although it is challenging to define a minimum value threshold for the mapping of plant communities and habitats, according to the above-mentioned references, we could consider that, with a number of target classes greater than five, an OA value of 80% represents a good result, while the range of 75–80% could be sufficient. When the number of classes is ≤5, a good result should exceed 85% OA, and it can be considered satisfactory if it exceeds the 80%.
- 4.
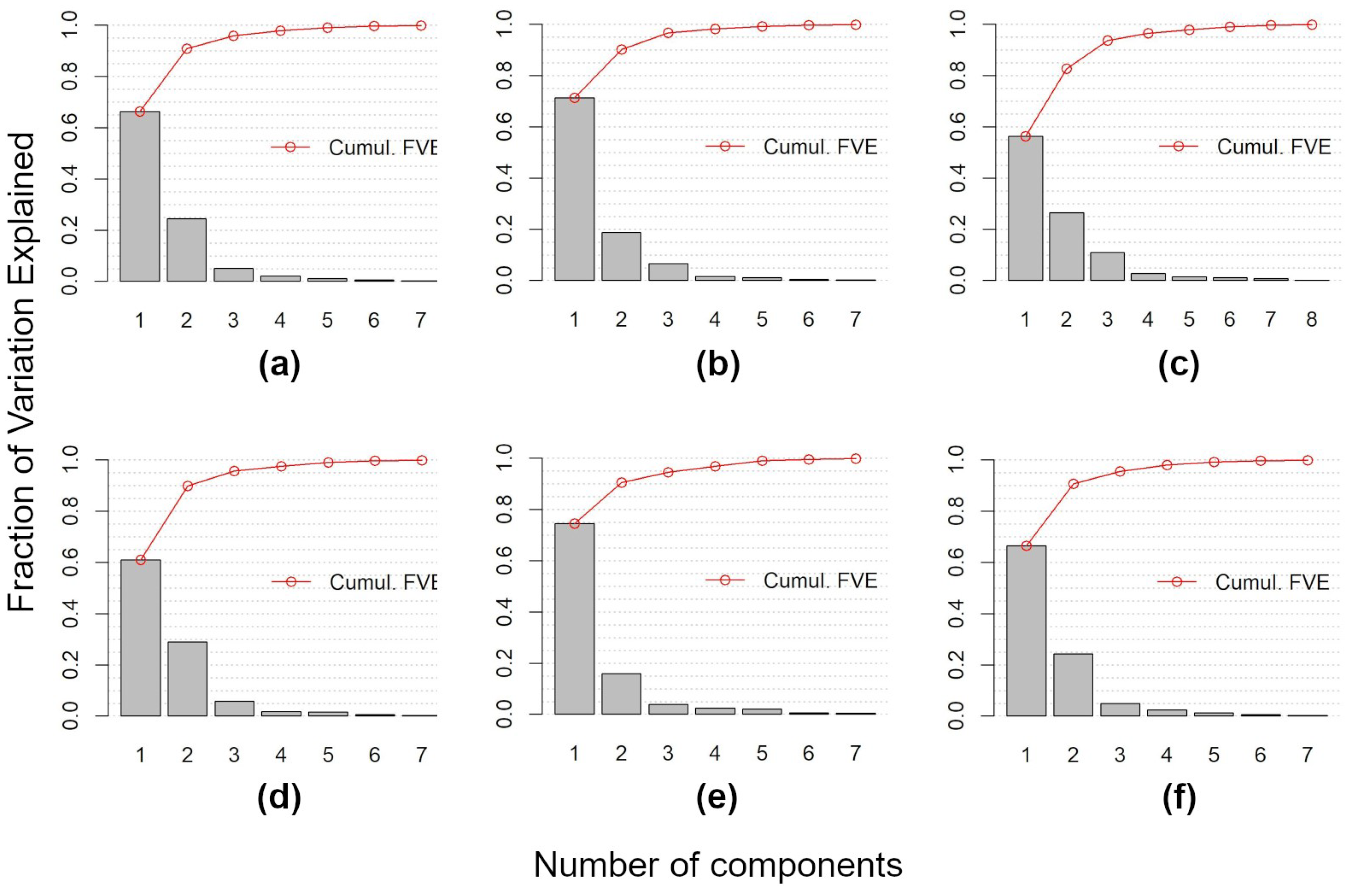
- Data Reduction using FPCA FPCA is an efficient input data reduction tool. First, it is the central idea of the FDA itself—compressing the input data. Indeed, it considers the pixel-based time-series as a function and as the (unique) object of analysis. Consequently, FPCA considers a stack of remote sensed images as a single container of pixel-based functions, regardless of the number of images that compose it.Then, FPCA extracts, from these pixel-based functions (considered as cohesive temporal record of pixel-based time-series), the main and orthogonal modalities of variation (i.e., functional components, numerically represented by the FPC scores that express the different seasonalities), preserving the order (i.e., chronology) of the data and facilitating ecological interpretation of the derived temporal and spatial patterns (see Figure A2; as well as [31,36,37]).In this work, for each vegetation index, we derived the time-series (set of values over 52 weeks) and then applied FPCA. This technique reduced the size to a total of 43 functional components (considering multi-spectral seasonality); see Figure A1 and Figure A2.Furthermore, functional analysis allows for the study of derivatives, thus, providing complementary information to describe the seasonal cycles derived from satellite data (see, e.g., [87,88]). The FPCA components with a low fraction of variance explained could also be evaluated by experts in order to derive local changes that are potential indicators of different plant communities [89].
- 5.
- Benefits for habitat directives, phyto-sociology, and landscape management The overall accuracy obtained (OA 85.58%) was greater than 80% and much higher than that obtained with the traditional method (photo-interpretation), thus, making the produced map (Figure 3) a reliable and usable tool in the main procedures of habitat directives (e.g., Appropriate Assessments and filling in of Standard Data Forms).In the study area, the high temporal resolution of the Sentinel-2 mission makes mapping (through the functional approach) of the vegetation and habitats up-to-date and repeatable, according to the timing required by the directive (six years), as well as with higher frequencies (e.g., every 2–3 years) [9]. Cyclical mapping (at least every six years, in accordance with the Habitats Directive) of the plant associations—and, therefore, of the habitats—with high accuracy (in terms of OA) could provide a valid tool for monitoring the transformation of habitats over time [10,90].For example, the conservation status of the grasslands in the studied area is strongly determined by the presence of invasive shrubs or species. The grasslands of habitat 6210, in fact, if not adequately managed, tend to be colonized by shrubs (e.g., Spartium junceum and Juniperus oxycedrus) and by a few perennial grasses, such as those of the genera Brachypodium, with a consequent loss of the floristic diversity of the grasslands [91,92,93,94].Any variation or modification of the phenological profile of the habitat (as identifiable through the use of the proposed methodology) can be used as a warning signal that, in certain areas, a transformation process is underway, therefore, highlighting the need for an inspection on the ground and, consequently, the need for corrective and timely management actions.The limitation for modeling and mapping plant communities is no longer the access and processing of satellite data, but rather the time it takes to access and generate reference data in the field. Therefore, it is crucial to disseminate the vegetation plots in databases (e.g., VegItaly [95]) [9]. The production of reference data at the plant associations level could benefit from the help of drones. An unmanned aerial vehicle could enable the recognition of the plant species (see, e.g., [96]) while reaching inaccessible places for human beings due to orography and many other factors.Recognizing few indicator species (by drone) could by key for the quick identification of vegetation types (plant association) in the field in complex environments [97] where the vegetation types and their species composition are available. Despite the high global accuracy of the map, the error matrix (Table 4) showed that black hornbeam wood and downy-oak wood (habitat 91AA*) could be misclassified.This was due to the fact that both woods share some species in the dominant tree layer, although the underlying, dominated layers are well-differentiated, in terms of mesophilic species dominating in the first case, with more thermophilic in the second. Therefore, from an ecological point of view, they are different woods, even if the tree structure and composition is partially similar. This confusion led to over-representation of downy-oak wood (PA 82.5% and UA of 65.9%) and under-representation of black hornbeam wood (PA 68.3% and UA 85.4%) in the predicted map (Figure 3). However, these accuracies were still much better than those obtained with traditional mapping.The methodology presented in this work enables a concrete link between the remote survey perspective and the (phyto-sociological) field-based one. Manual (traditional) approaches could be empowered by supervised ones, which, in any case, still rely on reference data generated by domain experts.Vegetation has unique spectral signatures that evolve along with the plant life cycle over the year [75], which represents an important trait of plant associations (Figure A3). The quantification of patterns in the seasonal behavior of the spectral reflectance can provide better characterization of plant communities [74]. Furthermore, FPCA scores represent reduced ordination spaces (i.e., useful tools for botanists and ecologists) [31,35,36] that are suitable for the ecological interpretation of the results and that contribute to the study of the relationships between the data observed in the field and those that are remotely sensed.
Limits and Challenges for Future Work
- 1.
- We found that the combined use of different spectral vegetation indices led to better results compared with using a single one. Of course, the six selected vegetation indices have been widely adopted by researchers; however, we cannot exclude that a different set of vegetation indices combining two or more bands could improve the overall performance, perhaps also fixing the misclassification between black hornbeam and downy-oak woods. The typical formula adopted for the NDVI is in the form of where a and b are spectral bands; this formula is only an example, and other formulas/functions may improve the final mapping.
- 2.
- If we need to consider multiple time-series derived from a wider set of vegetation indices, we must adapt an approach considering more advanced FDA techniques. A possible solution is to adopt the Multidimensional FPCA (MFPCA) [98], which enables reduction of the time-series (i.e., a lower number of components extracted from multiple time-series), thus, providing a single phenological ordering space to make the ecological interpretation easier [37]. Although harmonic and phenological features have recently been used to improve forest type mapping, as in [29,74], new vegetation indices could be derived, with novel formulas, in order to increase the spectral separation among target classes.
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Conflicts of Interest
Appendix A
| a | b | c | d | e | f | g | h | i | l | |
|---|---|---|---|---|---|---|---|---|---|---|
| OA | 59.4 | 59.2 | 73.1 | 69.5 | 63.7 | 69.5 | 67.9 | 67.3 | 82.1 | 85.5 |
| 8.3 | 7.2 | 7.6 | 7.2 | 8.7 | 7.1 | 7.6 | 6.3 | 5.2 | ||
| K | 0.54 | 0.53 | 0.69 | 0.65 | 0.58 | 0.65 | 0.63 | 0.62 | 0.79 | 0.83 |
| 0.09 | 0.08 | 0.08 | 0.08 | 0.10 | 0.08 | 0.08 | 0.07 | 0.06 | ||
| PA | ||||||||||
| 1 | 43.2 | 70.3 | 86.5 | 78.4 | 67.6 | 81.1 | 81.1 | 73.0 | 91.9 | 91.9 |
| 2 | 44.1 | 38.2 | 79.4 | 76.5 | 64.7 | 79.4 | 70.6 | 64.7 | 79.4 | 85.3 |
| 3 | 56.7 | 56.7 | 58.3 | 71.7 | 60.0 | 56.7 | 46.7 | 68.3 | 65.0 | 68.3 |
| 4 | 90.6 | 37.5 | 78.1 | 62.5 | 59.4 | 84.4 | 78.1 | 84.4 | 90.6 | 90.6 |
| 5 | 87.5 | 93.8 | 56.3 | 87.5 | 62.5 | 68.8 | 68.8 | 56.3 | 87.5 | 100.0 |
| 6 | 37.5 | 50.0 | 75.0 | 68.8 | 37.5 | 68.8 | 68.8 | 68.8 | 68.8 | 81.3 |
| 7 | 80.0 | 93.3 | 80.0 | 80.0 | 86.7 | 86.7 | 73.3 | 80.0 | 93.3 | 93.3 |
| 8 | 93.8 | 62.5 | 81.3 | 81.3 | 75.0 | 68.8 | 81.3 | 75.0 | 81.3 | 87.5 |
| 9 | 36.7 | 53.1 | 69.4 | 63.3 | 55.1 | 65.3 | 55.1 | 51.0 | 77.6 | 87.8 |
| 10 | 72.2 | 61.1 | 66.7 | 61.1 | 72.2 | 66.7 | 72.2 | 66.7 | 72.2 | 72.2 |
| 11 | 73.3 | 80.0 | 80.0 | 73.3 | 80.0 | 86.7 | 86.7 | 60.0 | 80.0 | 86.7 |
| UA | ||||||||||
| 1 | 47.1 | 68.4 | 78.0 | 72.5 | 64.1 | 83.3 | 85.7 | 64.3 | 82.9 | 85.0 |
| 2 | 36.6 | 44.8 | 57.4 | 68.4 | 47.8 | 57.4 | 49.0 | 51.2 | 60.0 | 65.9 |
| 3 | 57.6 | 64.2 | 79.5 | 86.0 | 75.0 | 82.9 | 68.3 | 77.4 | 83.0 | 85.4 |
| 4 | 63.0 | 44.4 | 75.8 | 60.6 | 57.6 | 71.1 | 78.1 | 84.4 | 90.6 | 90.6 |
| 5 | 100.0 | 75.0 | 60.0 | 87.5 | 47.6 | 68.8 | 37.9 | 60.0 | 77.8 | 88.9 |
| 6 | 40.0 | 42.1 | 57.1 | 45.8 | 35.3 | 45.8 | 68.8 | 61.1 | 64.7 | 76.5 |
| 7 | 100.0 | 66.7 | 52.2 | 60.0 | 68.4 | 61.9 | 47.8 | 66.7 | 70.0 | 87.5 |
| 8 | 55.6 | 43.5 | 76.5 | 86.7 | 85.7 | 78.6 | 81.3 | 75.0 | 81.3 | 82.4 |
| 9 | 78.3 | 65.0 | 91.9 | 86.1 | 71.1 | 84.2 | 87.1 | 71.4 | 88.4 | 93.5 |
| 10 | 50.0 | 61.1 | 85.7 | 64.7 | 86.7 | 75.0 | 65.0 | 63.2 | 100.0 | 92.9 |
| 11 | 100.0 | 60.0 | 75.0 | 57.9 | 66.7 | 76.5 | 81.3 | 52.9 | 75.0 | 81.3 |
| Month | Frequency |
|---|---|
| January | 4 |
| February | 8 |
| March | 7 |
| April | 7 |
| May | 8 |
| June | 9 |
| July | 12 |
| August | 14 |
| September | 9 |
| October | 7 |
| November | 3 |
| December | 5 |
| sum | 93 |
| Num | Date | Doy | Month | Num | Date | Doy | Month |
|---|---|---|---|---|---|---|---|
| 1 | 21 April 2017 | 111 | 4 | 48 | 3 October 2018 | 276 | 10 |
| 2 | 1 May 2017 | 121 | 5 | 49 | 13 October 2018 | 286 | 10 |
| 3 | 11 May 2017 | 131 | 5 | 50 | 12 November 2018 | 316 | 11 |
| 4 | 31 May 2017 | 151 | 5 | 51 | 7 December 2018 | 341 | 12 |
| 5 | 20 June 2017 | 171 | 6 | 52 | 12 December 2018 | 346 | 12 |
| 6 | 10 July 2017 | 191 | 7 | 53 | 27 December 2018 | 361 | 12 |
| 7 | 20 July 2017 | 201 | 7 | 54 | 26 January 2019 | 26 | 1 |
| 8 | 30 July 2017 | 211 | 7 | 55 | 5 February 2019 | 36 | 2 |
| 9 | 9 August 2017 | 221 | 8 | 56 | 15 February 2019 | 46 | 2 |
| 10 | 19 August 2017 | 231 | 8 | 57 | 20 February 2019 | 51 | 2 |
| 11 | 29 August 2017 | 241 | 8 | 58 | 25 February 2019 | 56 | 2 |
| 12 | 18 September 2017 | 261 | 9 | 59 | 2 March 2019 | 61 | 3 |
| 13 | 8 October 2017 | 281 | 10 | 60 | 12 March 2019 | 71 | 3 |
| 14 | 18 October 2017 | 291 | 10 | 61 | 17 March 2019 | 76 | 3 |
| 15 | 28 October 2017 | 301 | 10 | 62 | 22 March 2019 | 81 | 3 |
| 16 | 27 November 2017 | 331 | 11 | 63 | 1 April 2019 | 91 | 4 |
| 17 | 7 December 2017 | 341 | 12 | 64 | 16 April 2019 | 106 | 4 |
| 18 | 22 December 2017 | 356 | 12 | 65 | 31 May 2019 | 151 | 5 |
| 19 | 6 January 2018 | 6 | 1 | 66 | 5 June 2019 | 156 | 6 |
| 20 | 15 February 2018 | 46 | 2 | 67 | 15 June 2019 | 166 | 6 |
| 21 | 6 April 2018 | 96 | 4 | 68 | 25 June 2019 | 176 | 6 |
| 22 | 16 April 2018 | 106 | 4 | 69 | 30 June 2019 | 181 | 6 |
| 23 | 21 April 2018 | 111 | 4 | 70 | 5 July 2019 | 186 | 7 |
| 24 | 26 April 2018 | 116 | 4 | 71 | 20 July 2019 | 201 | 7 |
| 25 | 11 May 2018 | 131 | 5 | 72 | 25 July 2019 | 206 | 7 |
| 26 | 16 May 2018 | 136 | 5 | 73 | 30 July 2019 | 211 | 7 |
| 27 | 21 May 2018 | 141 | 5 | 74 | 4 August 2019 | 216 | 8 |
| 28 | 31 May 2018 | 151 | 5 | 75 | 9 August 2019 | 221 | 8 |
| 29 | 10 June 2018 | 161 | 6 | 76 | 14 August 2019 | 226 | 8 |
| 30 | 15 June 2018 | 166 | 6 | 77 | 19 August 2019 | 231 | 8 |
| 31 | 20 June 2018 | 171 | 6 | 78 | 24 August 2019 | 236 | 8 |
| 32 | 30 June 2018 | 181 | 6 | 79 | 29 August 2019 | 241 | 8 |
| 33 | 10 July 2018 | 191 | 7 | 80 | 8 September 2019 | 251 | 9 |
| 34 | 15 July 2018 | 196 | 7 | 81 | 13 September 2019 | 256 | 9 |
| 35 | 20 July 2018 | 201 | 7 | 82 | 18 September 2019 | 261 | 9 |
| 36 | 25 July 2018 | 206 | 7 | 83 | 8 October 2019 | 281 | 10 |
| 37 | 30 July 2018 | 211 | 7 | 84 | 23 October 2019 | 296 | 10 |
| 38 | 4 August 2018 | 216 | 8 | 85 | 7 November 2019 | 311 | 11 |
| 39 | 9 August 2018 | 221 | 8 | 86 | 1 January 2020 | 1 | 1 |
| 40 | 19 August 2018 | 231 | 8 | 87 | 6 January 2020 | 6 | 1 |
| 41 | 24 August 2018 | 236 | 8 | 88 | 5 February 2020 | 36 | 2 |
| 42 | 29 August 2018 | 241 | 8 | 89 | 15 February 2020 | 46 | 2 |
| 43 | 3 September 2018 | 246 | 9 | 90 | 20 February 2020 | 51 | 2 |
| 44 | 8 September 2018 | 251 | 9 | 91 | 11 March 2020 | 71 | 3 |
| 45 | 18 September 2018 | 261 | 9 | 92 | 16 March 2020 | 76 | 3 |
| 46 | 23 September 2018 | 266 | 9 | 93 | 21 March 2020 | 81 | 3 |
| 47 | 28 September 2018 | 271 | 9 |
References
- Braun–Blanquet, J.; Conard, H.S.; Fuller, G.D. Plant Sociology: The Study of Plant Communities, 1st ed.; McGraw-Hill: New York, NY, USA; London, UK, 1932; p. 439. [Google Scholar] [CrossRef] [Green Version]
- Chytrý, M.; Schaminée, J.H.J.; Schwabe, A. Vegetation survey: A new focus for Applied Vegetation Science. Appl. Veg. Sci. 2011, 14, 435–439. [Google Scholar] [CrossRef]
- Mucina, L. Europe, Ecosystems of. In Encyclopedia of Biodiversity, 2nd ed.; Levin, S.A., Ed.; Academic Press: Waltham, MA, USA, 2013; pp. 333–346. [Google Scholar] [CrossRef]
- Biondi, E. Phytosociology today: Methodological and conceptual evolution. Plant Biosyst. 2011, 145, 19–29. [Google Scholar] [CrossRef]
- Biondi, E.; Burrascano, S.; Casavecchia, S.; Copiz, R.; Del Vico, E.; Galdenzi, D.; Gigante, D.; Lasen, C.; Spampinato, G.; Venanzoni, R.; et al. Diagnosis and syntaxonomic interpretation of Annex I Habitats (Dir. 92/43/EEC) in Italy at the alliance level. Plant Sociol. 2012, 49, 5–37. [Google Scholar] [CrossRef]
- Rodwell, J.S.; Evans, D.; Schaminée, J.H. Phytosociological relationships in European Union policy-related habitat classifications. Rend. Lincei 2018, 29, 237–249. [Google Scholar] [CrossRef]
- CEC. Council Directive 92/43/EEC of 21 May 1992 on the conservation of natural habitats and of wild fauna and flora. Off. J. L 1992, 206, 7–50. [Google Scholar]
- Tarantino, C.; Forte, L.; Blonda, P.; Vicario, S.; Tomaselli, V.; Beierkuhnlein, C.; Adamo, M. Intra-annual sentinel-2 time-series supporting grassland habitat discrimination. Remote Sens. 2021, 13, 277. [Google Scholar] [CrossRef]
- Rapinel, S.; Rozo, C.; Delbosc, P.; Bioret, F.; Bouzillé, J.B.; Hubert-Moy, L. Contribution of free satellite time-series images to mapping plant communities in the Mediterranean Natura 2000 site: The example of Biguglia Pond in Corse (France). Mediterr. Bot. 2020, 41, 181–191. [Google Scholar] [CrossRef]
- Ichter, J.; Savio, L.; Evans, D.; Poncet, L. State-of-the-art of vegetation mapping in Europe: Results of a European survey and contribution to the French program CarHAB. Doc. Phytosociol. Ser. 3 2017, 6, 335–352. [Google Scholar]
- Feret, J.B.; Corbane, C.; Alleaume, S. Detecting the Phenology and Discriminating Mediterranean Natural Habitats with Multispectral Sensors-An Analysis Based on Multiseasonal Field Spectra. IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2015, 8, 2294–2305. [Google Scholar] [CrossRef]
- Corbane, C.; Güttler, F.; Alleaume, S.; Ienco, D.; Teisseire, M. Monitoring the phenology of mediterranean natural habitats with multispectral sensors—An analysis based on multiseasonal field spectra. In Proceedings of the 2014 IEEE Geoscience and Remote Sensing Symposium, Quebec City, QC, Canada, 13–18 July 2014; pp. 3934–3937. [Google Scholar] [CrossRef] [Green Version]
- Grignetti, A.; Salvatori, R.; Casacchia, R.; Manes, F. Mediterranean vegetation analysis by multi-temporal satellite sensor data. Int. J. Remote Sens. 1997, 18, 1307–1318. [Google Scholar] [CrossRef]
- Marzialetti, F.; Giulio, S.; Malavasi, M.; Sperandii, M.G.; Acosta, A.T.R.; Carranza, M.L. Capturing Coastal Dune Natural Vegetation Types Using a Phenology-Based Mapping Approach: The Potential of Sentinel-2. Remote Sens. 2019, 11, 1506. [Google Scholar] [CrossRef] [Green Version]
- Hoffmann, S.; Schmitt, T.M.; Chiarucci, A.; Irl, S.D.H.; Rocchini, D.; Vetaas, O.R.; Tanase, M.A.; Mermoz, S.; Bouvet, A.; Beierkuhnlein, C. Remote sensing of β-diversity: Evidence from plant communities in a semi-natural system. Appl. Veg. Sci. 2019, 22, 13–26. [Google Scholar] [CrossRef] [Green Version]
- Rapinel, S.; Mony, C.; Lecoq, L.; Clément, B.; Thomas, A.; Hubert-Moy, L. Evaluation of Sentinel-2 time-series for mapping floodplain grassland plant communities. Remote Sens. Environ. 2019, 223, 115–129. [Google Scholar] [CrossRef]
- Vanden Borre, J.; Paelinckx, D.; Mücher, C.A.; Kooistra, L.; Haest, B.; De Blust, G.; Schmidt, A.M. Integrating remote sensing in Natura 2000 habitat monitoring: Prospects on the way forward. J. Nat. Conserv. 2011, 19, 116–125. [Google Scholar] [CrossRef]
- Corbane, C.; Lang, S.; Pipkins, K.; Alleaume, S.; Deshayes, M.; García Millán, V.E.; Strasser, T.; Vanden Borre, J.; Toon, S.; Michael, F. Remote sensing for mapping natural habitats and their conservation status—New opportunities and challenges. Int. J. Appl. Earth Obs. Geoinf. 2015, 37, 7–16. [Google Scholar] [CrossRef]
- Cabello, J.; Mairota, P.; Alcaraz-Segura, D.; Arenas-Castro, S.; Escribano, P.; Leitão, P.J.; Martínez-López, J.; Regos, A.; Requena-Mullor, J.M. Satellite remote sensing of ecosystem functions: Opportunities and challenges for reporting obligations of the EU habitat directive. Int. Geosci. Remote Sens. Symp. 2018, 2018, 6604–6607. [Google Scholar] [CrossRef]
- Schmidt, J.; Fassnacht, F.E.; Förster, M.; Schmidtlein, S. Synergetic use of Sentinel-1 and Sentinel-2 for assessments of heathland conservation status. Remote Sens. Ecol. Conserv. 2018, 4, 225–239. [Google Scholar] [CrossRef] [Green Version]
- Marzialetti, F.; Di Febbraro, M.; Malavasi, M.; Giulio, S.; Acosta, A.T.R.; Carranza, M.L. Mapping Coastal Dune Landscape through Spectral Rao’s Q Temporal Diversity. Remote Sens. 2020, 12, 2315. [Google Scholar] [CrossRef]
- Stendardi, L.; Karlsen, S.R.; Niedrist, G.; Gerdol, R.; Zebisch, M.; Rossi, M.; Notarnicola, C. Exploiting time series of Sentinel-1 and Sentinel-2 imagery to detect meadow phenology in mountain regions. Remote Sens. 2019, 11, 542. [Google Scholar] [CrossRef] [Green Version]
- Bajocco, S.; Ferrara, C.; Alivernini, A.; Bascietto, M.; Ricotta, C. Remotely-sensed phenology of Italian forests: Going beyond the species. Int. J. Appl. Earth Obs. Geoinf. 2019, 74, 314–321. [Google Scholar] [CrossRef]
- Chignell, S.M.; Luizza, M.W.; Skach, S.; Young, N.E.; Evangelista, P.H. An integrative modeling approach to mapping wetlands and riparian areas in a heterogeneous Rocky Mountain watershed. Remote Sens. Ecol. Conserv. 2018, 4, 150–165. [Google Scholar] [CrossRef] [Green Version]
- Schmidt, T.; Schuster, C.; Kleinschmit, B.; Förster, M. Evaluating an intra-annual time series for grassland classification—How many acquisitions and what seasonal origin are optimal? IEEE J. Sel. Top. Appl. Earth Obs. Remote Sens. 2014, 7, 3428–3439. [Google Scholar] [CrossRef]
- Lopes, M.; Frison, P.L.; Durant, S.M.; Schulte to Bühne, H.; Ipavec, A.; Lapeyre, V.; Pettorelli, N. Combining optical and radar satellite image time series to map natural vegetation: Savannas as an example. Remote Sens. Ecol. Conserv. 2020, 6, 316–326. [Google Scholar] [CrossRef]
- Young, N.E.; Anderson, R.S.; Chignell, S.M.; Vorster, A.G.; Lawrence, R.; Evangelista, P.H. A survival guide to Landsat preprocessing. Ecology 2017, 98, 920–932. [Google Scholar] [CrossRef] [Green Version]
- Pasquarella, V.J.; Holden, C.E.; Kaufman, L.; Woodcock, C.E. From imagery to ecology: Leveraging time series of all available Landsat observations to map and monitor ecosystem state and dynamics. Remote Sens. Ecol. Conserv. 2016, 2, 152–170. [Google Scholar] [CrossRef]
- Adams, B.; Iverson, L.; Matthews, S.; Peters, M.; Prasad, A.; Hix, D.M. Mapping Forest Composition with Landsat Time Series: An Evaluation of Seasonal Composites and Harmonic Regression. Remote Sens. 2020, 12, 610. [Google Scholar] [CrossRef] [Green Version]
- Kennedy, R.E.; Andréfouët, S.; Cohen, W.B.; Gómez, C.; Griffiths, P.; Hais, M.; Healey, S.P.; Helmer, E.H.; Hostert, P.; Lyons, M.B.; et al. Bringing an ecological view of change to landsat-based remote sensing. Front. Ecol. Environ. 2014, 12, 339–346. [Google Scholar] [CrossRef]
- Hurley, M.A.; Hebblewhite, M.; Gaillard, J.; Dray, S.; Taylor, K.A.; Smith, W.K.; Zager, P.; Bonenfant, C. Functional analysis of normalized difference vegetation index curves reveals overwinter mule deer survival is driven by both spring and autumn phenology. Philos. Trans. R. Soc. Lond. Ser. B Biol. Sci. 2014, 369, 20130196. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ramsay, J. Functional Data Analysis; John Wiley & Sons: Hoboken, NJ, USA, 2005. [Google Scholar] [CrossRef]
- Di Salvo, F.; Ruggieri, M.; Plaia, A. Functional principal component analysis for multivariate multidimensional environmental data. Environ. Ecol. Stat. 2015, 22, 739–757. [Google Scholar] [CrossRef]
- Shang, H.L. A survey of functional principal component analysis. Adv. Stat. Anal. 2014, 98, 121–142. [Google Scholar] [CrossRef] [Green Version]
- Li, H.; Xiao, G.; Xia, T.; Tang, Y.Y.; Li, L. Hyperspectral image classification using functional data analysis. IEEE Trans. Cybern. 2014, 44, 1544–1555. [Google Scholar] [CrossRef] [PubMed]
- Pesaresi, S.; Mancini, A.; Casavecchia, S. Recognition and Characterization of Forest Plant Communities through Remote-Sensing NDVI Time Series. Diversity 2020, 12, 313. [Google Scholar] [CrossRef]
- Pesaresi, S.; Mancini, A.; Quattrini, G.; Casavecchia, S. Mapping mediterranean forest plant associations and habitats with functional principal component analysis using Landsat 8 NDVI time series. Remote Sens. 2020, 12, 1132. [Google Scholar] [CrossRef] [Green Version]
- Rivas-Martínez, S.; Sáenz, S.R.; Penas, A. Worldwide bioclimatic classification system. Glob. Geobot. 2011, 1, 1–634. [Google Scholar]
- Pesaresi, S.; Biondi, E.; Casavecchia, S. Bioclimates of Italy. J. Maps 2017, 13, 955–960. [Google Scholar] [CrossRef]
- Biondi, E.; Casavecchia, S.; Gigante, D. Contribution to the syntaxonomic knowledge of the Quercus ilex L. woods of the Central European Mediterranean Basin. Fitosociologia 2003, 40, 129–156. [Google Scholar]
- Blasi, C.; Feoli, E.; Avena, G.C. Due nuove associazioni dei Quercetalia pubescentis dell’Appennino centrale. Stud. Geobot. 1982, 2, 155–167. [Google Scholar]
- Blasi, C.; Di Pietro, R.; Filesi, L. Syntaxonomical revision of Quercetalia pubescenti-petraeae in the Italian Peninsula. Fitosociologia 2004, 41, 87–164. [Google Scholar]
- Allegrezza, M.; Pesaresi, S.; Ballelli, S.; Tesei, G.; Ottaviani, C. Influences of mature pinus nigra plantations on the floristic-vegetational composition along an altitudinal gradient in the central apennines, Italy. IForest 2020, 13, 279–285. [Google Scholar] [CrossRef]
- Biondi, E.; Casavecchia, S. Inquadramento fitosociologico della vegetazione arbustiva di un settore dell’Appennino settentrionale. Fitosociologia 2002, 39, 65–73. [Google Scholar]
- Biondi, E.; Allegrezza, M.; Zuccarello, V. Syntaxonomic revision of the Apennine grasslands belonging to Brometalia erecti, and an analysis of their relationships with the xerophilous vegetation of Rosmarinetea officinalis (Italy). Phytocoenologia 2005, 35, 129–164. [Google Scholar] [CrossRef]
- Allegrezza, M.; Biondi, E.; Ballelli, S.; Formica, E. La vegetazione dei settori rupestri calcarei dell’Italia centrale. Fitosociologia 1997, 32, 91–120. [Google Scholar]
- Geobotanic Group at Università Politecnica delle Marche. Dataset and Code Related to the Habitat Mapping of SAC of Frasassi. Available online: https://github.com/geobotany/habitatmapfrasassi (accessed on 10 February 2022).
- Franklin, J. Predictive vegetation mapping: Geographic modelling of biospatial patterns in relation to environmental gradients. Prog. Phys. Geogr. Earth Environ. 1995, 19, 474–499. [Google Scholar] [CrossRef]
- Regione Marche. La Carta Geologica Della Regione Marche in Scala 1:10.000. 2001. Available online: http://www.regione.marche.it/Regione-Utile/Paesaggio-Territorio-Urbanistica/Cartografia/Repertorio/Cartageologicaregionale10000 (accessed on 10 February 2022).
- Dray, S.; Dufour, A.B. The ade4 Package: Implementing the Duality Diagram for Ecologists. J. Stat. Softw. 2007, 22, 1–20. [Google Scholar] [CrossRef] [Green Version]
- Ranghetti, L.; Boschetti, M.; Nutini, F.; Busetto, L. “sen2r”: An R toolbox for automatically downloading and preprocessing Sentinel-2 satellite data. Comput. Geosci. 2020, 139, 104473. [Google Scholar] [CrossRef]
- Rouse, J.W.; Haas, R.H.; Schell, J.A.; Deering, D.W. Monitoring vegetation systems in the Great Plains with ERTS 10–14 December. In Proceedings of the Third ERTS Symposium; NASA: Washington, DC, USA, 1973; pp. 309–317. [Google Scholar]
- Buschmann, C.; Nagel, E. In vivo spectroscopy and internal optics of leaves as basis for remote sensing of vegetation. Int. J. Remote Sens. 1993, 14, 711–722. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Kaufman, Y.J.; Merzlyak, M.N. Use of a green channel in remote sensing of global vegetation from EOS-MODIS. Remote Sens. Environ. 1996, 58, 289–298. [Google Scholar] [CrossRef]
- Daughtry, C. Estimating Corn Leaf Chlorophyll Concentration from Leaf and Canopy Reflectance. Remote Sens. Environ. 2000, 74, 229–239. [Google Scholar] [CrossRef]
- Barnes, E.; Clarke, T.; Richards, S.; Colaizzi, P.; Haberland, J.; Kostrzewski, M.; Waller, P.; Choi, C.; Riley, E.; Thompson, T.; et al. Coincident detection of crop water stress, nitrogen status and canopy density using ground based multispectral data. In Proceedings of the Fifth International Conference on Precision Agriculture, Bloomington, MN, USA, 16–19 July 2000; Volume 1619. [Google Scholar]
- Gao, B.C. NDWI—A normalized difference water index for remote sensing of vegetation liquid water from space. Remote Sens. Environ. 1996, 58, 257–266. [Google Scholar] [CrossRef]
- Xu, H. Modification of normalised difference water index (NDWI) to enhance open water features in remotely sensed imagery. Int. J. Remote Sens. 2006, 27, 3025–3033. [Google Scholar] [CrossRef]
- Fisher, J.I.; Mustard, J.F.; Vadeboncoeur, M.A. Green leaf phenology at Landsat resolution: Scaling from the field to the satellite. Remote Sens. Environ. 2006, 100, 265–279. [Google Scholar] [CrossRef]
- Schuster, C.; Schmidt, T.; Conrad, C.; Kleinschmit, B.; Förster, M. Grassland habitat mapping by intra-annual time series analysis -Comparison of RapidEye and TerraSAR-X satellite data. Int. J. Appl. Earth Obs. Geoinf. 2015, 34, 25–34. [Google Scholar] [CrossRef]
- Hyndman, R.J.; Khandakar, Y. Automatic Time Series Forecasting: The forecast Package for R. J. Stat. Softw. 2008, 27, 1–22. [Google Scholar] [CrossRef] [Green Version]
- Hyndman, R.; Athanasopoulos, G.; Bergmeir, C.; Caceres, G.; Chhay, L.; O’Hara-Wild, M.; Petropoulos, F.; Razbash, S.; Wang, E.; Yasmeen, F. Forecast: Forecasting Functions for Time Series and Linear Models, R Package Version 8.15; 2021. Available online: https://pkg.robjhyndman.com/forecast/ (accessed on 10 February 2022).
- Jacques, J.; Preda, C. Functional data clustering: A survey. Adv. Data Anal. Classif. 2014, 8, 231–255. [Google Scholar] [CrossRef] [Green Version]
- Dai, X.; Hadjipantelis, P.Z.; Han, K.; Ji, H. Fdapace: Functional Data Analysis and Empirical Dynamics, R Package Version 0.4.0; 2018. Available online: https://CRAN.R-project.org/package=fdapace/ (accessed on 10 February 2022).
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Drăguţ, L. Random forest in remote sensing: A review of applications and future directions. ISPRS J. Photogramm. Remote Sens. 2016, 114, 24–31. [Google Scholar] [CrossRef]
- Evans, J.S.; Cushman, S.A. Gradient modeling of conifer species using random forests. Landsc. Ecol. 2009, 24, 673–683. [Google Scholar] [CrossRef]
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, R Package Version 2.8-4; 2018. Available online: https://CRAN.R-project.org/package=raster/ (accessed on 10 February 2022).
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. Artic. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Congalton, R.G. A review of assessing the accuracy of classifications of remotely sensed data. Remote Sens. Environ. 1991, 37, 35–46. [Google Scholar] [CrossRef]
- Cohen, J. A Coefficient of Agreement for Nominal Scales. Educ. Psychol. Meas. 1960, 20, 37–46. [Google Scholar] [CrossRef]
- Frontoni, E.; Mancini, A.; Zingaretti, P.; Malinverni, E.S.; Pesaresi, S.; Biondi, E.; Pandolfi, M.; Marseglia, M.; Sturari, M.; Zabaglia, C. SIT-REM: An Interoperable and Interactive Web Geographic Information System for Fauna, Flora and Plant Landscape Data Management. ISPRS Int. J. Geo-Inf. 2014, 3, 853–867. [Google Scholar] [CrossRef] [Green Version]
- Zhu, X.; Liu, D. Accurate mapping of forest types using dense seasonal Landsat time-series. ISPRS J. Photogramm. Remote Sens. 2014, 96, 1–11. [Google Scholar] [CrossRef]
- Pasquarella, V.J.; Holden, C.E.; Woodcock, C.E. Improved mapping of forest type using spectral-temporal Landsat features. Remote Sens. Environ. 2018, 210, 193–207. [Google Scholar] [CrossRef]
- Barrett, B.; Raab, C.; Cawkwell, F.; Green, S. Upland vegetation mapping using Random Forests with optical and radar satellite data. Remote Sens. Ecol. Conserv. 2016, 2, 212–231. [Google Scholar] [CrossRef] [PubMed]
- Huang, S.; Tang, L.; Hupy, J.P.; Wang, Y.; Shao, G. A commentary review on the use of normalized difference vegetation index (NDVI) in the era of popular remote sensing. J. For. Res. 2021, 32, 1–6. [Google Scholar] [CrossRef]
- Kopel, D.; Michalska-Hejduk, D.; Sfawik, F.; Berezowski, T.; Borowski, M.; Rosadzifski, S.; Chormafski, J. Application of multisensoral remote sensing data in the mapping of alkaline fens Natura 2000 habitat. Ecol. Indic. 2016, 70, 196–208. [Google Scholar] [CrossRef]
- Haest, B.; Vanden Borre, J.; Spanhove, T.; Thoonen, G.; Delalieux, S.; Kooistra, L.; Mücher, C.A.; Paelinckx, D.; Scheunders, P.; Kempeneers, P. Habitat Mapping and Quality Assessment of NATURA 2000 Heathland Using Airborne Imaging Spectroscopy. Remote Sens. 2017, 9, 266. [Google Scholar] [CrossRef] [Green Version]
- Marcinkowska-Ochtyra, A.; Gryguc, K.; Ochtyra, A.; Kopeć, D.; Jarocińska, A.; Slawik, L. Multitemporal Hyperspectral Data Fusion with Topographic Indices—Improving Classification of Natura 2000 Grassland Habitats. Remote Sens. 2019, 11, 2264. [Google Scholar] [CrossRef] [Green Version]
- Simonson, W.D.; Allen, H.D.; Coomes, D.A. Remotely sensed indicators of forest conservation status: Case study from a Natura 2000 site in southern Portugal. Ecol. Indic. 2013, 24, 636–647. [Google Scholar] [CrossRef]
- Álvarez Martínez, J.M.; Jiménez-Alfaro, B.; Barquín, J.; Ondiviela, B.; Recio, M.; Silió-Calzada, A.; Juanes, J.A. Modelling the area of occupancy of habitat types with remote sensing. Methods Ecol. Evol. 2018, 9, 580–593. [Google Scholar] [CrossRef]
- Zlinszky, A.; Schroiff, A.; Kania, A.; Deák, B.; Mücke, W.; Vári, Á.; Székely, B.; Pfeifer, N. Categorizing grassland vegetation with full-waveform airborne laser scanning: A feasibility study for detecting natura 2000 habitat types. Remote Sens. 2014, 6, 8056–8087. [Google Scholar] [CrossRef] [Green Version]
- Chan, J.C.W.; Beckers, P.; Spanhove, T.; Borre, J.V. An evaluation of ensemble classifiers for mapping Natura 2000 heathland in Belgium using spaceborne angular hyperspectral (CHRIS/Proba) imagery. Int. J. Appl. Earth Obs. Geoinf. 2012, 18, 13–22. [Google Scholar] [CrossRef]
- Sanchez-Hernandez, C.; Boyd, D.S.; Foody, G.M. Mapping specific habitats from remotely sensed imagery: Support vector machine and support vector data description based classification of coastal saltmarsh habitats. Ecol. Inform. 2007, 2, 83–88. [Google Scholar] [CrossRef]
- Foody, G. Harshness in image classification accuracy assessment. Int. J. Remote Sens. 2008, 29, 3137–3158. [Google Scholar] [CrossRef] [Green Version]
- Stehman, S.V.; Foody, G.M. Key issues in rigorous accuracy assessment of land cover products. Remote Sens. Environ. 2019, 231, 111199. [Google Scholar] [CrossRef]
- Hmimina, G.; Dufrêne, E.; Pontailler, J.Y.; Delpierre, N.; Aubinet, M.; Caquet, B.; de Grandcourt, A.; Burban, B.; Flechard, C.; Granier, A.; et al. Evaluation of the potential of MODIS satellite data to predict vegetation phenology in different biomes: An investigation using ground-based NDVI measurements. Remote Sens. Environ. 2013, 132, 145–158. [Google Scholar] [CrossRef]
- Norman, S.P.; Hargrove, W.W.; Christie, W.M. Spring and Autumn Phenological Variability across Environmental Gradients of Great Smoky Mountains National Park, USA. Remote Sens. 2017, 9, 407. [Google Scholar] [CrossRef] [Green Version]
- Hall-Beyer, M. Comparison of single-year and multiyear NDVI time series principal components in cold temperate biomes. IEEE Trans. Geosci. Remote Sens. 2003, 41, 2568–2574. [Google Scholar] [CrossRef] [Green Version]
- Gigante, D.; Attorre, F.; Venanzoni, R.; Acosta, A.T.; Agrillo, E.; Aleffi, M.; Alessi, N.; Allegrezza, M.; Angelini, P.; Angiolini, C.; et al. A methodological protocol for Annex I Habitats monitoring: The contribution of vegetation science. Plant Sociol. 2016, 53, 77–87. [Google Scholar] [CrossRef]
- Bonanomi, G.; Caporaso, S.; Allegrezza, M. Short-term effects of nitrogen enrichment, litter removal and cutting on a Mediterranean grassland. Acta Oecologica 2006, 30, 419–425. [Google Scholar] [CrossRef]
- Bonanomi, G.; Caporaso, S.; Allegrezza, M. Effects of nitrogen enrichment, plant litter removal and cutting on a species-rich Mediterranean calcareous grassland. Plant Biosyst. 2009, 143, 443–455. [Google Scholar] [CrossRef]
- Catorci, A.; Cesaretti, S.; Gatti, R.; Ottaviani, G. Abiotic and biotic changes due to spread of Brachypodium genuense (DC.) Roem. & Schult. in sub-Mediterranean meadows. Community Ecol. 2011, 12, 117–125. [Google Scholar] [CrossRef]
- De Simone, W.; Allegrezza, M.; Frattaroli, A.R.; Montecchiari, S.; Tesei, G.; Zuccarello, V.; Di Musciano, M. From Remote Sensing to Species Distribution Modelling: An Integrated Workflow to Monitor Spreading Species in Key Grassland Habitats. Remote Sens. 2021, 13, 1904. [Google Scholar] [CrossRef]
- Landucci, F.; Acosta, A.T.R.; Agrillo, E.; Attorre, F.; Biondi, E.; Cambria, V.E.; Chiarucci, A.; Vico, E.D.; Sanctis, M.D.; Facioni, L.; et al. VegItaly: The Italian collaborative project for a national vegetation database. Plant Biosyst. 2012, 146, 756–763. [Google Scholar] [CrossRef] [Green Version]
- Onishi, M.; Ise, T. Explainable identification and mapping of trees using UAV RGB image and deep learning. Sci. Rep. 2021, 11, 903. [Google Scholar] [CrossRef]
- Tichý, L.; Chytrý, M. Probabilistic key for identifying vegetation types in the field: A new method and Android application. J. Veg. Sci. 2019, 30, 1035–1038. [Google Scholar] [CrossRef] [Green Version]
- Happ, C.; Greven, S. Multivariate Functional Principal Component Analysis for Data Observed on Different (Dimensional) Domains. J. Am. Stat. Assoc. 2018, 113, 649–659. [Google Scholar] [CrossRef] [Green Version]
- Whetten, A.B.; Demler, H.J. Detection of Multidecadal Changes in Vegetation Dynamics and Association with Intra-Annual Climate Variability in the Columbia River Basin. Remote Sens. 2022, 14, 569. [Google Scholar] [CrossRef]
| Class | Plant Association (Syntaxa) | Habitat Code | Plots |
|---|---|---|---|
| Woodland | |||
| 1 | Holm-oak wood (Cephalanthero longifoliae-Quercetum ilicis) | 9340 | 37 |
| 2 | Downy-oak wood (Cytiso sessilifolii-Quercetum pubescentis) | 91AA * | 34 |
| 3 | Black hornbeam wood (Scutellario columnae-Ostryetum carpinifoliae) | - | 60 |
| 4 | Pinus sp. plantations | - | 32 |
| 5 | Riparian woods (Rubo ulmifolii-Salicetum albae and Salici-Popolutem nigrae) | 92A0 | 16 |
| Shrublands | |||
| 6 | Spartium junceum shrub (Spartio juncei-Cytisetum sessilifolii Spartium junceum variant) | - | 16 |
| 7 | Juniperus oxycedrus shrub (Spartio juncei-Cytisetum sessilifolii Juniperus oxycedrus variant) | - | 15 |
| Grasslands | |||
| 8 | Bromus erectus grassland (Asperulo purpurei-Brometum erecti) | 6210 (*) | 16 |
| Mosaic of garrigues and chasmophytic vegetation | |||
| 9 | Garrigues and vegetation of rock and scree (Cephalario leucanthae-Saturejetum montanae and Moehringio papulosae-Potentilletum caulescentis) | 6110, 6220, 8210 | 49 |
| Other | |||
| 10 | Crop land | - | 18 |
| 11 | Urban land | - | 15 |
| Type | Description |
|---|---|
| Terrain Parameters | Altitude (m a.s.l) Slope of the terrain (°) Topographic Position Index (TPI) Topographic Wetness Index (TWI) Northness Eastness Incoming solar radiation |
| Lithology | “Calcare Massiccio” formation; micritic limestone:“Bugarone” and “Maiolica formation”; marly-calcareous formation; landslide deposits; slope deposits; alluvial deposits |
| Vegetation Index | Acronym | References |
|---|---|---|
| Normalized Difference Vegetation Index | NDVI | [52] |
| Green Normalized Difference Vegetation Index | GNDVI | [53,54] |
| Modified Chlorophyll Absorption in Reflectance Index | MCARI | [55] |
| Normalized Difference Red-Edge | NDRE | [56] |
| Normalized Difference Water Index | NDWI | [57] |
| Modified Normalized Difference Water Index | MNDWI | [58] |
| Reference Data | |||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 | UA | ||
| Prediction | 1 | 34 | 0 | 5 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 85.0 |
| 2 | 0 | 29 | 12 | 0 | 0 | 2 | 0 | 0 | 1 | 0 | 0 | 65.9 | |
| 3 | 1 | 4 | 41 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 85.4 | |
| 4 | 2 | 0 | 1 | 29 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 90.6 | |
| 5 | 0 | 0 | 1 | 0 | 16 | 0 | 0 | 0 | 0 | 1 | 0 | 88.9 | |
| 6 | 0 | 0 | 0 | 0 | 0 | 13 | 0 | 1 | 1 | 2 | 0 | 76.5 | |
| 7 | 0 | 0 | 0 | 1 | 0 | 1 | 14 | 0 | 0 | 0 | 0 | 87.5 | |
| 8 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 14 | 1 | 1 | 0 | 82.4 | |
| 9 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 43 | 1 | 1 | 93.5 | |
| 10 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 13 | 0 | 92.9 | |
| 11 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 13 | 81.3 | |
| PA | 91.9 | 82.5 | 68.3 | 90.6 | 100.0 | 81.3 | 93.3 | 87.5 | 87.8 | 72.2 | 86.7 | ||
| OA | 85.58 (±5.28) | ||||||||||||
| K | 0.83 (±0.06) | ||||||||||||
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Pesaresi, S.; Mancini, A.; Quattrini, G.; Casavecchia, S. Functional Analysis for Habitat Mapping in a Special Area of Conservation Using Sentinel-2 Time-Series Data. Remote Sens. 2022, 14, 1179. https://doi.org/10.3390/rs14051179
Pesaresi S, Mancini A, Quattrini G, Casavecchia S. Functional Analysis for Habitat Mapping in a Special Area of Conservation Using Sentinel-2 Time-Series Data. Remote Sensing. 2022; 14(5):1179. https://doi.org/10.3390/rs14051179
Chicago/Turabian StylePesaresi, Simone, Adriano Mancini, Giacomo Quattrini, and Simona Casavecchia. 2022. "Functional Analysis for Habitat Mapping in a Special Area of Conservation Using Sentinel-2 Time-Series Data" Remote Sensing 14, no. 5: 1179. https://doi.org/10.3390/rs14051179
APA StylePesaresi, S., Mancini, A., Quattrini, G., & Casavecchia, S. (2022). Functional Analysis for Habitat Mapping in a Special Area of Conservation Using Sentinel-2 Time-Series Data. Remote Sensing, 14(5), 1179. https://doi.org/10.3390/rs14051179






