Applying a Portable Backpack Lidar to Measure and Locate Trees in a Nature Forest Plot: Accuracy and Error Analyses
Abstract
1. Introduction
2. Materials and Methods
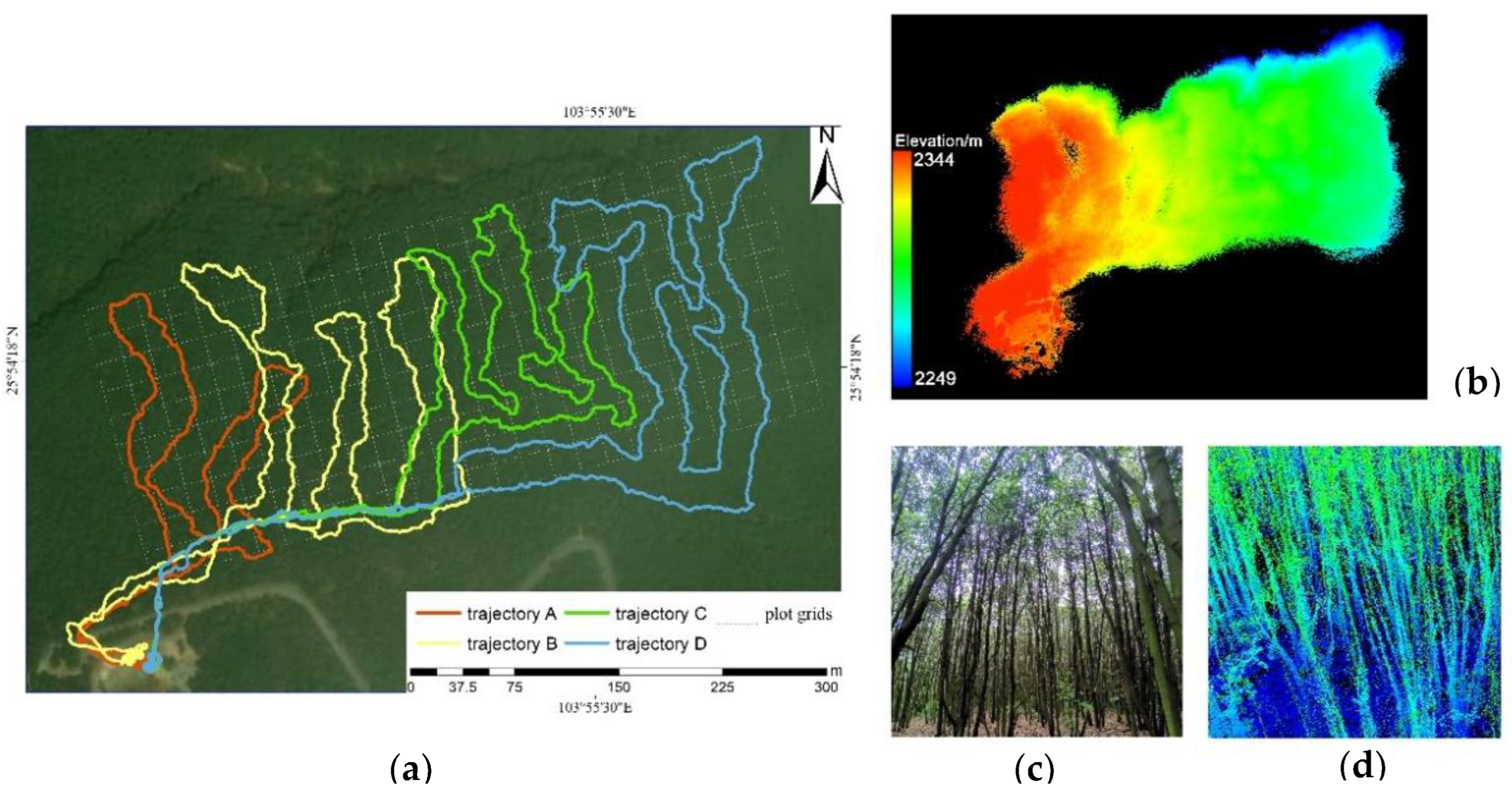
2.1. Study Area
2.2. Field Measurement
2.3. Lidar Data Collection
2.4. Data Processing
2.4.1. Preprocessing of Lidar Point Clouds
2.4.2. DBH Fitting and Trees Positioning
2.4.3. Paired-Tree Distance
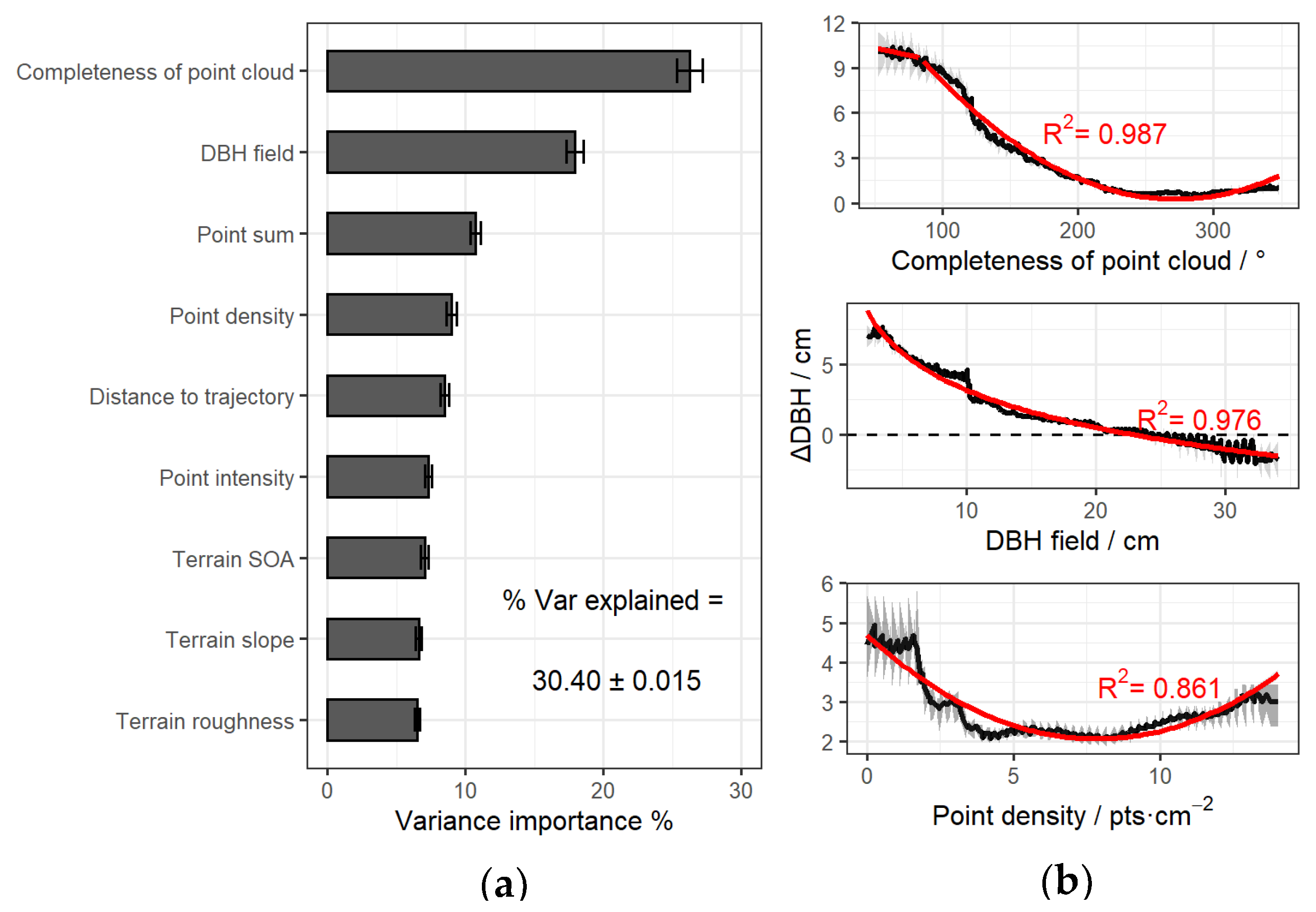
2.4.4. Extraction of Influencing Factors
2.5. Data Analysis
3. Result
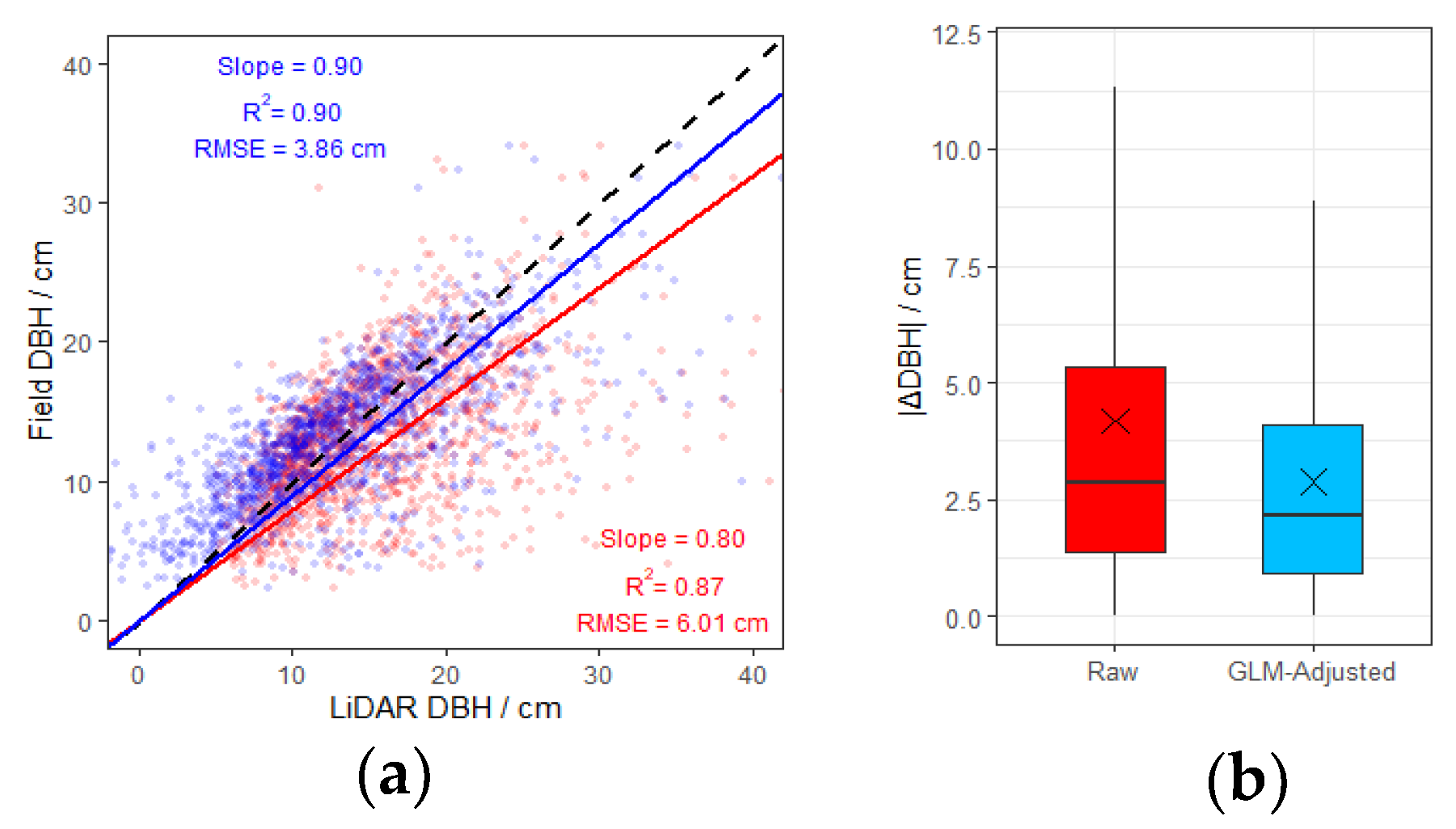
3.1. DBH Measurement Error and Correction
3.2. Tree Positioning Error and Correction
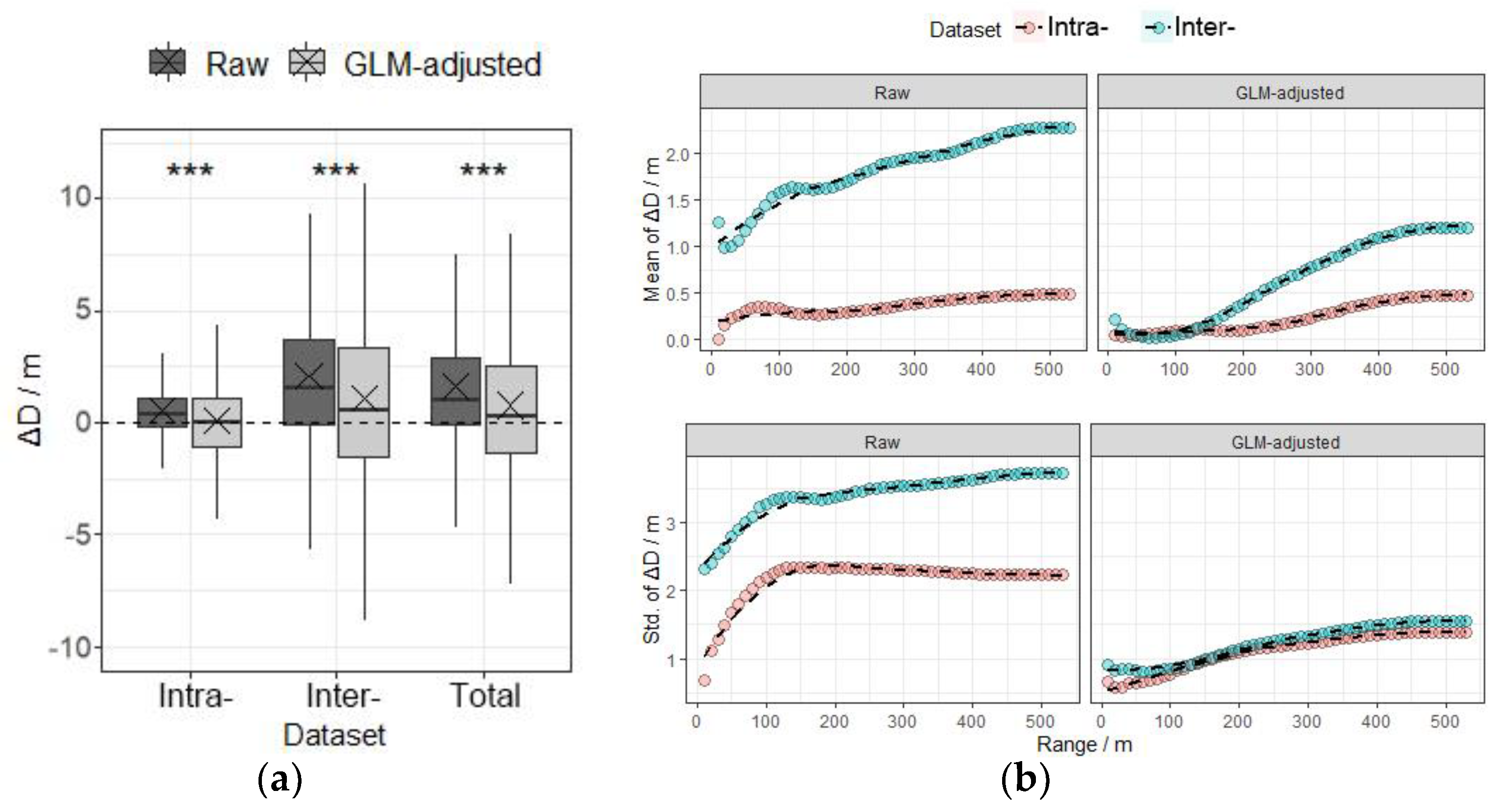
3.3. Measurement Error and Correction of Relative Distance between Trees
4. Discussion
4.1. Efficiency of Backpack Lidar in Forest Inventory
4.2. Factors Affecting DBH Measurement Accuracy
4.3. Factors Influencing Tree Positioning
4.4. Paired-Tree Distance
5. Conclusions
Author Contributions
Funding
Institutional Review Board Statement
Informed Consent Statement
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Feng, G.; Mi, X.; Yan, H.; Li, F.Y.; Svenning, J.-C.; Ma, K. CForBio: A network monitoring Chinese forest biodiversity. Sci. Bull. 2016, 61, 1163–1170. [Google Scholar] [CrossRef]
- Liang, X.; Juha, H.; Harri, K.; Matti, L.; Jiri, P.; Norbert, P.; Markus, H.; Gábor, B.; Pirotti, F.; Jan, H. International benchmarking of terrestrial laser scanning approaches for forest inventories. ISPRS J. Photogramm. Remote Sens. 2018, 144, 137–179. [Google Scholar] [CrossRef]
- Su, Y.; Guo, Q.; Jin, S.; Guan, H.; Sun, X.; Ma, Q.; Hu, T.; Wang, R.; Li, Y. The Development and Evaluation of a Backpack LiDAR System for Accurate and Efficient Forest Inventory. IEEE Geosci. Remote Sens. Lett. 2021, 18, 1660–1664. [Google Scholar] [CrossRef]
- Guo, Q.; Su, Y.; Hu, T.; Guan, H.; Jin, S.; Zhang, J.; Zhao, X.; Xu, K.; Wei, D.; Kelly, M.; et al. Lidar Boosts 3D Ecological Observations and Modelings: A Review and Perspective. IEEE Geosci. Remote Sens. Mag. 2021, 9, 232–257. [Google Scholar] [CrossRef]
- Calders, K.; Newnham, G.; Burt, A.; Murphy, S.; Raumonen, P.; Herold, M.; Culvenor, D.; Avitabile, V.; Disney, M.; Armston, J. Nondestructive estimates of above-ground biomass using terrestrial laser scanning. Methods Ecol. Evol. 2015, 6, 198–208. [Google Scholar] [CrossRef]
- Liang, X.; Kankare, V.; Hyypp, J.; Wang, Y.; Kukko, A.; Haggrén, H.; Yu, X.; Kaartinen, H.; Jaakkola, A.; Guan, F. Terrestrial laser scanning in forest inventories. ISPRS J. Photogramm. Remote Sens. 2016, 115, 63–77. [Google Scholar] [CrossRef]
- Disney, M.I. Terrestrial LiDAR: A three dimensional revolution in how we look at trees. New Phytol. 2019, 222, 1736–1741. [Google Scholar] [CrossRef]
- Xie, Y.; Zhang, J.; Chen, X.; Pang, S.; Shen, Z. Accuracy assessment and error analysis for diameter at breast height measurement of trees obtained using a novel backpack LiDAR system. For. Ecosyst. 2020, 7, 1–11. [Google Scholar] [CrossRef]
- Su, Y.; Guan, H.; Hu, T.; Guo, Q. The Integration of Uavand Backpack Lidar Systems for Forest Inventory. In Proceedings of the IGARSS 2018—2018 IEEE International Geoscience and Remote Sensing Symposium, Valencia, Spain, 22–27 July 2018. [Google Scholar]
- Olofsson, K.; Holmgren, J.; Olsson, H. Tree Stem and Height Measurements using Terrestrial Laser Scanning and the RANSAC Algorithm. Remote Sens. 2014, 6, 4323–4344. [Google Scholar] [CrossRef]
- Oveland, I.; Hauglin, M.; Gobakken, T.; Naesset, E.; Maalen-Johansen, I. Automatic Estimation of Tree Position and Stem Diameter Using a Moving Terrestrial Laser Scanner. Remote Sens. 2017, 9, 350. [Google Scholar] [CrossRef]
- Oveland, I.; Hauglin, M.; Giannetti, F.; Kjorsvik, N.S.; Gobakken, T. Comparing Three Different Ground Based Laser Scanning Methods for Tree Stem Detection. Remote Sens. 2018, 10, 538. [Google Scholar] [CrossRef]
- Almeida, D.R.A.; Stark, S.C.; Chazdon, R.; Nelson, B.W.; Cesar, R.G.; Meli, P.; Gorgens, E.B.; Duarte, M.M.; Valbuena, R.; Moreno, V.S.; et al. The effectiveness of lidar remote sensing for monitoring forest cover attributes and landscape restoration. For. Ecol. Manag. 2019, 438, 34–43. [Google Scholar] [CrossRef]
- Szostak, M. Automated Land Cover Change Detection and Forest Succession Monitoring Using LiDAR Point Clouds and GIS Analyses. Geosciences 2020, 10, 321. [Google Scholar] [CrossRef]
- Szostak, M.; Likus-Cieslik, J.; Pietrzykowski, M. PlanetScope Imageries and LiDAR Point Clouds Processing for Automation Land Cover Mapping and Vegetation Assessment of a Reclaimed Sulfur Mine. Remote Sens. 2021, 13, 2717. [Google Scholar] [CrossRef]
- Versace, S.; Gianelle, D.; Frizzera, L.; Tognetti, R.; Garfi, V.; Dalponte, M. Prediction of Competition Indices in a Norway Spruce and Silver Fir-Dominated Forest Using Lidar Data. Remote Sens. 2019, 11, 2734. [Google Scholar] [CrossRef]
- Qian, C.; Liu, H.; Tang, J.; Chen, Y.; Kaartinen, H.; Kukko, A.; Zhu, L.; Liang, X.; Chen, L.; Hyyppa, J. An Integrated GNSS/INS/LiDAR-SLAM Positioning Method for Highly Accurate Forest Stem Mapping. Remote Sens. 2017, 9, 3. [Google Scholar] [CrossRef]
- Liang, X.; Hyyppa, J.; Kukko, A.; Kaartinen, H.; Jaakkola, A.; Yu, X. The Use of a Mobile Laser Scanning System for Mapping Large Forest Plots. IEEE Geosci. Remote Sens. Lett. 2014, 11, 1504–1508. [Google Scholar] [CrossRef]
- Shao, J.; Zhang, W.; Mellado, N.; Wang, N.; Jin, S.; Cai, S.; Luo, L.; Lejemble, T.; Yan, G. SLAM-aided forest plot mapping combining terrestrial and mobile laser scanning. ISPRS J. Photogramm. Remote Sens. 2020, 163, 214–230. [Google Scholar] [CrossRef]
- Li, Y.; Olson, E.B. Extracting general-purpose features from LIDAR data. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Anchorage, AK, USA, 3–8 May 2010; pp. 1388–1393. [Google Scholar]
- Hess, W.; Kohler, D.; Rapp, H.; Andor, D. Real-Time Loop Closure in 2D LIDAR SLAM. In Proceedings of the IEEE International Conference on Robotics and Automation (ICRA), Royal Inst Technol, Ctr Autonomous Syst, Stockholm, Sweden, 16–21 May 2016; pp. 1271–1278. [Google Scholar]
- Chudá, J.; Hunčaga, M.; Tuček, J.; Mokroš, M. The Handheld Mobile Laser Scanners as a Tool for Accurate Positioning under Forest Canopy. Int. Arch. Photogramm. Remote Sens. Spat. Inf. Sci. 2020, XLIII-B1-2020, 211–218. [Google Scholar] [CrossRef]
- Pommerening, A. Evaluating structural indices by reversing forest structural analysis. For. Ecol. Manag. 2006, 224, 266–277. [Google Scholar] [CrossRef]
- Hegyi, F. A simulation model for managing jack-pine stands. Growth Models for Tree and Stand Simulation. For. Res. 1974, 30, 74–90. [Google Scholar]
- Newnham, R.M.; Smith, J. Development and Testing of Stand Models for Douglas Fir and Lodgepole Pine. J. Jpn. For. Soc. 1964, 40, 494–504. [Google Scholar] [CrossRef][Green Version]
- Zhao, X.; Guo, Q.; Su, Y.; Xue, B. Improved progressive TIN densification filtering algorithm for airborne LiDAR data in forested areas. ISPRS J. Photogramm. Remote Sens. 2016, 117, 79–91. [Google Scholar] [CrossRef]
- Clark, J.S.; Bell, D.M.; Kwit, M.C.; Zhu, K. Competition-interaction landscapes for the joint response of forests to climate change. Glob. Chang. Biol. 2014, 20, 1979–1991. [Google Scholar] [CrossRef]
- Ford, K.R.; Breckheimer, I.K.; Franklin, J.F.; Freund, J.A.; Kroiss, S.J.; Larson, A.J.; Theobald, E.J.; Hillerislambers, J. Competition alters tree growth responses to climate at individual and stand scales. Can. J. For. Res. 2017, 47, 53–62. [Google Scholar] [CrossRef]
- Zhou, M.; Lei, X.; Duan, G.; Lu, J.; Zhang, H. The effect of the calculation method, plot size, and stand density on the top height estimation in natural spruce-fir-broadleaf mixed forests. For. Ecol. Manag. 2019, 453, 117574. [Google Scholar] [CrossRef]
- Liu, C.; Xing, Y.; Duanmu, J.; Tian, X. Evaluating Different Methods for Estimating Diameter at Breast Height from Terrestrial Laser Scanning. Remote Sens. 2018, 10, 513. [Google Scholar] [CrossRef]
- Tao, S.; Wu, F.; Guo, Q.; Wang, Y.; Li, W.; Xue, B.; Hu, X.; Li, P.; Tian, D.; Li, C.; et al. Segmenting tree crowns from terrestrial and mobile LiDAR data by exploring ecological theories. ISPRS J. Photogramm. Remote Sens. 2015, 110, 66–76. [Google Scholar] [CrossRef]
- Campos, M.B.; Litkey, P.; Wang, Y.; Chen, Y.; Hyyti, H.; Hyyppa, J.; Puttonen, E. A Long-Term Terrestrial Laser Scanning Measurement Station to Continuously Monitor Structural and Phenological Dynamics of Boreal Forest Canopy. Front. Plant Sci. 2021, 11, 2132. [Google Scholar] [CrossRef]
- Hyyppä, E.; Hyyppä, J.; Hakala, T.; Kukko, A.; Wulder, M.A.; White, J.C.; Pyörälä, J.; Yu, X.; Wang, Y.; Virtanen, J.-P.; et al. Under-canopy UAV laser scanning for accurate forest field measurements. ISPRS J. Photogramm. Remote Sens. 2020, 164, 41–60. [Google Scholar] [CrossRef]
- Miller, J.; Morgenroth, J.; Gomez, C. 3D modelling of individual trees using a handheld camera: Accuracy of height, diameter and volume estimates. Urban For. Urban Green. 2015, 14, 932–940. [Google Scholar] [CrossRef]
- Dalla Corte, A.P.; Rex, F.E.; Alves de Almeida, D.R.; Sanquetta, C.R.; Silva, C.A.; Moura, M.M.; Wilkinson, B.; Almeyda Zambrano, A.M.; da Cunha Neto, E.M.; Veras, H.F.P.; et al. Measuring Individual Tree Diameter and Height Using GatorEye High-Density UAV-Lidar in an Integrated Crop-Livestock-Forest System. Remote Sens. 2020, 12, 863. [Google Scholar] [CrossRef]
- Zarco-Tejada, P.J.; Diaz-Varela, R.; Angileri, V.; Loudjani, P. Tree height quantification using very high resolution imagery acquired from an unmanned aerial vehicle (UAV) and automatic 3D photo-reconstruction methods. Eur. J. Agron. 2014, 55, 89–99. [Google Scholar] [CrossRef]
- Trochta, J.; Krucek, M.; Vrska, T.; Kral, K. 3D Forest: An application for descriptions of three-dimensional forest structures using terrestrial LiDAR. PLoS ONE 2017, 12, e176871. [Google Scholar] [CrossRef] [PubMed]
- Vauhkonen, J.; Ene, L.; Gupta, S.; Heinzel, J.; Holmgren, J.; Pitkanen, J.; Solberg, S.; Wang, Y.; Weinacker, H.; Hauglin, K.M.; et al. Comparative testing of single-tree detection algorithms under different types of forest. Forestry 2012, 85, 27–40. [Google Scholar] [CrossRef]
- Kaartinen, H.; Hyyppa, J.; Yu, X.; Vastaranta, M.; Hyyppa, H.; Kukko, A.; Holopainen, M.; Heipke, C.; Hirschmugl, M.; Morsdorf, F.; et al. An International Comparison of Individual Tree Detection and Extraction Using Airborne Laser Scanning. Remote Sens. 2012, 4, 950–974. [Google Scholar] [CrossRef]
- Li, W.; Guo, Q.; Jakubowski, M.K.; Kelly, M. A New Method for Segmenting Individual Trees from the Lidar Point Cloud. Photogramm. Eng. Remote Sens. 2012, 78, 75–84. [Google Scholar] [CrossRef]
- Xie, Y.; Wang, B.; Yao, Y.; Yang, L.; Gao, Y.; Zhang, Z.; Lin, L. Quantification of vertical community structure of subtropical evergreen broadleaved forest community using UAV-Lidar data. Acta Ecol. Sin. 2020, 40, 940–951. [Google Scholar]
- Tao, S. Radar_Rainforest. Figshare 2021. Available online: https://figshare.com/articles/dataset/Radar_Rainforest/14061428 (accessed on 30 April 2021).
- Mur-Artal, R.; Tardos, J.D. Visual-Inertial Monocular SLAM with Map Reuse. IEEE Rob. Autom. Lett. 2017, 2, 796–803. [Google Scholar] [CrossRef]
- Endres, F.; Hess, J.; Sturm, J.; Cremers, D.; Burgard, W. 3-D Mapping with an RGB-D Camera. IEEE Trans. Rob. 2014, 30, 177–187. [Google Scholar] [CrossRef]
- Dong, Z.; Liang, F.; Yang, B.; Xu, Y.; Zang, Y.; Li, J.; Wang, Y.; Dai, W.; Fan, H.; Hyyppä, J.; et al. Registration of large-scale terrestrial laser scanner point clouds: A review and benchmark. ISPRS J. Photogramm. Remote Sens. 2020, 163, 327–342. [Google Scholar] [CrossRef]
- Besl, P.J.; Mckay, H.D. A method for registration of 3-D shapes. IEEE Trans. Pattern Anal. Mach. Intell. 1992, 14, 239–256. [Google Scholar] [CrossRef]



| Variable | Linear | Logarithmic | Quadratic | Exponential | Logistic | |
|---|---|---|---|---|---|---|
| Completeness of point cloud (CPC) | F | 1274.358 | 3577.328 | 7804.132 | 2182.511 | 2033.291 |
| R2 | 0.786 | 0.912 | 0.978 | 0.863 | 0.854 | |
| DBH field (DBHfield) | F | 3572.559 | 16,223.59 | 6269.308 | — | — |
| R2 | 0.901 | 0.976 | 0.97 | — | — | |
| Point density (PD) | F | 44.255 | 204.248 | 598.36 | 32.151 | 45.977 |
| R2 | 0.183 | 0.508 | 0.861 | 0.14 | 0.188 |
| Variable | Linear | Logarithmic | Quadratic | Exponential | Logistic | |
|---|---|---|---|---|---|---|
| Distance to start (DS) | F | 2202.59 | 658.00 | 3244.53 | 2380.83 | 1767.65 |
| R2 | 0.83 | 0.60 | 0.94 | 0.84 | 0.80 | |
| Route from start (RS) | F | 563.65 | 1044.69 | 426.50 | 495.14 | 788.05 |
| R2 | 0.62 | 0.75 | 0.71 | 0.59 | 0.69 |
Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Xie, Y.; Yang, T.; Wang, X.; Chen, X.; Pang, S.; Hu, J.; Wang, A.; Chen, L.; Shen, Z. Applying a Portable Backpack Lidar to Measure and Locate Trees in a Nature Forest Plot: Accuracy and Error Analyses. Remote Sens. 2022, 14, 1806. https://doi.org/10.3390/rs14081806
Xie Y, Yang T, Wang X, Chen X, Pang S, Hu J, Wang A, Chen L, Shen Z. Applying a Portable Backpack Lidar to Measure and Locate Trees in a Nature Forest Plot: Accuracy and Error Analyses. Remote Sensing. 2022; 14(8):1806. https://doi.org/10.3390/rs14081806
Chicago/Turabian StyleXie, Yuyang, Tao Yang, Xiaofeng Wang, Xi Chen, Shuxin Pang, Juan Hu, Anxian Wang, Ling Chen, and Zehao Shen. 2022. "Applying a Portable Backpack Lidar to Measure and Locate Trees in a Nature Forest Plot: Accuracy and Error Analyses" Remote Sensing 14, no. 8: 1806. https://doi.org/10.3390/rs14081806
APA StyleXie, Y., Yang, T., Wang, X., Chen, X., Pang, S., Hu, J., Wang, A., Chen, L., & Shen, Z. (2022). Applying a Portable Backpack Lidar to Measure and Locate Trees in a Nature Forest Plot: Accuracy and Error Analyses. Remote Sensing, 14(8), 1806. https://doi.org/10.3390/rs14081806







