Combining Environmental, Multispectral, and LiDAR Data Improves Forest Type Classification: A Case Study on Mapping Cool Temperate Rainforests and Mixed Forests
Abstract
:1. Introduction
2. Materials and Methods
2.1. Study Area
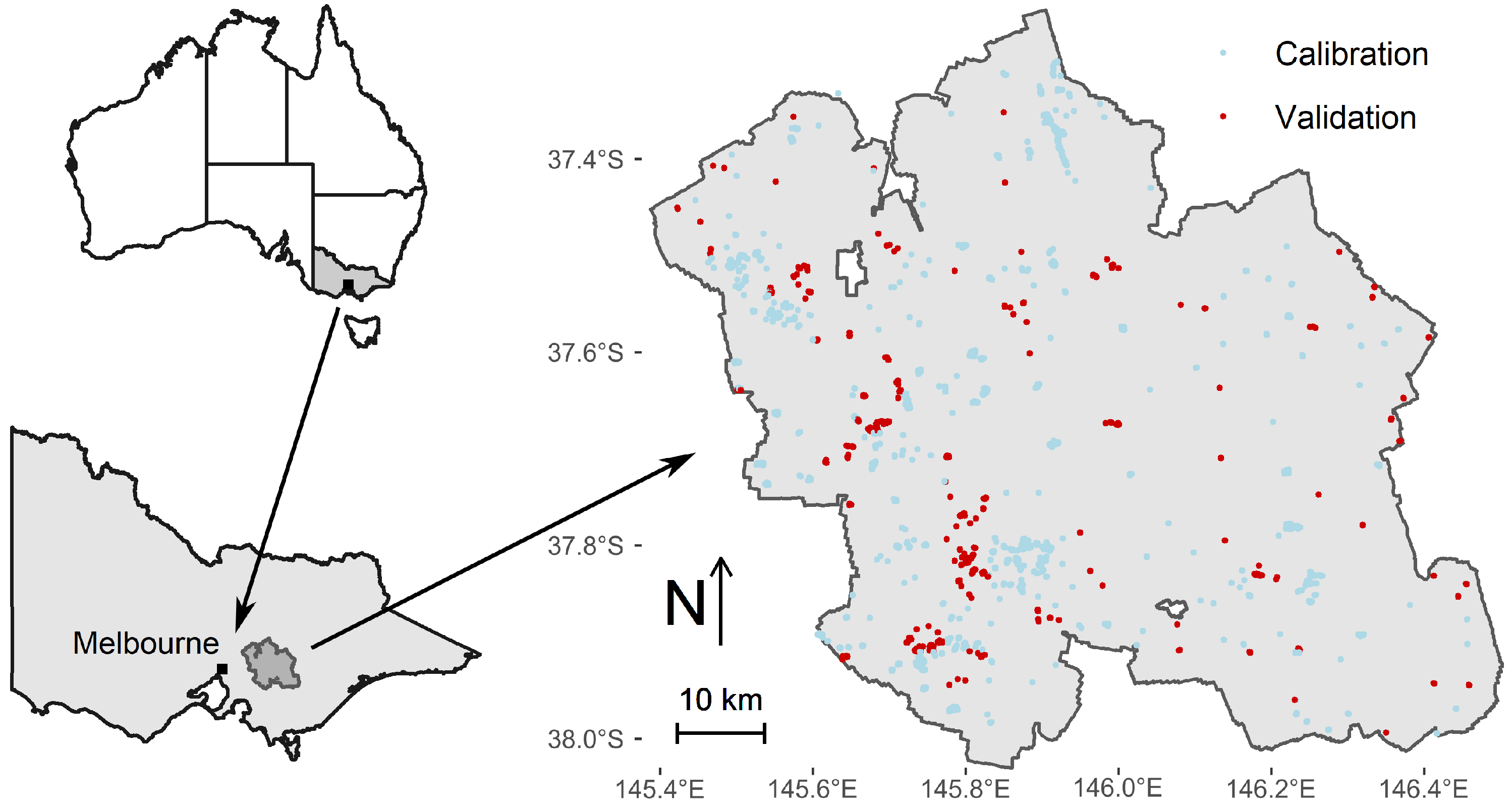
2.1.1. The Central Highlands of Victoria
2.1.2. Cool Temperate Rainforests
2.1.3. Cool Temperate Mixed Forests
2.1.4. Other Forest Types
2.2. Field-Based Forest Type Assessment
- 1.
- Plot data were collected from several previous research projects. Based on the biometric inventory of the plots (i.e., species, diameter at breast height, crown width, crown cover, etc.), we categorized plots as Euc, Mixed Forest, and Rainforest (None of these Stage 1 plots met the criteria for the Fern type). We used Nearmap (https://www.nearmap.com/au/en (accessed on 8 December 2021)) and LiDAR canopy height models to address potential GPS inaccuracies (the GPS coordinates had ∼20 m accuracy) associated with plot location. Plots that had a mismatch between field observations and nearmap were relocated within their 20 m surrounding area to match the forest type that was observed in the field. At this step, we also filtered plots from the same forest type that were less than 100 m from each other to avoid small-scale spatial autocorrelation issues. Stage 1 plots are the dominant source of ground-based data for this research.
- 2.
- Under-sampled areas of the Central Highlands (e.g., this was the case in the eastern part of the region in Figure 1) were identified and additional field data were collected. Clusters of three plots were identified in a prototype model as Rainforest, Mixed Forest, and Euc forest types in areas that lacked samples and confirmed by site visits.
- 3.
- To avoid classifying areas with only tree fern canopies as Rainforest areas, we used high resolution (7.5 cm) aerial photos to create data points for the Fern forest type category. The star shape and bright green colouration of tree fern stands made them easy to distinguish from the other forest types based on aerial photography alone.
2.3. Environmental Predictors
2.4. Multispectral Predictors
2.5. LiDAR Predictors
2.5.1. LiDAR Raw Data
2.5.2. Standard Area-Based LiDAR Metrics
2.5.3. LiDAR Metrics Based on Individual Tree Detection
2.5.4. Plant Area Volume Density Predictors
2.6. Statistical Models
2.7. Model Evaluation
2.7.1. Training and Testing Datasets
2.7.2. Goodness-of-Fit Metrics
3. Results
3.1. Prediction Abilities of the Different Models
3.2. Which Variables Are Best at Discriminating Amongst Forest Types?
4. Discussion
4.1. Strengths and Weaknesses of Different Predictor Types
4.2. Challenges and Opportunities for Classifying Complex Forested Landscapes
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Fedrigo, M.; Stewart, S.B.; Roxburgh, S.H.; Kasel, S.; Bennett, L.T.; Vickers, H.; Nitschke, C.R. Predictive Ecosystem Mapping of South-Eastern Australian Temperate Forests Using Lidar-Derived Structural Profiles and Species Distribution Models. Remote Sens. 2019, 11, 93. [Google Scholar] [CrossRef] [Green Version]
- Wood, S.W.; Murphy, B.P.; Bowman, D.M.J.S. Firescape ecology: How topography determines the contrasting distribution of fire and rain forest in the south-west of the Tasmanian Wilderness World Heritage Area. J. Biogeogr. 2011, 38, 1807–1820. [Google Scholar] [CrossRef]
- Fairman, T.A.; Nitschke, C.R.; Bennett, L.T. Too much, too soon? A review of the effects of increasing wildfire frequency on tree mortality and regeneration in temperate eucalypt forests. Int. J. Wildland Fire 2016, 25, 831–848. [Google Scholar] [CrossRef]
- Busby, J.; Brown, M. Southern Rainforests Chapter. In Australian Vegetation; Cambridge University Press: Cambridge, UK, 1994; pp. 131–155. [Google Scholar]
- Leonard, S.W.J.; Bennett, A.F.; Clarke, M.F. Determinants of the occurrence of unburnt forest patches: Potential biotic refuges within a large, intense wildfire in south-eastern Australia. For. Ecol. Manag. 2014, 314, 85–93. [Google Scholar] [CrossRef]
- Gilbert, J.M. Forest succession in the Florentine valley, Tasmania. Proc. R. Soc. Tasman. 1959, 93, 129–152. [Google Scholar]
- Fedrigo, M.; Kasel, S.; Bennett, L.T.; Roxburgh, S.H.; Nitschke, C.R. Carbon stocks in temperate forests of south-eastern Australia reflect large tree distribution and edaphic conditions. For. Ecol. Manag. 2014, 334, 129–143. [Google Scholar] [CrossRef]
- White, M.; Batpurev, K.; Salkin, O.; Newell, G. Primary Rainforest Mapping in Victoria 2018—Extent and Type; Technical Report; Arthur Rylah Institute for Environmental Research: Heidelberg, Australia, 2019. [Google Scholar]
- Fedrigo, M.; Newnham, G.J.; Coops, N.C.; Culvenor, D.S.; Bolton, D.K.; Nitschke, C.R. Predicting temperate forest stand types using only structural profiles from discrete return airborne lidar. ISPRS J. Photogramm. Remote Sens. 2018, 136, 106–119. [Google Scholar] [CrossRef]
- Guisan, A.; Zimmermann, N.E. Predictive habitat distribution models in ecology. Ecol. Model. 2000, 135, 147–186. [Google Scholar] [CrossRef]
- Araujo, M.B.; Guisan, A. Five (or so) challenges for species distribution modelling. J. Biogeogr. 2006, 33, 1677–1688. [Google Scholar] [CrossRef]
- Williams, J.N.; Seo, C.; Thorne, J.; Nelson, J.K.; Erwin, S.; O’Brien, J.M.; Schwartz, M.W. Using species distribution models to predict new occurrences for rare plants. Divers. Distrib. 2009, 15, 565–576. [Google Scholar] [CrossRef]
- Gogol-Prokurat, M. Predicting habitat suitability for rare plants at local spatial scales using a species distribution model. Ecol. Appl. 2011, 21, 33–47. [Google Scholar] [CrossRef] [PubMed]
- Lannuzel, G.; Balmot, J.; Dubos, N.; Thibault, M.; Fogliani, B. High-resolution topographic variables accurately predict the distribution of rare plant species for conservation area selection in a narrow-endemism hotspot in New Caledonia. Biodivers. Conserv. 2021, 30, 963–990. [Google Scholar] [CrossRef]
- Austin, M.P.; Meyers, J.A. Current approaches to modelling the environmental niche of eucalypts: Implication for management of forest biodiversity. For. Ecol. Manag. 1996, 85, 95–106. [Google Scholar] [CrossRef]
- Guisan, A.; Tingley, R.; Baumgartner, J.B.; Naujokaitis-Lewis, I.; Sutcliffe, P.R.; Tulloch, A.I.T.; Regan, T.J.; Brotons, L.; McDonald-Madden, E.; Mantyka-Pringle, C.; et al. Predicting species distributions for conservation decisions. Ecol. Lett. 2013, 16, 1424–1435. [Google Scholar] [CrossRef] [PubMed]
- Sinclair, S.J.; White, M.D.; Newell, G.R. How useful are species distribution models for managing biodiversity under future climates? Ecol. Soc. 2010, 15, 8. [Google Scholar] [CrossRef]
- Booth, T.H. Species distribution modelling tools and databases to assist managing forests under climate change. For. Ecol. Manag. 2018, 430, 196–203. [Google Scholar] [CrossRef]
- Dyderski, M.K.; Paź, S.; Frelich, L.E.; Jagodziński, A.M. How much does climate change threaten European forest tree species distributions? Glob. Chang. Biol. 2018, 24, 1150–1163. [Google Scholar] [CrossRef]
- Pecchi, M.; Marchi, M.; Burton, V.; Giannetti, F.; Moriondo, M.; Bernetti, I.; Bindi, M.; Chirici, G. Species distribution modelling to support forest management. A literature review. Ecol. Model. 2019, 411, 108817. [Google Scholar] [CrossRef]
- Nitschke, C.R.; Amoroso, M.; Coates, K.D.; Astrup, R. The influence of climate change, site type, and disturbance on stand dynamics in northwest British Columbia, Canada. Ecosphere 2012, 3, art11. [Google Scholar] [CrossRef]
- Araujo, M.B.; Luoto, M. The importance of biotic interactions for modelling species distributions under climate change. Glob. Ecol. Biogeogr. 2007, 16, 743–753. [Google Scholar] [CrossRef]
- Phiri, D.; Morgenroth, J. Developments in Landsat Land Cover Classification Methods: A Review. Remote Sens. 2017, 9, 967. [Google Scholar] [CrossRef]
- Carrao, H.; Goncalves, P.; Caetano, M. Contribution of multispectral and multitemporal information from MODIS images to land cover classification. Remote Sens. Environ. 2008, 112, 986–997. [Google Scholar] [CrossRef]
- Xie, Y.; Sha, Z.; Yu, M. Remote sensing imagery in vegetation mapping: A review. J. Plant Ecol. 2008, 1, 9–23. [Google Scholar] [CrossRef]
- Zhu, X.; Liu, D. Accurate mapping of forest types using dense seasonal Landsat time-series. ISPRS J. Photogramm. Remote Sens. 2014, 96, 1–11. [Google Scholar] [CrossRef]
- Xiao, Q.; Ustin, S.L.; McPherson, E.G. Using AVIRIS data and multiple-masking techniques to map urban forest tree species. Int. J. Remote Sens. 2004, 25, 5637–5654. [Google Scholar] [CrossRef] [Green Version]
- Eriksson, H.M.; Eklundh, L.; Kuusk, A.; Nilson, T. Impact of understory vegetation on forest canopy reflectance and remotely sensed LAI estimates. Remote Sens. Environ. 2006, 103, 408–418. [Google Scholar] [CrossRef]
- Naesset, E. Predicting forest stand characteristics with airborne scanning laser using a practical two-stage procedure and field data. Remote Sens. Environ. 2002, 80, 88–99. [Google Scholar] [CrossRef]
- Coops, N.C.; Tompalski, P.; Goodbody, T.R.H.; Queinnec, M.; Luther, J.E.; Bolton, D.K.; White, J.C.; Wulder, M.A.; van Lier, O.R.; Hermosilla, T. Modelling lidar-derived estimates of forest attributes over space and time: A review of approaches and future trends. Remote Sens. Environ. 2021, 260, 112477. [Google Scholar] [CrossRef]
- Korpela, I.; Orka, H.; Maltamo, M.; Tokola, T.; Hyyppa, J. Tree species classification using airborne LiDAR—Effects of stand and tree parameters, downsizing of training set, intensity normalization, and sensor type. Silva Fenn. 2010, 44, 319–339. [Google Scholar] [CrossRef] [Green Version]
- Schulte to Buhne, H.; Pettorelli, N. Better together: Integrating and fusing multispectral and radar satellite imagery to inform biodiversity monitoring, ecological research and conservation science. Methods Ecol. Evol. 2018, 9, 849–865. [Google Scholar] [CrossRef]
- Sarndal, C.; Thomsen, I.; Hoem, J.; Lindley, D.; Barndorff-Nielsen, O.; Dalenius, T. Design-Based and Model-Based Inference in Survey Sampling [with Discussion and Reply]. Scand. J. Stat. 1978, 5, 27–52. [Google Scholar]
- Pulsford, S.A.; Lindenmayer, D.B.; Driscoll, D.A. A succession of theories: Purging redundancy from disturbance theory. Biol. Rev. 2016, 91, 148–167. [Google Scholar] [CrossRef]
- Kasel, S.; Bennett, L.T.; Aponte, C.; Fedrigo, M.; Nitschke, C.R. Environmental heterogeneity promotes floristic turnover in temperate forests of south-eastern Australia more than dispersal limitation and disturbance. Landsc. Ecol. 2017, 32, 1613–1629. [Google Scholar] [CrossRef]
- Ashton, D.; Attiwill, P. Tall open-forests Chapter. In Australian Vegetation, 2nd ed.; Cambridge University Press: Cambridge, UK, 1994; pp. 157–219. [Google Scholar]
- Ashton, D. Fire in Tall Open-Forests (Wet Sclerophyll) Chapter. In Fire and the Australian Biota; Australian Academy of Science: Canberra, Australia, 1981; pp. 339–366. [Google Scholar]
- White, M.; Sutter, G.; Lucas, A.; Downe, J. Ecological Vegetation Class Mapping for the Goolengook Forest Management Block. A Report to the Victorian Environmental Assessment Council; Technical Report; Arthur Rylah Institute, Department of Sustainability and Environment: Heidelberg, Australia, 2006. [Google Scholar]
- DNRE. Forest Management plan for the Central Highlands; Technical Report; Department of Natural Resources and Environment: East Melbourne, Australia, 1998. [Google Scholar]
- Ashton, D.H. Ecology of bryophytic communities in mature Eucalyptus regnans F Muell forest at Wallaby Creek, Victoria. Aust. J. Bot. 1986, 34, 107–129. [Google Scholar] [CrossRef]
- Floyed, A.B.; Gibson, M. Epiphytic bryophytes of Dicksonia antarctica Labill. from selected pockets of cool temperate rainforest, central highlands, Victoria. Victorian Nat. 2006, 123, 229–235. [Google Scholar]
- Donoghue, S.; Turner, P.A.M. A review of Australian tree fern ecology in forest communities. Austral Ecol. 2021, 47, 145–165. [Google Scholar] [CrossRef]
- Flora and Fauna Guarantee, Final Recommendation of the Scientific Advisory Commitee on a Nomination for Listing of Cool Temperate Mixed Forest Community; Technical Report; FFG, S.A.C.: Roseville, CO, USA, 2012.
- Hijmans, R.J. Raster: Geographic Data Analysis and Modeling, R package version 3.5.21; 2022. [Google Scholar]
- Stewart, S.B.; Nitschke, C.R. Improving temperature interpolation using MODIS LST and local topography: A comparison of methods in south east Australia. Int. J. Climatol. 2017, 37, 3098–3110. [Google Scholar] [CrossRef]
- Ruizhu, J. Using LiDAR for Landscape-Scale Mapping of Potential Habitat for the Critically Endangered Leadbeater’s Possum. Ph.D. Thesis, The University of Melbourne, School of Ecosystem and Forest Sciences, Melbourne, Australia, 2020. [Google Scholar]
- McGaughey, R.J. FUSION/LDV: Software for LIDAR Data Analysis and Visualization, version 3.50; 2015. [Google Scholar]
- Holmgren, J.; Persson, A.; Soderman, U. Species identification of individual trees by combining high resolution LiDAR data with multi-spectral images. Int. J. Remote Sens. 2008, 29, 1537–1552. [Google Scholar] [CrossRef]
- Carvalho, C.M.; Polson, N.G.; Scott, J.G. Handling Sparsity via the Horseshoe. In Proceedings of the Twelfth International Conference on Artificial Intelligence and Statistics, Clearwater Beach, FL, USA, 16–18 April 2009; van Dyk, D., Welling, M., Eds.; PMLR: London, UK, 2009; Volume 5, pp. 73–80. [Google Scholar]
- Piironen, J.; Vehtari, A. Sparsity information and regularization in the horseshoe and other shrinkage priors. Electron. J. Statist. 2017, 11, 5018–5051. [Google Scholar] [CrossRef]
- Bürkner, P.C. brms: An R Package for Bayesian Multilevel Models Using Stan. J. Stat. Softw. 2017, 80, 1–28. [Google Scholar] [CrossRef] [Green Version]
- R Foundation for Statistical Computing. R: A Language and Environment for Statistical Computing. Version 4.0.2; R Foundation for Statistical Computing: Vienna, Austria, 2020; Available online: http://www.R-project.org (accessed on 10 October 2022).
- Valavi, R.; Elith, J.; Lahoz-Monfort, J.J.; Guillera-Arroita, G. blockCV: An r package for generating spatially or environmentally separated folds for k-fold cross-validation of species distribution models. Methods Ecol. Evol. 2019, 10, 225–232. [Google Scholar] [CrossRef] [Green Version]
- Kuhn, M. Building Predictive Models in R Using the caret Package. J. Stat. Softw. Artic. 2008, 28, 1–26. [Google Scholar] [CrossRef] [Green Version]
- Foody, G.M. Status of land cover classification accuracy assessment. Remote Sens. Environ. 2002, 80, 185–201. [Google Scholar] [CrossRef]
- Landis, J.R.; Koch, G.G. The Measurement of Observer Agreement for Categorical Data. Biometrics 1977, 33, 159–174. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Guisan, A.; Thuiller, W. Predicting species distribution: Offering more than simple habitat models. Ecol. Lett. 2005, 8, 993–1009. [Google Scholar] [CrossRef] [PubMed]
- Bauer, M.E.; Burk, T.E.; Ek, A.R.; Chopin, P.R.; Lime, S.D.; Walsh, T.A.; Walters, D.K.; Befort, W.; Heinzen, D.F. Satellite inventory of Minnesota forest resources. Photogramm. Eng. Remote Sens. 1994, 60, 287–298. [Google Scholar]
- Hemmerling, J.; Pflugmacher, D.; Hostert, P. Mapping temperate forest tree species using dense Sentinel-2 time series. Remote Sens. Environ. 2021, 267, 112743. [Google Scholar] [CrossRef]
- Blazquez-Casado, A.; Calama, R.; Valbuena, M.; Vergarechea, M.; Rodriguez, F. Combining low-density LiDAR and satellite images to discriminate species in mixed Mediterranean forest. Ann. For. Sci. 2019, 76, 57. [Google Scholar] [CrossRef]
- Scholl, V.M.; Cattau, M.E.; Joseph, M.B.; Balch, J.K. Integrating national ecological observatory network (NEON) airborne remote sensing and in-situ data for optimal tree species classification. Remote Sens. 2020, 12, 1414. [Google Scholar] [CrossRef]
- Li, Q.; Wong, F.K.K.; Fung, T. Mapping multi-layered mangroves from multispectral, hyperspectral, and LiDAR data. Remote Sens. Environ. 2021, 258, 112403. [Google Scholar] [CrossRef]
- Lindenmayer, D.; Blair, D.; McBurney, L.; Banks, S.; Bowd, E. Ten years on—A decade of intensive biodiversity research after the 2009 Black Saturday wildfires in Victoria’s Mountain Ash forest. Aust. Zool. 2020, 41, 220–230. [Google Scholar] [CrossRef]
- Turner, P.A.M.; Balmer, J.; Kirkpatrick, J.B. Stand-replacing wildfires? The incidence of multi-cohort and single-cohort Eucalyptus regnans and E. obliqua forests in southern Tasmania. For. Ecol. Manag. 2009, 258, 366–375. [Google Scholar] [CrossRef]
- Shokirov, S.; Jucker, T.; Levick, S.R.; Manning, A.D.; Bonnet, T.; Yebra, M.; Youngentob, K.N. Habitat highs and lows: Using terrestrial and UAV LiDAR for modelling avian species richness and abundance in a restored woodland. Remote Sens. Environ. 2023, 285, 113326. [Google Scholar] [CrossRef]
- Graler, B.; Pebesma, E.; Heuvelink, G. Spatio-temporal interpolation using gstat. R J. 2016, 8, 204–218. [Google Scholar] [CrossRef]
- Roberts, D.; Bahn, V.; Ciuti, S.; Boyce, M.; Elith, J.; Guillera-Arroita, G.; Hauenstein, S.; Lahoz-Monfort, J.J.; Schröder, B.; Thuiller, W.; et al. Cross-validation strategies for data with temporal, spatial, hierarchical, or phylogenetic structure. Ecography 2017, 40, 913–929. [Google Scholar] [CrossRef]





| Forest Type Code | Definition |
|---|---|
| Rainforest | Cool temperate rainforest. Area with more than 70% projective foliage cover of rainforest species and less than 10% of Eucalyptus spp. |
| Mixed Forest | Cool temperate mixed forest. Area with more than 70% projective foliage cover of rainforest species and more than 10% Eucalyptus spp. |
| Fern | Fern-dominated stands. Fern stands are sometimes accompanied by scrubs. |
| Euc | Primarily Eucalyptus-dominated stands but includes everything that is not classified as Rainforest, Mixed Forest, or Fern. |
| Group | Predictor | Description | Resolution (m) |
|---|---|---|---|
| Environmental | Creek index | Gully index. Higher values indicate the presence of gullies | 20 |
| bio01 | Mean annual temperature | 20 | |
| bio01_2 | Squared bio01 | 20 | |
| bio03 | Isothermality (ratio of mean diurnal range (Mean of monthly (max temp-min temp)) to temperature annual range) | 20 | |
| bio03_2 | Squared bio03 | 20 | |
| bio04 | Temperature Seasonality (standard deviation of monthly mean temperature) | 20 | |
| bio04_2 | Squared bio04 | 20 | |
| bio12 | Mean annual precipitation | 20 | |
| bio11_2 | Squared bio12 | 20 | |
| Multispectral | Red_edge | Pan-sharpened (with red band as hi-res) red-edge (Sentinel band 5, 705 nm) | 20 |
| Green/Red | Simple ratio of Sentinel Band 3 (green, 560 nm) with Sentinel Band 4 (red, 665 nm) | 20 | |
| Green/NIR1 | Simple ratio of Sentinel Band 3 (green, 560 nm) with sentinel Band 6 (near IR1, 740 nm) | 20 | |
| Green/NIR2 | Simple ratio of Sentinel Band 3 (green, 560 nm) with sentinel Band 7 (Near IR 2, 783 nm) | 20 | |
| Green/NIR3 | Simple ratio of Sentinel Band 3 (green, 560 nm) with Sentinel Band 8 (Near IR 3, 842 nm) | 20 | |
| Green/NIR4 | Simple ratio of Sentinel Band 3 (green, 560 nm) with sentinel Band 8a (Near IR 4, 865 nm) | 20 | |
| Blue/Green | Simple ratio of Sentinel Band 2 (blue, 490 nm) with sentinel Band 3 (Green, 560 nm) | 20 | |
| TNGRDI | Normalized green-red difference index 32 years Landsat composite: | 25 | |
| LiDAR | p10, p25, p50, and p90 | 10th, 25th, 50th and 90th percentiles of point return height (m) | 20 |
| s00 to s60 | Percentage of all LiDAR point returns in 5 m height classes (%) | 20 | |
| pfc | Percentage of first returns (i.e., percentage forest cover) | 20 | |
| ovnum | Number of overstorey trees with crown width > 8 m | 20 | |
| ovcwavg | Average overstorey tree crown width (in m) | 20 | |
| mscover | Percentage cover of midstorey trees | 20 | |
| msnum | Number of misdtorey trees | 20 | |
| mshtavg | Average midstorey tree height | 20 | |
| pavd_PC1–pavd_PC9 | First to ninth PCA component of PAVD | 20 |
| EVC (%) | Total (%) | Total (ha) | ||||||
|---|---|---|---|---|---|---|---|---|
| 29 | 30 | 31 | 38 | 39 | Others | |||
| Euc | 27.7 | 22.5 | 1.9 | 4.5 | 10.6 | 32.8 | 92.6 | 430,385 |
| Fern | 29.0 | 56.6 | 2.1 | 0.4 | 0.6 | 11.2 | 2.0 | 9164 |
| Mixed Forest | 16.2 | 36.4 | 14.4 | 2.8 | 16.0 | 14.3 | 3.9 | 18,054 |
| Rainforest | 8.8 | 40.3 | 23.6 | 1.3 | 13.8 | 12.2 | 1.5 | 7114 |
| Total | 27.0 | 24.0 | 2.7 | 4.3 | 10.6 | 31.4 | 100.0 | 464,716 |
| Model | Acc | Kappa | Sensitivity | Specificity | Precision | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Euc | Fern | Mixed Forest | Rain Forest | Euc | Fern | Mixed Forest | Rain Forest | Euc | Fern | Mixed Forest | Rain Forest | |||
| Environmental | 0.55 | 0.32 | 0.89 | 0.21 | 0.25 | 0.39 | 0.66 | 0.99 | 0.85 | 0.84 | 0.65 | 0.79 | 0.29 | 0.46 |
| Multispectral | 0.70 | 0.56 | 0.89 | 0.58 | 0.37 | 0.71 | 0.75 | 0.98 | 0.92 | 0.90 | 0.72 | 0.79 | 0.55 | 0.72 |
| Lidar | 0.81 | 0.73 | 0.82 | 0.96 | 0.81 | 0.73 | 0.83 | 0.99 | 0.92 | 0.98 | 0.78 | 0.94 | 0.72 | 0.92 |
| Multi. & LiDAR | 0.87 | 0.81 | 0.85 | 0.94 | 0.79 | 0.92 | 0.89 | 1.00 | 0.92 | 0.99 | 0.85 | 1.00 | 0.73 | 0.97 |
| Full | 0.88 | 0.83 | 0.94 | 0.87 | 0.75 | 0.90 | 0.87 | 0.99 | 0.97 | 0.98 | 0.83 | 0.96 | 0.85 | 0.95 |
| Actual | ||||||
|---|---|---|---|---|---|---|
| Euc | Fern | Mixed Forest | Rainforest | |||
| Predicted | Environmental | Euc | 168 | 31 | 32 | 29 |
| Fern | 3 | 11 | 0 | 0 | ||
| Mixed Forest | 11 | 0 | 23 | 45 | ||
| Rainforest | 6 | 10 | 39 | 47 | ||
| Multispectral | Euc | 167 | 1 | 52 | 13 | |
| Fern | 0 | 30 | 0 | 8 | ||
| Mixed Forest | 14 | 1 | 35 | 14 | ||
| Rainforest | 7 | 20 | 7 | 86 | ||
| Lidar | Euc | 155 | 1 | 17 | 27 | |
| Fern | 3 | 50 | 0 | 0 | ||
| Mixed Forest | 24 | 0 | 76 | 6 | ||
| Rainforest | 6 | 1 | 1 | 88 | ||
| Multispectral and LiDAR | Euc | 160 | 3 | 19 | 7 | |
| Fern | 0 | 49 | 0 | 0 | ||
| Mixed Forest | 25 | 0 | 74 | 3 | ||
| Rainforest | 3 | 0 | 1 | 111 | ||
| Full | Euc | 176 | 5 | 21 | 9 | |
| Fern | 2 | 45 | 0 | 0 | ||
| Mixed Forest | 9 | 0 | 70 | 3 | ||
| Rainforest | 1 | 2 | 3 | 109 | ||
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2022 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Trouvé, R.; Jiang, R.; Fedrigo, M.; White, M.D.; Kasel, S.; Baker, P.J.; Nitschke, C.R. Combining Environmental, Multispectral, and LiDAR Data Improves Forest Type Classification: A Case Study on Mapping Cool Temperate Rainforests and Mixed Forests. Remote Sens. 2023, 15, 60. https://doi.org/10.3390/rs15010060
Trouvé R, Jiang R, Fedrigo M, White MD, Kasel S, Baker PJ, Nitschke CR. Combining Environmental, Multispectral, and LiDAR Data Improves Forest Type Classification: A Case Study on Mapping Cool Temperate Rainforests and Mixed Forests. Remote Sensing. 2023; 15(1):60. https://doi.org/10.3390/rs15010060
Chicago/Turabian StyleTrouvé, Raphael, Ruizhu Jiang, Melissa Fedrigo, Matt D. White, Sabine Kasel, Patrick J. Baker, and Craig R. Nitschke. 2023. "Combining Environmental, Multispectral, and LiDAR Data Improves Forest Type Classification: A Case Study on Mapping Cool Temperate Rainforests and Mixed Forests" Remote Sensing 15, no. 1: 60. https://doi.org/10.3390/rs15010060
APA StyleTrouvé, R., Jiang, R., Fedrigo, M., White, M. D., Kasel, S., Baker, P. J., & Nitschke, C. R. (2023). Combining Environmental, Multispectral, and LiDAR Data Improves Forest Type Classification: A Case Study on Mapping Cool Temperate Rainforests and Mixed Forests. Remote Sensing, 15(1), 60. https://doi.org/10.3390/rs15010060








