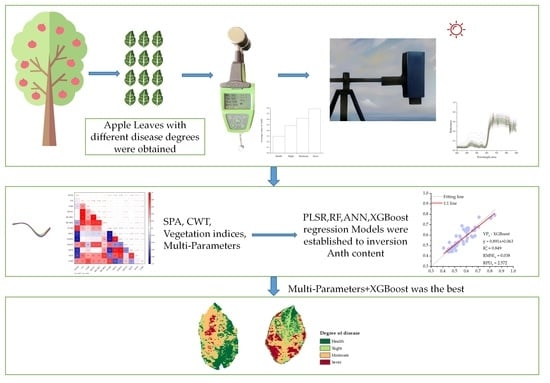
Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images
Abstract
:1. Introduction
2. Materials and Methods
2.1. Sample Collection and Data Acquisition
2.2. Data Pre–Processing
2.2.1. Hyperspectral Data Preprocessing
2.2.2. Vegetation Indices
2.3. Variable Selection Methods
2.3.1. Successive Projections Algorithm (SPA)
2.3.2. Continuous Wavelet Transform
2.4. Regression Models
2.4.1. Partial Least-Square Regression (PLSR)
2.4.2. Random Forest (RF) Regression
2.4.3. Artificial Neural Network (ANN)
2.4.4. Extreme-Gradient Boost (XGBoost) Regression
2.5. Test of Accuracy
3. Results
3.1. Spectral Characteristics of Leaves
3.2. Correlation between Spectral Characteristics and Anthocyanin and Select Modeling Parameters
3.2.1. Correlation between Spectral Reflectance and Anthocyanin Content
3.2.2. Characteristic Bands Selected by SPA
3.2.3. Correlation between Vegetation Indices and Anthocyanin Content
3.2.4. Correlation between Wavelet Coefficients and Anthocyanin
3.3. Regression Models and Accuracy Evaluation
3.3.1. Models Based on SPA Selected Bands
3.3.2. Models Based on Vegetation Index
3.3.3. Construction of Wavelet Transform Model
3.3.4. Multi-Parameter Model
3.4. Inversion of Degree of Mosaic Disease in Hyperspectral Images
4. Discussion
4.1. Spectral Reflectance of Leaves Closely Relates to Degree of Mosaic Disease
4.2. Vegetation Index and Wavelet Coefficients of any Two Bands have Higher Accuracy for Monitoring Mosaic Disease
4.3. Application of Machine Learning Algorithm to Monitoring Mosaic Disease
5. Conclusions
- The spectral difference between the healthy and diseased leaves was concentrated in the range of 470–750 nm, with the largest difference appearing near 702 nm. With the increase in the severity of mosaic disease, the anthocyanin content increased, the absorption characteristics gradually disappeared at 500–560 nm, and the phenomenon called “blue shift” appeared at the reflection spectrum of the red edge.
- Wavelets transformed the decomposed spectral information and effectively improved the correlation between the reflectance spectrum and anthocyanin content. Moreover, the accuracy of the anthocyanin regression models constructed using wavelet coefficients was significantly improved compared to the anthocyanin regression models constructed using characteristic bands and vegetation indices.
- The VPs-XGBoost estimation model based on multiple parameters (R2v = 0.849, RPD = 2.572) was more accurate and reliable than the other methods. The VPs-XGBoost method, based on hyperspectral images, may be a rapid, accurate, and simple method to monitor the degree of mosaic disease in apple leaves.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- FAOSTAT. Available online: https://www.fao.org/faostat/en/#data/QCL/visualize (accessed on 6 February 2023).
- Hu, G.-J.; Dong, Y.-F.; Zhang, Z.-P.; Fan, X.-D.; Ren, F. Molecular characterization of Apple necrotic mosaic virus identified in crabapple (Malus spp.) tree of China. J. Integr. Agric. 2019, 18, 698–701. [Google Scholar] [CrossRef]
- Shi, W.; Yao, R.; Sunwu, R.; Huang, K.; Liu, Z.; Li, X.; Yang, Y.; Wang, J. Incidence and Molecular Identification of Apple Necrotic Mosaic Virus (ApNMV) in Southwest China. Plants 2020, 9, 415. [Google Scholar] [CrossRef] [PubMed]
- Noda, H.; Yamagishi, N.; Yaegashi, H.; Xing, F.; Xie, J.; Li, S.; Zhou, T.; Ito, T.; Yoshikawa, N. Apple necrotic mosaic virus, a novel ilarvirus from mosaic-diseased apple trees in Japan and China. J. Gen. Plant Pathol. 2017, 83, 83–90. [Google Scholar] [CrossRef]
- Chalker-Scott, L. Environmental Significance of Anthocyanins in Plant Stress Responses. Photochem. Photobiol. 1999, 70, 1–9. [Google Scholar] [CrossRef]
- Sullivan, C.N.; Koski, M.H. The effects of climate change on floral anthocyanin polymorphisms. Proc. Biol. Sci. 2021, 288, 20202693. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Merzlyak, M.N.; Chivkunova, O.B. Optical Properties and Nondestructive Estimation of Anthocyanin Content in Plant Leaves. Photochem. Photobiol. 2001, 74, 38–45. [Google Scholar] [CrossRef]
- Gitelson, A.A. Non-Destructive Assessment of Chlorophyll Carotenoid and Anthocyanin Content in Higher Plant Leaves: Principles and Algorithms. 2004. Available online: https://digitalcommons.unl.edu/natrespapers/263/ (accessed on 23 April 2023).
- Steele, M.R.; Gitelson, A.A.; Rundquist, D.C.; Merzlyak, M.N. Nondestructive Estimation of Anthocyanin Content in Grapevine Leaves. Am. J. Enol. Vitic. 2009, 60, 87–92. [Google Scholar] [CrossRef]
- Qin, J.; Rundquist, D.; Gitelson, A.; Tan, Z.; Steele, M. A Non-linear Model of Nondestructive Estimation of Anthocyanin Content in Grapevine Leaves with Visible/Red-Infrared Hyperspectral. In Proceedings of the Computer and Computing Technologies in Agriculture IV, Nanchang, China, 22–25 October 2010; Volume 3, pp. 47–62. [Google Scholar]
- Jiang, J.; Zhang, Z.; Cao, Q.; Liang, Y.; Krienke, B.; Tian, Y.; Zhu, Y.; Cao, W.; Liu, X. Use of an Active Canopy Sensor Mounted on an Unmanned Aerial Vehicle to Monitor the Growth and Nitrogen Status of Winter Wheat. Remote Sens. 2020, 12, 3684. [Google Scholar] [CrossRef]
- Terentev, A.; Dolzhenko, V.; Fedotov, A.; Eremenko, D. Current State of Hyperspectral Remote Sensing for Early Plant Disease Detection: A Review. Sensors 2022, 22, 757. [Google Scholar] [CrossRef]
- Wang, J.; Ren, G.; Lin, Z.; Wang, A.; Hu, Y.; Li, X.; Wu, P.; Ma, Y.; Zhang, J. Estimation of Aboveground Vegetation Nitrogen Contents in the Yellow River Estuary Wetland Using GaoFen-1 Remote Sensing Data. J. Coastal Res. 2020, 102, 1–10. [Google Scholar] [CrossRef]
- Deng, X.; Huang, Z.; Zheng, Z.; Lan, Y.; Dai, F. Field detection and classification of citrus Huanglongbing based on hyperspectral reflectance. Comput. Electron. Agric. 2019, 167, 105006. [Google Scholar] [CrossRef]
- Haboudane, D. Hyperspectral vegetation indices and novel algorithms for predicting green LAI of crop canopies: Modeling and validation in the context of precision agriculture. Remote Sens. Environ. 2004, 90, 337–352. [Google Scholar] [CrossRef]
- Haboudane, D.; Miller, J.R.; Tremblay, N.; Zarco-Tejada, P.J.; Dextraze, L. Integrated narrow-band vegetation indices for prediction of crop chlorophyll content for application to precision agriculture. Remote Sens. Environ. 2002, 81, 416–426. [Google Scholar] [CrossRef]
- Pal, T.; Jaiswal, V.; Chauhan, R.S. DRPPP: A machine learning based tool for prediction of disease resistance proteins in plants. Comput. Biol. Med. 2016, 78, 42–48. [Google Scholar] [CrossRef] [PubMed]
- Shafri, H.Z.M.; Hamdan, N. Hyperspectral Imagery for Mapping Disease Infection in Oil Palm Plantation Using Vegetation Indices and Red Edge Techniques. Am. J. Appl. Sci. 2009, 6, 1031–1035. [Google Scholar]
- Sun, Q.; Jiao, Q.; Qian, X.; Liu, L.; Liu, X.; Dai, H. Improving the Retrieval of Crop Canopy Chlorophyll Content Using Vegetation Index Combinations. Remote Sens. 2021, 13, 470. [Google Scholar] [CrossRef]
- Liu, X.; Ji, L.; Zhang, C.; Liu, Y. A method for reconstructing NDVI time-series based on envelope detection and the Savitzky-Golay filter. Int. J. Digit. Earth 2022, 15, 553–584. [Google Scholar] [CrossRef]
- Ruffin, C.; King, R.L.; Younan, N.H. A Combined Derivative Spectroscopy and Savitzky-Golay Filtering Method for the Analysis of Hyperspectral Data. GISci. Remote Sens. 2013, 45, 1–15. [Google Scholar] [CrossRef]
- Wang, J.; Shi, T.; Liu, H.; Wu, G. Successive projections algorithm-based three-band vegetation index for foliar phosphorus estimation. Ecol. Indic. 2016, 67, 12–20. [Google Scholar] [CrossRef]
- Ding, D.; Yu, H.; Yin, Y.; Yuan, Y.; Li, Z.; Li, F. Determination of Chlorophyll and Hardness in Cucumbers by Raman Spectroscopy with Successive Projections Algorithm (SPA)—Extreme Learning Machine (ELM). Anal. Lett. 2022, 56, 1216–1228. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Keydan, G.P.; Merzlyak, M.N. Three-band model for noninvasive estimation of chlorophyll, carotenoids, and anthocyanin contents in higher plant leaves. Geophys. Res. Lett. 2006, 33, L11402. [Google Scholar] [CrossRef]
- Luo, L.; Chang, Q.; Wang, Q.; Huang, Y. Identification and Severity Monitoring of Maize Dwarf Mosaic Virus Infection Based on Hyperspectral Measurements. Remote Sens. 2021, 13, 4560. [Google Scholar] [CrossRef]
- Ma, H.; Huang, W.; Jing, Y.; Pignatti, S.; Laneve, G.; Dong, Y.; Ye, H.; Liu, L.; Guo, A.; Jiang, J. Identification of Fusarium Head Blight in Winter Wheat Ears Using Continuous Wavelet Analysis. Sensors 2019, 20, 20. [Google Scholar] [CrossRef] [PubMed]
- Zhang, J.; Sun, H.; Gao, D.; Qiao, L.; Liu, N.; Li, M.; Zhang, Y. Detection of Canopy Chlorophyll Content of Corn Based on Continuous Wavelet Transform Analysis. Remote Sens. 2020, 12, 2741. [Google Scholar] [CrossRef]
- Shi, Y.; Huang, W.; González-Moreno, P.; Luke, B.; Dong, Y.; Zheng, Q.; Ma, H.; Liu, L. Wavelet-Based Rust Spectral Feature Set (WRSFs): A Novel Spectral Feature Set Based on Continuous Wavelet Transformation for Tracking Progressive Host–Pathogen Interaction of Yellow Rust on Wheat. Remote Sens. 2018, 10, 525. [Google Scholar] [CrossRef]
- Wang, Z.; Chen, J.; Fan, Y.; Cheng, Y.; Wu, X.; Zhang, J.; Wang, B.; Wang, X.; Yong, T.; Liu, W.; et al. Evaluating photosynthetic pigment contents of maize using UVE-PLS based on continuous wavelet transform. Comput. Electron. Agric. 2020, 169, 105160. [Google Scholar] [CrossRef]
- Ferwerda, J.G.; Jones, S.D. Continuous Wavelet Transformations for Hyperspectral Feature Detection. In Progress in Spatial Data Handling: 12th International Symposium on Spatial Data Handling; Riedl, A., Kainz, W., Elmes, G.A., Eds.; Springer: Berlin, Heidelberg, 2006; pp. 167–178. [Google Scholar]
- Zhao, L.; Li, Q.; Zhang, Y.; Wang, H.; Du, X. Integrating the Continuous Wavelet Transform and a Convolutional Neural Network to Identify Vineyard Using Time Series Satellite Images. Remote Sens. 2019, 11, 2641. [Google Scholar] [CrossRef]
- He, R.; Li, H.; Qiao, X.; Jiang, J. Using wavelet analysis of hyperspectral remote-sensing data to estimate canopy chlorophyll content of winter wheat under stripe rust stress. Int. J. Remote Sens. 2018, 39, 4059–4076. [Google Scholar] [CrossRef]
- An, G.; Xing, M.; Liao, C.; He, B. Estimating Chlorophyll Content of Rice Based on UAV-Based Hyperspectral Imagery and Continuous Wavelet Transform. In Proceedings of the IGARSS 2020—2020 IEEE International Geoscience and Remote Sensing Symposium, Waikoloa, Hawaii, 26 September–2 October 2020; pp. 5270–5273. [Google Scholar]
- Bagheri, N.; Mohamadi-Monavar, H.; Azizi, A.; Ghasemi, A. Detection of Fire Blight disease in pear trees by hyperspectral data. Eur. J. Remote Sens. 2017, 51, 1–10. [Google Scholar] [CrossRef]
- Gold, K.M.; Townsend, P.A.; Chlus, A.; Herrmann, I.; Couture, J.J.; Larson, E.R.; Gevens, A.J. Hyperspectral Measurements Enable Pre-Symptomatic Detection and Differentiation of Contrasting Physiological Effects of Late Blight and Early Blight in Potato. Remote Sens. 2020, 12, 286. [Google Scholar] [CrossRef]
- Huang, W.; Lamb, D.W.; Niu, Z.; Zhang, Y.; Liu, L.; Wang, J. Identification of yellow rust in wheat using in-situ spectral reflectance measurements and airborne hyperspectral imaging. Precis. Agric. 2007, 8, 187–197. [Google Scholar] [CrossRef]
- Xie, C.; He, Y. Spectrum and Image Texture Features Analysis for Early Blight Disease Detection on Eggplant Leaves. Sensors 2016, 16, 676. [Google Scholar] [CrossRef] [PubMed]
- Nagasubramanian, K.; Jones, S.; Singh, A.K.; Sarkar, S.; Singh, A.; Ganapathysubramanian, B. Plant disease identification using explainable 3D deep learning on hyperspectral images. Plant Methods 2019, 15, 98. [Google Scholar] [CrossRef] [PubMed]
- Polder, G.; Blok, P.M.; de Villiers, H.A.C.; van der Wolf, J.M.; Kamp, J. Potato Virus Y Detection in Seed Potatoes Using Deep Learning on Hyperspectral Images. Front. Plant Sci. 2019, 10, 209. [Google Scholar] [CrossRef]
- Yuan, L.; Yan, P.; Han, W.; Huang, Y.; Wang, B.; Zhang, J.; Zhang, H.; Bao, Z. Detection of anthracnose in tea plants based on hyperspectral imaging. Comput. Electron. Agric. 2019, 167, 105039. [Google Scholar] [CrossRef]
- Wu, Y.; Cao, Y.; Zhai, Z. Early Detection of Bacterial Blight in Hyperspectral Images Based on Random Forest and Adaptive Coherence Estimator. Sustainability 2022, 14, 13168. [Google Scholar] [CrossRef]
- Xu, S.; Xu, X.; Blacker, C.; Gaulton, R.; Zhu, Q.; Yang, M.; Yang, G.; Zhang, J.; Yang, Y.; Yang, M.; et al. Estimation of Leaf Nitrogen Content in Rice Using Vegetation Indices and Feature Variable Optimization with Information Fusion of Multiple-Sensor Images from UAV. Remote Sens. 2023, 15, 854. [Google Scholar] [CrossRef]
- Tao, H.; Feng, H.; Xu, L.; Miao, M.; Yang, G.; Yang, X.; Fan, L. Estimation of the Yield and Plant Height of Winter Wheat Using UAV-Based Hyperspectral Images. Sensors 2020, 20, 1231. [Google Scholar] [CrossRef]
- Ta, N.; Chang, Q.; Zhang, Y. Estimation of Apple Tree Leaf Chlorophyll Content Based on Machine Learning Methods. Remote Sens. 2021, 13, 3902. [Google Scholar] [CrossRef]
- Wei, M.; Wang, H.; Zhang, Y.; Li, Q.; Du, X.; Shi, G.; Ren, Y. Investigating the Potential of Crop Discrimination in Early Growing Stage of Change Analysis in Remote Sensing Crop Profiles. Remote Sens. 2023, 15, 853. [Google Scholar] [CrossRef]
- Couture, J.J.; Singh, A.; Charkowski, A.O.; Groves, R.L.; Gray, S.M.; Bethke, P.C.; Townsend, P.A. Integrating Spectroscopy with Potato Disease Management. Plant Dis. 2018, 102, 2233–2240. [Google Scholar] [CrossRef] [PubMed]
- Li, F.; Wang, L.; Liu, J.; Wang, Y.; Chang, Q. Evaluation of Leaf N Concentration in Winter Wheat Based on Discrete Wavelet Transform Analysis. Remote Sens. 2019, 11, 1331. [Google Scholar] [CrossRef]
- Qi, H.; Zhu, B.; Kong, L.; Yang, W.; Zou, J.; Lan, Y.; Zhang, L. Hyperspectral Inversion Model of Chlorophyll Content in Peanut Leaves. Appl. Sci. 2020, 10, 2259. [Google Scholar] [CrossRef]
- Lussem, U.; Bolten, A.; Gnyp, M.L.; Jasper, J.; Bareth, G. Evaluation of Rgb-Based Vegetation Indices from Uav Imagery To Estimate Forage Yield in Grassland. In Proceedings of the International Archives of the Photogrammetry, Remote Sensing and Spatial Information Sciences, Beijing, China, 7–10 May 2018; pp. 1215–1219. [Google Scholar]
- Sharifi, A. Remotely sensed vegetation indices for crop nutrition mapping. J. Sci. Food. Agric. 2020, 100, 5191–5196. [Google Scholar] [CrossRef] [PubMed]
- Rondeaux, G.; Steven, M.; Baret, F. Optimization of soil-adjusted vegetation indices. Remote Sens. Environ. 1996, 55, 95–107. [Google Scholar] [CrossRef]
- Nagai, S.; Ishii, R.; Suhaili, A.B.; Kobayashi, H.; Matsuoka, M.; Ichie, T.; Motohka, T.; Kendawang, J.J.; Suzuki, R. Usability of noise-free daily satellite-observed green–red vegetation index values for monitoring ecosystem changes in Borneo. Int. J. Remote Sens. 2014, 35, 7910–7926. [Google Scholar] [CrossRef]
- Chen, F.; Chen, C.; Li, W.; Xiao, M.; Yang, B.; Yan, Z.; Gao, R.; Zhang, S.; Han, H.; Chen, C.; et al. Rapid detection of seven indexes in sheep serum based on Raman spectroscopy combined with DOSC-SPA-PLSR-DS model. Spectrochim. Acta Part A Mol. Biomol. Spectrosc. 2021, 248, 119260. [Google Scholar] [CrossRef]
- The Successive Projections Algorithm (SPA) Homepage. Available online: http://www.ele.ita.br/~kawakami/spa/ (accessed on 23 April 2023).
- Kim, M.S.; Daughtry, C.S.T.; Chappelle, E.W.; McMurtrey, J.E. The use of high spectral resolution bands for estimating absorbed photosynthetically active radiation (APAR). In Proceedings of the ISPRS’94, Val d’Isere, France, 17–21 January 1994. [Google Scholar]
- Rivard, B.; Feng, J.; Gallie, A.; Sanchez-Azofeifa, A. Continuous wavelets for the improved use of spectral libraries and hyperspectral data. Remote Sens. Environ. 2008, 112, 2850–2862. [Google Scholar] [CrossRef]
- Blackburn, G.; Ferwerda, J. Retrieval of chlorophyll concentration from leaf reflectance spectra using wavelet analysis. Remote Sens. Environ. 2008, 112, 1614–1632. [Google Scholar] [CrossRef]
- Guo, Y.; Zhang, L.; Wang, D.; Ma, M. Application of Wavelete Analysis for Determining Chlorophyll Concentration in Vegetation by Hyperspectral Reflectance. Bull. Surv. Mapp. 2010, 8, 31–33,53. [Google Scholar]
- Li, X.; Li, Z.; Chen, G.; Qiu, H.; Hou, G.; Fan, P. Prediction of Tidal Flat Sediment Moisture Content Based on Wavelet Transform. Spectrosc. Spect. Anal. 2022, 42, 1156–1161. [Google Scholar]
- Cheng, T.; Rivard, B.; Sánchez-Azofeifa, G.A.; Feng, J.; Calvo-Polanco, M. Continuous wavelet analysis for the detection of green attack damage due to mountain pine beetle infestation. Remote Sens. Environ. 2010, 114, 899–910. [Google Scholar] [CrossRef]
- Zhu Wen, Y.; Zhou, J.; Wu, Y.f.; Wang, M.J. Iris Feature Extraction based on Haar Wavelet Transform. Int. J. Secur. Its Appl. 2014, 8, 265–272. [Google Scholar] [CrossRef]
- Koger, C. Wavelet analysis of hyperspectral reflectance data for detecting pitted morningglory (Ipomoea lacunosa) in soybean (Glycine max). Remote Sens. Environ. 2003, 86, 108–119. [Google Scholar] [CrossRef]
- Cheng, J.-H.; Sun, D.-W. Partial Least Squares Regression (PLSR) Applied to NIR and HSI Spectral Data Modeling to Predict Chemical Properties of Fish Muscle. Food Eng. Rev. 2016, 9, 36–49. [Google Scholar] [CrossRef]
- Lin, L.; Liu, X. Soil-moisture-index spectrum reconstruction improves partial least squares regression of spectral analysis of soil organic carbon. Precis. Agric. 2022, 23, 1707–1719. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Soltanikazemi, M.; Minaei, S.; Shafizadeh-Moghadam, H.; Mahdavian, A. Field-scale estimation of sugarcane leaf nitrogen content using vegetation indices and spectral bands of Sentinel-2: Application of random forest and support vector regression. Comput. Electron. Agric. 2022, 200, 107130. [Google Scholar] [CrossRef]
- Estrada Zúñiga, A.C.; Cárdenas, J.; Víctor Bejar, J.; Ñaupari, J. Biomass estimation of a high Andean plant community with multispectral images acquired using UAV remote sensing and Multiple Linear Regression, Support Vector Machine and Random Forests models. Sci. Agropecu. 2022, 13, 301–310. [Google Scholar] [CrossRef]
- Feng, H.; Tao, H.; Fan, Y.; Liu, Y.; Li, Z.; Yang, G.; Zhao, C. Comparison of Winter Wheat Yield Estimation Based on Near-Surface Hyperspectral and UAV Hyperspectral Remote Sensing Data. Remote Sens. 2022, 14, 4158. [Google Scholar] [CrossRef]
- Yuan, H.; Yang, G.; Li, C.; Wang, Y.; Liu, J.; Yu, H.; Feng, H.; Xu, B.; Zhao, X.; Yang, X. Retrieving Soybean Leaf Area Index from Unmanned Aerial Vehicle Hyperspectral Remote Sensing: Analysis of RF, ANN, and SVM Regression Models. Remote Sens. 2017, 9, 309. [Google Scholar] [CrossRef]
- Chen, T.; Guestrin, C. XGBoost. In Proceedings of the 22nd ACM SIGKDD International Conference on Knowledge Discovery and Data Mining, San Francisco, CA, USA, 13–17 August 2016; pp. 785–794. [Google Scholar]
- Zhang, N.; Zhang, X.; Wang, C.; Li, L.; Bai, T. Cotton LAI Estimation Based on Hyperspectral and Successive Projection Algorithm. Trans. Chin. Soc. Agric. Mach. 2022, 53, 257–262. [Google Scholar]
- Mutanga, O.; Skidmore, A.K. Red edge shift and biochemical content in grass canopies. ISPRS-J. Photogramm. Remote Sens. 2007, 62, 34–42. [Google Scholar] [CrossRef]
- Li, L.; Ren, T.; Ma, Y.; Wei, Q.; Wang, S.; Li, X.; Cong, R.; Liu, S.; Lu, J. Evaluating chlorophyll density in winter oilseed rape (Brassica napus L.) using canopy hyperspectral red-edge parameters. Comput. Electron. Agric. 2016, 126, 21–31. [Google Scholar] [CrossRef]
- Mahlein, A.K.; Rumpf, T.; Welke, P.; Dehne, H.W.; Plümer, L.; Steiner, U.; Oerke, E.C. Development of spectral indices for detecting and identifying plant diseases. Remote Sens. Environ. 2013, 128, 21–30. [Google Scholar] [CrossRef]
- Wang, L.; Liu, J.; Shao, J.; Yang, F.; Gao, J. Remote sensing index selection of leaf blight disease in spring maize based on hyperspectral data. Trans. CSAE 2017, 33, 170–177. [Google Scholar]
- Ménard, R.; Deshaies-Jacques, M. Evaluation of Analysis by Cross-Validation. Part I: Using Verification Metrics. Atmosphere 2018, 9, 86. [Google Scholar] [CrossRef]








| Vegetation Indices | Bands | Equation | Reference |
|---|---|---|---|
| NDSI | Any two bands | [48] | |
| RSI | [48] | ||
| DSI | [48] | ||
| TVI | Three specific bands | [14] | |
| VARI | [45] | ||
| MTVI1 | [15] | ||
| MCARI1 | )) | [15] | |
| MCARI2 | [15] | ||
| TCARI | [40] | ||
| MTCI | [49] | ||
| GNDVI | Two specific bands | [25] | |
| OSAVI | [50] | ||
| GRVI | [51] | ||
| SAVI | [50] | ||
| CARI | [52] |
| Mother Wavelet Functions | Applications | Reference |
|---|---|---|
| Gaussian 1 | Chlorophyll content | [57] |
| Rbio 5.5 | Chlorophyll content | [56] |
| Mor l | Chlorophyll content | [58] |
| Db 5 | Nitrogen content and classification | [28] |
| Bior 3.3 | Pigment content | [27] |
| Sym 8 | Water content | [59] |
| Mexh | Water and chlorophyll content | [60] |
| Meyr | Classification | [29] |
| Haar | Chlorophyll content | [61] |
| Coif 2 | Chlorophyll content | [62] |
| Vegetation Index | Bands | Coefficient of Determination | Vegetation Index | Bands | Coefficient of Determination |
|---|---|---|---|---|---|
| TVI | Three | 0.06 | MTCI | Three | 0.85 ** |
| VARI | 0.16 * | GNDVI | Tow | 0.90 ** | |
| MTVI1 | 0.05 | OSAVI | 0.45 ** | ||
| MCARI1 | 0.05 | GRVI | 0.83 ** | ||
| MCARI2 | 0.35 ** | SAVI | 0.77 ** | ||
| TCARI | 0.83 ** | CARI | 0.46 ** |
| Model | Modeling Set | Verification Set | ||||
|---|---|---|---|---|---|---|
| R2 | RMSEc | RPD | R2 | RMSEv | RPD | |
| PLSR | 0.713 | 0.055 | 1.873 | 0.828 | 0.042 | 2.345 |
| RF | 0.965 | 0.022 | 4.717 | 0.736 | 0.051 | 1.918 |
| ANN | 0.788 | 0.047 | 2.183 | 0.828 | 0.045 | 2.196 |
| XGBoost | 0.900 | 0.033 | 3.124 | 0.757 | 0.053 | 1.865 |
| Model | Modeling Set | Verification Set | |||||
|---|---|---|---|---|---|---|---|
| R2 | RMSEc | RPD | R2 | RMSEv | RPD | ||
| VIA | PLSR | 0.843 | 0.040 | 2.536 | 0.840 | 0.040 | 2.440 |
| RF | 0.970 | 0.019 | 5.448 | 0.805 | 0.044 | 2.257 | |
| ANN | 0.844 | 0.040 | 2.544 | 0.861 | 0.039 | 2.542 | |
| XGBoost | 0.890 | 0.034 | 3.014 | 0.840 | 0.039 | 2.496 | |
| VIS | PLSR | 0.823 | 0.043 | 2.387 | 0.757 | 0.053 | 1.846 |
| RF | 0.974 | 0.018 | 5.676 | 0.730 | 0.057 | 1.719 | |
| ANN | 0.834 | 0.042 | 2.463 | 0.749 | 0.055 | 1.778 | |
| XGBoost | 0.897 | 0.033 | 3.099 | 0.735 | 0.058 | 1.711 | |
| VI (VIA + VIS) | PLSR | 0.833 | 0.042 | 2.460 | 0.800 | 0.048 | 2.049 |
| RF | 0.977 | 0.017 | 6.075 | 0.838 | 0.041 | 2.415 | |
| ANN | 0.857 | 0.039 | 2.651 | 0.831 | 0.041 | 2.379 | |
| XGBoost | 0.918 | 0.029 | 3.483 | 0.853 | 0.038 | 2.568 | |
| Model | Modeling Set | Verification Set | ||||
|---|---|---|---|---|---|---|
| R2 | RMSEc | RPD | R2 | RMSEv | RPD | |
| PLSR | 0.839 | 0.041 | 2.505 | 0.795 | 0.048 | 2.048 |
| RF | 0.975 | 0.017 | 5.919 | 0.827 | 0.042 | 2.316 |
| ANN | 0.832 | 0.042 | 2.451 | 0.853 | 0.043 | 2.301 |
| XGBoost | 0.904 | 0.032 | 3.22 | 0.818 | 0.042 | 2.328 |
| Model | Modeling Set | Verification Set | ||||
|---|---|---|---|---|---|---|
| R2 | RMSEc | RPD | R2 | RMSEv | RPD | |
| PLSR | 0.852 | 0.039 | 2.606 | 0.829 | 0.042 | 2.348 |
| RF | 0.977 | 0.017 | 5.979 | 0.842 | 0.040 | 2.471 |
| ANN | 0.854 | 0.039 | 2.626 | 0.836 | 0.044 | 2.254 |
| XGBoost | 0.923 | 0.026 | 3.875 | 0.849 | 0.038 | 2.572 |
| Min | Max | Average | Number of Pixels | Healthy Pixels % | Slight Pixels % | Moderate Pixels % | Severe Pixels % | |
|---|---|---|---|---|---|---|---|---|
| (a) | 0.429 | 0.550 | 0.490 | 11949 | 56.87 | 41.98 | 1.15 | 0.00 |
| (b) | 0.442 | 0.764 | 0.581 | 6880 | 29.03 | 25.23 | 40.36 | 5.52 |
| (c) | 0.423 | 0.860 | 0.612 | 10432 | 13.60 | 31.93 | 34.16 | 20.30 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Jiang, D.; Chang, Q.; Zhang, Z.; Liu, Y.; Zhang, Y.; Zheng, Z. Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images. Remote Sens. 2023, 15, 2504. https://doi.org/10.3390/rs15102504
Jiang D, Chang Q, Zhang Z, Liu Y, Zhang Y, Zheng Z. Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images. Remote Sensing. 2023; 15(10):2504. https://doi.org/10.3390/rs15102504
Chicago/Turabian StyleJiang, Danyao, Qingrui Chang, Zijuan Zhang, Yanfu Liu, Yu Zhang, and Zhikang Zheng. 2023. "Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images" Remote Sensing 15, no. 10: 2504. https://doi.org/10.3390/rs15102504
APA StyleJiang, D., Chang, Q., Zhang, Z., Liu, Y., Zhang, Y., & Zheng, Z. (2023). Monitoring the Degree of Mosaic Disease in Apple Leaves Using Hyperspectral Images. Remote Sensing, 15(10), 2504. https://doi.org/10.3390/rs15102504