1. Introduction
Hyperspectral imagery (HSIs) represents a unique category of images captured using advanced sensors tailored to gather data across a continuous and broad spectral range. Each pixel in an HSI carries a distinctive signature spanning multiple spectral bands [
1]. Owing to their data richness, HSIs find utility in diverse fields such as agricultural monitoring [
2], ecological assessments [
3], mineral detection [
4], medical diagnostics [
5], and military tactics [
6]. With hyperspectral image classification (HSIC) gaining prominence in remote sensing research, the precise interpretation and correct analysis of both the spatial and spectral components of these images have become paramount [
7].
In the initial stages of HSIC, the focus was primarily on spectral feature-based techniques. Key methods encompassed Support Vector Machines (SVM) [
8], a Random Forest (RF) [
9], and k-Nearest Neighbors (KNN) [
10]. Although these conventional algorithms possess certain advantages, they often overlook spatial characteristics, yielding classification outcomes that do not consistently meet practical requirements. A compounding challenge is that HSIs frequently demonstrate substantial variation within a class while presenting considerable resemblance among distinct classes. For instance, external environmental variables might cause shifts in the spectral signatures of consistent ground entities. Conversely, unrelated terrestrial groups might share similar spectral patterns, especially when contamination exists between neighboring areas, complicating accurate object classification [
11]. Hence, an excessive reliance on merely spectral characteristics is likely to result in the misclassification of the intended subjects [
12].
In recent years, hyperspectral image classification (HSIC) has benefitted from the integration of deep learning algorithms, noted for their outstanding feature extraction prowess [
13]. Convolutional neural networks (CNNs), in particular, have established themselves as a dominant approach within this field [
14]. Initial endeavors, exemplified by the study of Hu et al., utilized a one-dimensional CNN to transform each pixel in the hyperspectral image into a 1D vector, aiming for spectral feature extraction [
15]. In a similar vein, Cao et al. harnessed a two-dimensional CNN in conjunction with Markov Random Fields to amalgamate spatial and spectral data, thus augmenting classification accuracy [
16]. Zhao et al. incorporated Principal Component Analysis (PCA) for dimensionality reduction, followed by the application of a 2D-CNN for spatial feature extraction from hyperspectral images [
17]. However, while these studies primarily employ 1D and 2D CNN architectures, they face inherent constraints. Namely, 1D CNNs present simplicity issues, whereas 2D CNNs do not fully exploit both spatial and spectral features, creating impediments to potential enhancements in HSIC accuracy [
18].
Three-dimensional Convolutional Neural Networks (3D-CNNs) have become instrumental in extracting both spatial and spectral features concurrently, enhancing the precision of hyperspectral image classification (HSIC). Zhang et al. proposed a unique architecture incorporating parallel 3D Inception layers. This model leverages cross-entropy-based dimensionality reduction techniques to adaptively select spectral bands, thus refining classification results [
19]. Similarly, Zhang et al. introduced the End-to-End Spectral Spatial Residual Network (SSRN), a design that integrates 3D-CNN layers within a residual network structure. It employs dual consecutive residual blocks to independently learn spatial and spectral features, confirming the efficacy of dual-dimension feature learning in refining classification precision [
20]. Additionally, Roy et al. crafted a network architecture named HybridSN, which sequences three 3D-CNN layers for initial feature fusion and subsequent 2D-CNN layers for spatial feature extraction, highlighting the benefits of mixed convolutional dimensions in HSIC contexts [
21].
Despite the precision of 3D-CNN-based classifications, they are often associated with considerable computational demands, especially during fully connected mapping stages, attributable to the extensive parameter generation [
22]. Addressing this, Howard et al. substituted 2D-CNN layers in their design with depthwise separable convolutions, significantly reducing parameter counts during convolutional operations [
23]. Similarly, Zhao et al. incorporated depthwise separable convolutions into a multi-residual framework to balance computational efficiency with classification accuracy [
22]. Wang et al. developed a resource-optimized convolutional model, blending rapid 3D-CNN algorithms with depthwise separable convolutions, facilitating spatial–spectral feature extraction without parameter escalation [
24]. A subsequent analysis indicated the superior descriptive capacity of multi-scale features for the intricacies of HSIs. Thus, integrating features across various scales can elevate classification performance [
25]. Correspondingly, Shi et al. conceptualized a dual-branch, multi-scale spectral attention network utilizing diverse convolutional kernel sizes to extract features over multiple scales [
26]. Gong et al. devised a Hybrid 2D-3D CNN Multi-Scale Information Fusion Network (MSPN) to deepen network structures vertically and expand multi-scale spatial and spectral data horizontally [
27]. Wang et al. also introduced the Multi-Scale Dense Connection Attention Network (MSDAN) adept at capturing variegated features from HSIs across different scales, mitigating issues such as model overfitting and gradient disappearance [
28].
The attention mechanism has recently gained significant traction in computer vision, paving the way for enhanced feature extraction [
29]. Fang et al. introduced a 3D Dense Convolutional Network, fortified with a spectral attention mechanism. This network utilizes 3D dilated convolutions to apprehend spatial and spectral features across various scales. Moreover, spectral attention is employed to enhance the discriminative capabilities of high-spectral images (HSIs) [
30]. Adding to the body of work on attention mechanisms, Li et al. presented the Dual-Branch Double-Attention Mechanism Network (DBDA). This innovative model introduces self-attention in both spectral and spatial dimensions, thus augmenting the capacity of images to articulate nuanced details [
31]. Liu’s team, on the other hand, designed a Modified Dense Attention Network (MDAN) specifically for Hyperspectral Image Classification (HSIC). Their model incorporates a Convolutional Block Self-Attention Module (CBSM), which is an adaptation of the Convolutional Block Attention Module (CBAM), and improves classification performance by refining the connection patterns among attention blocks [
32].
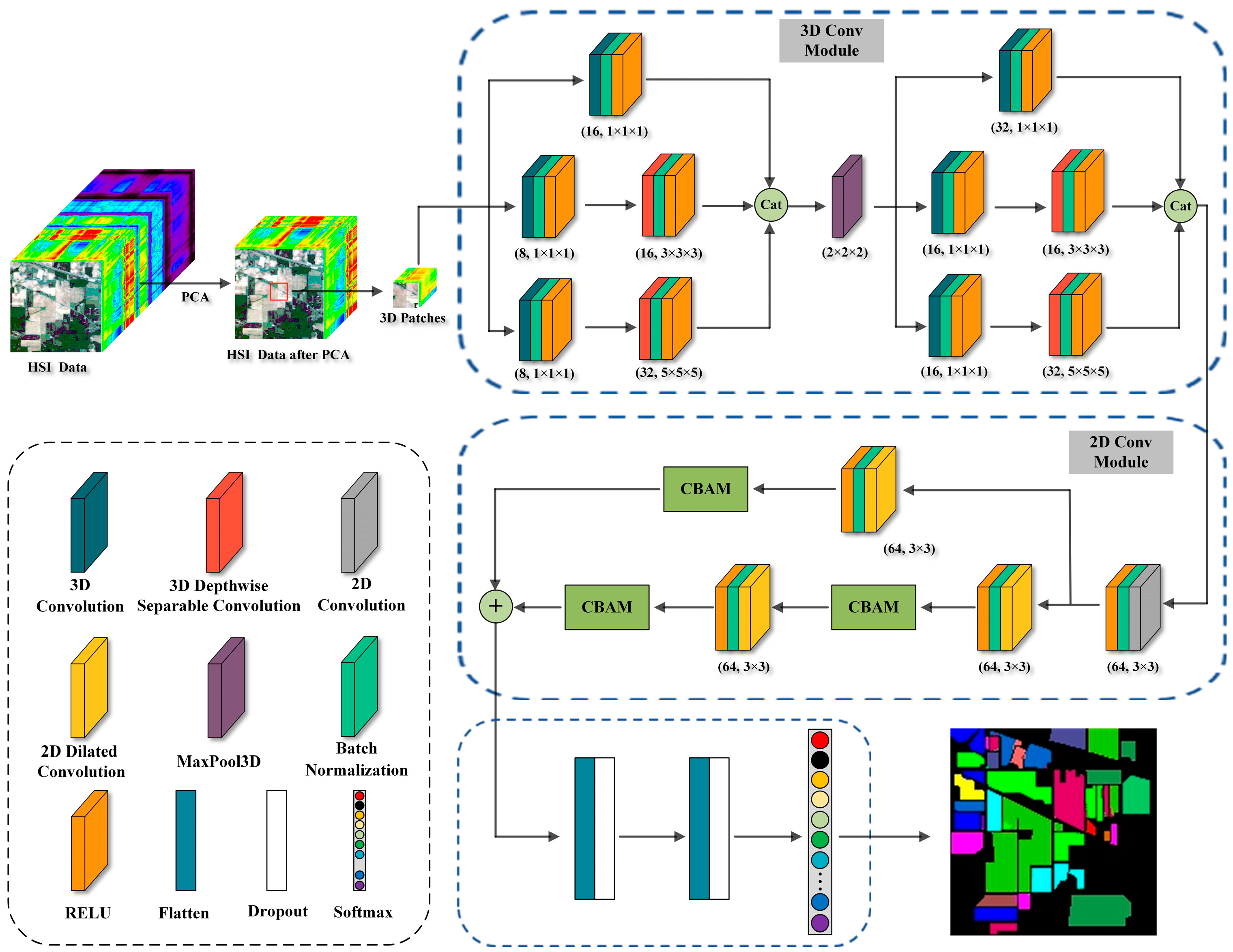
The existing literature underscores that while 3D convolution excels in spatial–spectral feature extraction, it induces significant computational overhead. Further, the integration of multi-scale features and attention mechanisms markedly enhances classification model performance. Given these insights, this study presents an end-to-end hybrid convolutional network, comprising both 2D-CNN and 3D-CNN modules, tailored for Hyperspectral Image Classification (HSIC). Within this framework, the 3D-CNN modules adeptly discern spatial–spectral features across diverse scales, and the 2D-CNN modules accentuate spatial characteristics.
The paper makes the following contributions:
We employ a multi-scale convolutional fusion technique within the 3D-CNN module, enhancing the comprehensive extraction of hyperspectral image features in both the spatial and spectral domains.
Replacing standard 3D convolutional layers with 3D depth separable convolutional layers, we optimize training efficiency and mitigate the risk of overfitting, while maintaining high classification accuracy.
Incorporating the Convolutional Block Attention Module (CBAM) within the residual network architecture, we substitute standard 2D convolutional layers with dilated versions, thus expanding the receptive field and boosting feature extraction for ground object identification.
The subsequent sections of this manuscript are structured in the following manner:
Section 2 explores the intricacies of the MDRDNet’s design and its associated features; in
Section 3, we shed light on the results of the experiments conducted; in
Section 4, we engage in a discussion regarding their implications; and finally,
Section 5 serves to wrap up the findings and conclusion of this paper.