Mapping Large-Scale Bamboo Forest Based on Phenology and Morphology Features
Abstract
:1. Introduction
2. Materials and Methods
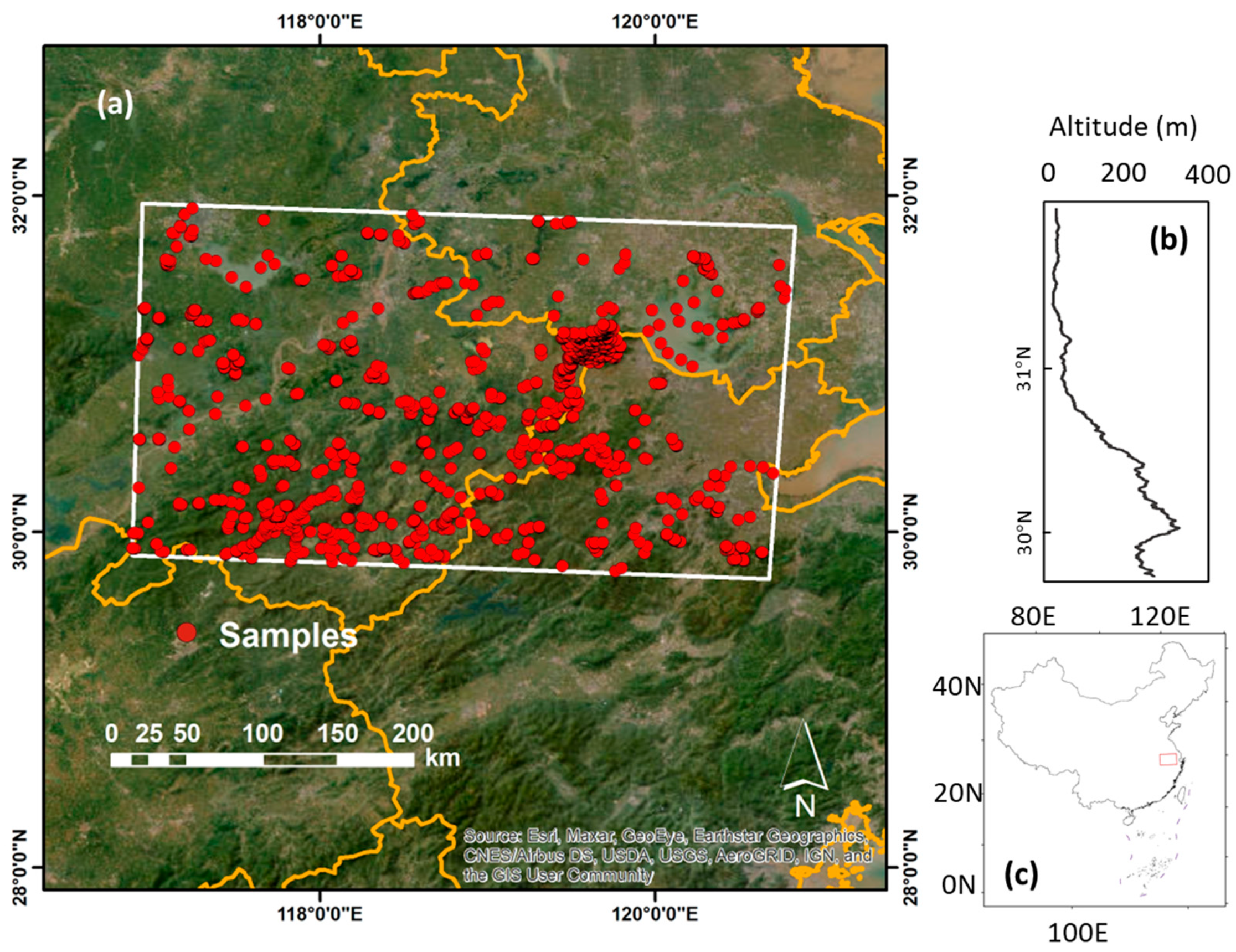
2.1. Study Area
2.2. Data
2.2.1. Field Samples
2.2.2. Remote Sensing Data
2.2.3. Statistical Data
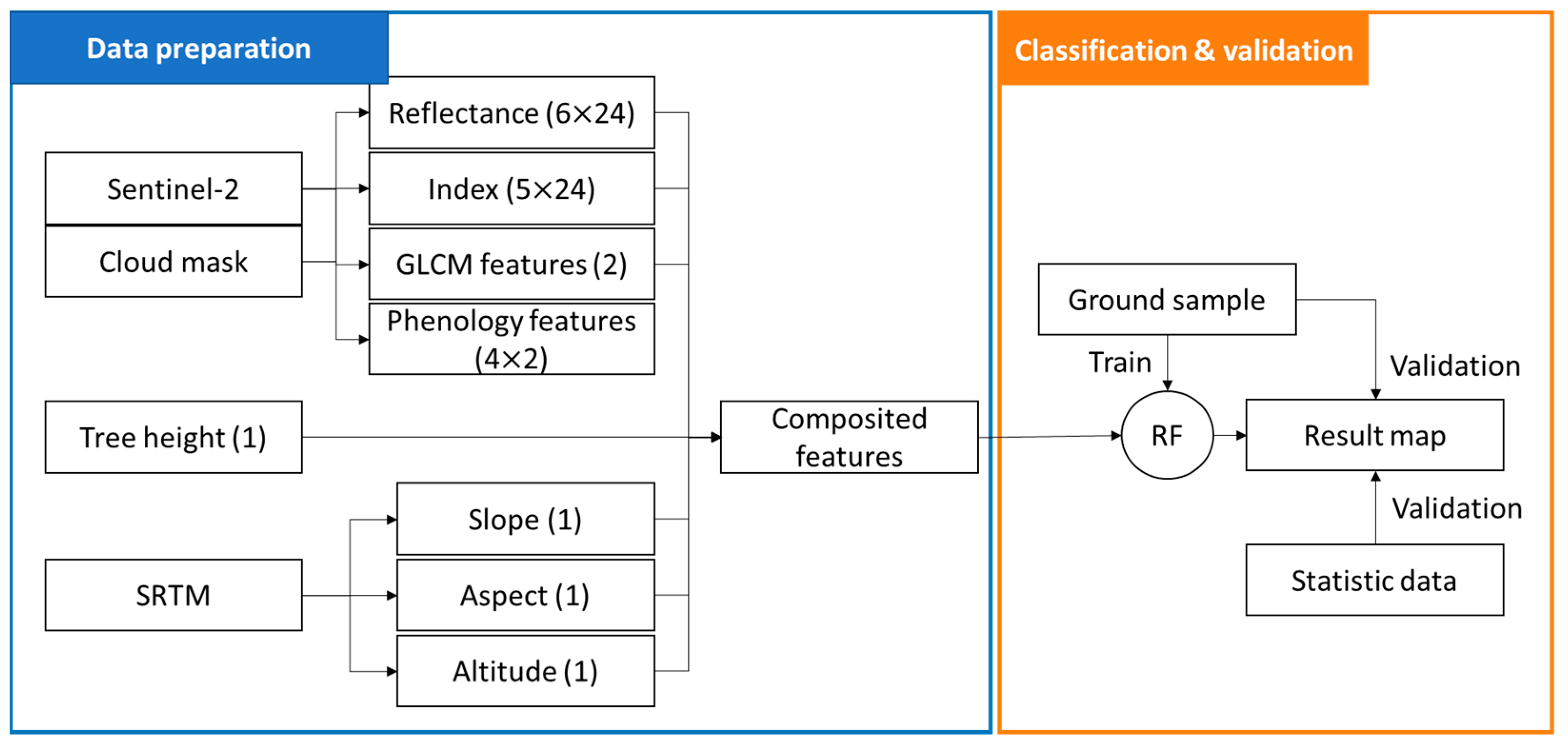
2.3. Workflow
3. Results
3.1. Patterns for Time-Series and Static Features
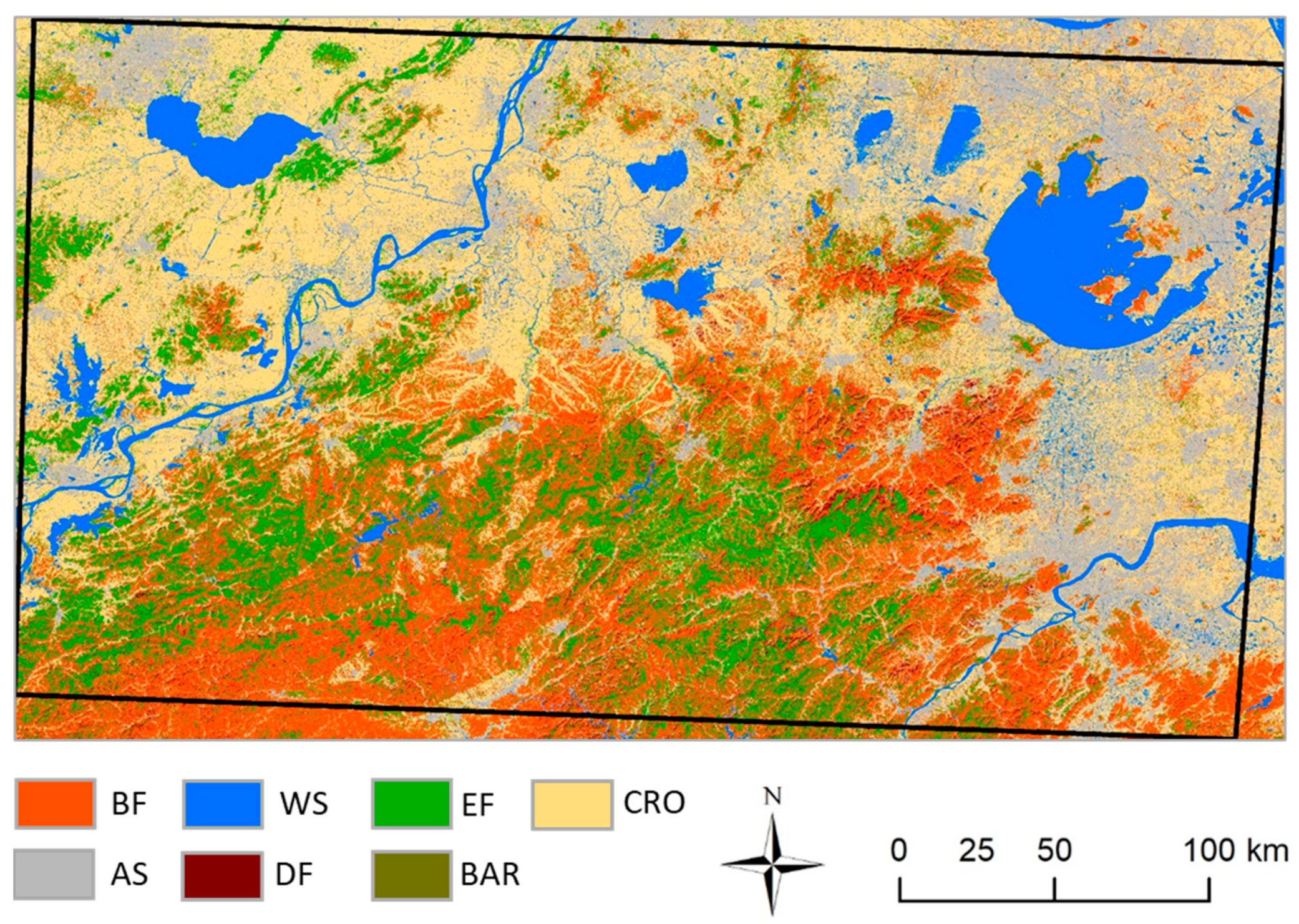
3.2. Classification Results
3.3. Feature Contribution
4. Discussion
4.1. Success of Mapping Bamboo Forests Based on Phenology and Morphology Features
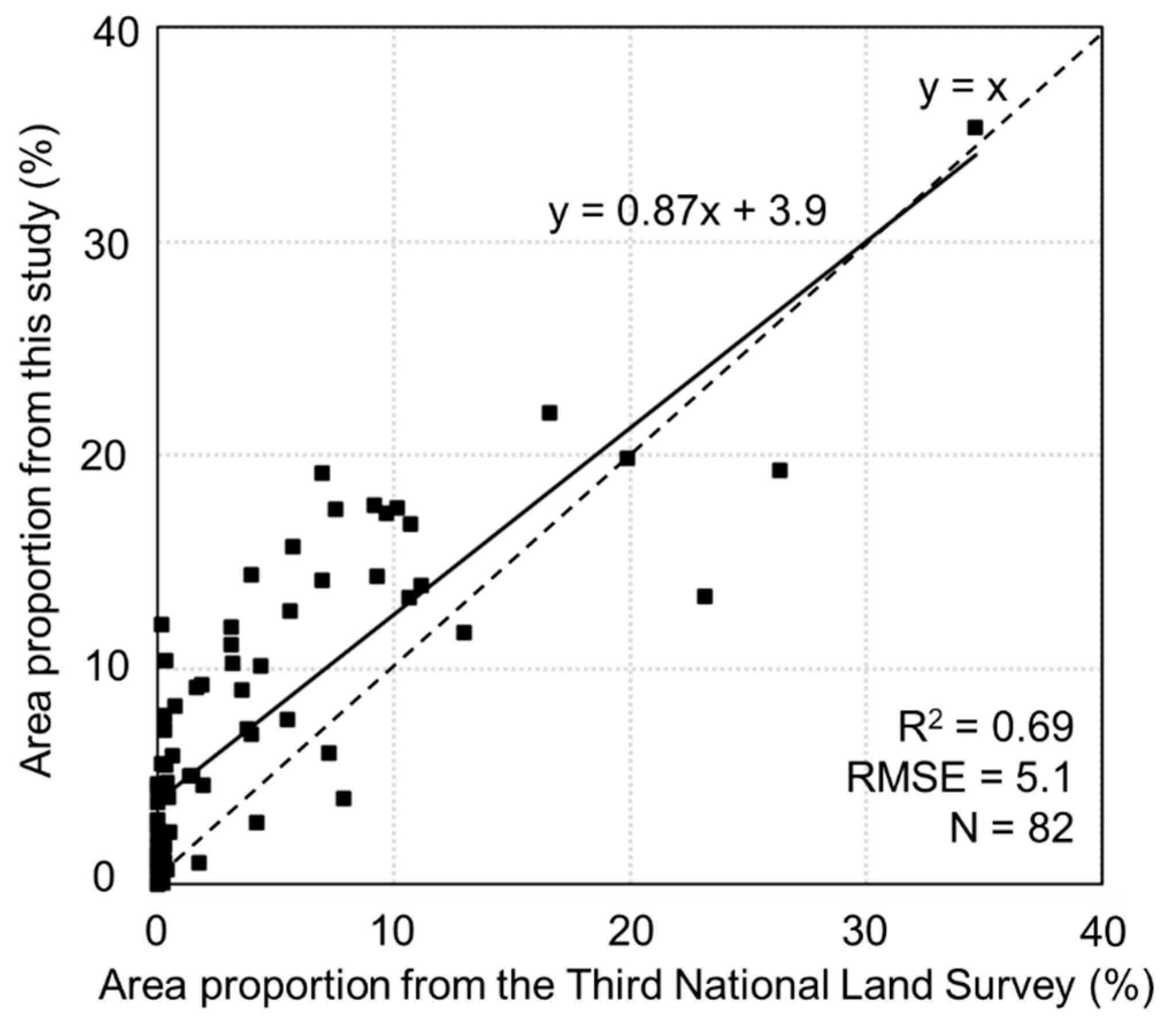
4.2. Mapping Bamboo at a Larger Scale
4.3. The Optimal Time Window for Bamboo Classification
5. Conclusions
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Qi, S.; Song, B.; Liu, C.; Gong, P.; Luo, J.; Zhang, M.; Xiong, T. Bamboo Forest Mapping in China Using the Dense Landsat 8 Image Archive and Google Earth Engine. Remote Sens. 2022, 14, 762. [Google Scholar] [CrossRef]
- Jian, J.; Jiang, H.; Zhou, G.; Jiang, Z.; Yu, S.; Peng, S.; Liu, S.; Wang, J. Mapping the vegetation changes in giant panda habitat using Landsat remotely sensed data. Int. J. Remote Sens. 2011, 32, 1339–1356. [Google Scholar] [CrossRef]
- Manandhar, R.; Kim, J.-H.; Kim, J.-T. Environmental, social and economic sustainability of bamboo and bamboo-based construction materials in buildings. J. Asian Archit. Build. Eng. 2019, 18, 49–59. [Google Scholar] [CrossRef] [Green Version]
- Ben-Zhi, Z.; Mao-Yi, F.; Jin-Zhong, X.; Xiao-Sheng, Y.; Zheng-Cai, L. Ecological functions of bamboo forest: Research and application. J. For. Res. 2005, 16, 143–147. [Google Scholar] [CrossRef]
- Zhou, G.; Meng, C.; Jiang, P.; Xu, Q. Review of carbon fixation in bamboo forests in China. Bot. Rev. 2011, 77, 262–270. [Google Scholar] [CrossRef]
- Liu, Z.; Deng, Z.; He, G.; Wang, H.; Zhang, X.; Lin, J.; Qi, Y.; Liang, X. Challenges and opportunities for carbon neutrality in China. Nat. Rev. Earth Environ. 2022, 3, 141–155. [Google Scholar] [CrossRef]
- Churkina, G.; Organschi, A.; Reyer, C.P.; Ruff, A.; Vinke, K.; Liu, Z.; Reck, B.K.; Graedel, T.; Schellnhuber, H.J. Buildings as a global carbon sink. Nat. Sustain. 2020, 3, 269–276. [Google Scholar] [CrossRef]
- Dong, J.; Kuang, W.; Liu, J. Continuous land cover change monitoring in the remote sensing big data era. Sci. China Earth Sci. 2017, 60, 2223–2224. [Google Scholar] [CrossRef]
- Sonobe, R.; Yamaya, Y.; Tani, H.; Wang, X.; Kobayashi, N.; Mochizuki, K.-I. Crop classification from Sentinel-2-derived vegetation indices using ensemble learning. J. Appl. Remote Sens. 2018, 12, 026019. [Google Scholar] [CrossRef] [Green Version]
- Zhang, X.; Wu, B.; Ponce-Campos, G.E.; Zhang, M.; Chang, S.; Tian, F. Mapping up-to-date paddy rice extent at 10 m resolution in china through the integration of optical and synthetic aperture radar images. Remote Sens. 2018, 10, 1200. [Google Scholar] [CrossRef]
- Yeom, J.; Jung, J.; Chang, A.; Ashapure, A.; Maeda, M.; Maeda, A.; Landivar, J. Comparison of vegetation indices derived from UAV data for differentiation of tillage effects in agriculture. Remote Sens. 2019, 11, 1548. [Google Scholar] [CrossRef] [Green Version]
- Xiao, X.; Boles, S.; Liu, J.; Zhuang, D.; Frolking, S.; Li, C.; Salas, W.; Moore III, B. Mapping paddy rice agriculture in southern China using multi-temporal MODIS images. Remote Sens. Environ. 2005, 95, 480–492. [Google Scholar] [CrossRef]
- Campos, J.C.; Sillero, N.; Brito, J.C. Normalized difference water indexes have dissimilar performances in detecting seasonal and permanent water in the Sahara–Sahel transition zone. J. Hydrol. 2012, 464, 438–446. [Google Scholar] [CrossRef]
- Tian, F.; Wu, B.; Zeng, H.; Zhang, X.; Xu, J. Efficient identification of corn cultivation area with multitemporal synthetic aperture radar and optical images in the google earth engine cloud platform. Remote Sens. 2019, 11, 629. [Google Scholar] [CrossRef] [Green Version]
- Belgiu, M.; Csillik, O. Sentinel-2 cropland mapping using pixel-based and object-based time-weighted dynamic time warping analysis. Remote Sens. Environ. 2018, 204, 509–523. [Google Scholar] [CrossRef]
- Vuolo, F.; Neuwirth, M.; Immitzer, M.; Atzberger, C.; Ng, W.-T. How much does multi-temporal Sentinel-2 data improve crop type classification? Int. J. Appl. Earth Obs. Geoinf. 2018, 72, 122–130. [Google Scholar] [CrossRef]
- Chen, Z.; White, L.; Banks, S.; Behnamian, A.; Montpetit, B.; Pasher, J.; Duffe, J.; Bernard, D. Characterizing marsh wetlands in the Great Lakes Basin with C-band InSAR observations. Remote Sens. Environ. 2020, 242, 111750. [Google Scholar] [CrossRef]
- Hill, R.; Wilson, A.; George, M.; Hinsley, S. Mapping tree species in temperate deciduous woodland using time-series multi-spectral data. Appl. Veg. Sci. 2010, 13, 86–99. [Google Scholar] [CrossRef]
- Gilmore, M.S.; Wilson, E.H.; Barrett, N.; Civco, D.L.; Prisloe, S.; Hurd, J.D.; Chadwick, C. Integrating multi-temporal spectral and structural information to map wetland vegetation in a lower Connecticut River tidal marsh. Remote Sens. Environ. 2008, 112, 4048–4060. [Google Scholar] [CrossRef]
- Madonsela, S.; Cho, M.A.; Mathieu, R.; Mutanga, O.; Ramoelo, A.; Kaszta, Ż.; Van De Kerchove, R.; Wolff, E. Multi-phenology WorldView-2 imagery improves remote sensing of savannah tree species. Int. J. Appl. Earth Obs. Geoinf. 2017, 58, 65–73. [Google Scholar] [CrossRef]
- Liu, H.; Gong, P.; Wang, J.; Clinton, N.; Bai, Y.; Liang, S. Annual dynamics of global land cover and its long-term changes from 1982 to 2015. Earth Syst. Sci. Data 2020, 12, 1217–1243. [Google Scholar] [CrossRef]
- Petitjean, F.; Inglada, J.; Gançarski, P. Satellite image time series analysis under time warping. IEEE Trans. Geosci. Remote Sens. 2012, 50, 3081–3095. [Google Scholar] [CrossRef]
- Hansen, M.C.; Potapov, P.V.; Moore, R.; Hancher, M.; Turubanova, S.A.; Tyukavina, A.; Thau, D.; Stehman, S.V.; Goetz, S.J.; Loveland, T.R. High-resolution global maps of 21st-century forest cover change. Science 2013, 342, 850–853. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Sun, X.; Han, N.; Ge, H.; Gu, C. Multi-scale segmentation, object-based extraction of moso bamboo forest from SPOT5 imagery. Sci. Silvae Sin. 2013, 49, 80–87. [Google Scholar]
- Gorelick, N.; Hancher, M.; Dixon, M.; Ilyushchenko, S.; Thau, D.; Moore, R. Google Earth Engine: Planetary-scale geospatial analysis for everyone. Remote Sens. Environ. 2017, 202, 18–27. [Google Scholar] [CrossRef]
- Liu, C.; Xiong, T.; Gong, P.; Qi, S. Improving large-scale moso bamboo mapping based on dense Landsat time series and auxiliary data: A case study in Fujian Province, China. Remote Sens. Lett. 2018, 9, 1–10. [Google Scholar] [CrossRef]
- Rußwurm, M.; Wang, S.; Korner, M.; Lobell, D. Meta-learning for few-shot land cover classification. In Proceedings of the IEEE/CVF Conference on Computer Vision and Pattern Recognition Workshops, Virtual, 14–19 June 2020; pp. 200–201. [Google Scholar]
- Li, L.; Li, N.; Lu, D.; Chen, Y. Mapping Moso bamboo forest and its on-year and off-year distribution in a subtropical region using time-series Sentinel-2 and Landsat 8 data. Remote Sens. Environ. 2019, 231, 111265. [Google Scholar] [CrossRef]
- Pelletier, C.; Webb, G.I.; Petitjean, F. Temporal convolutional neural network for the classification of satellite image time series. Remote Sens. 2019, 11, 523. [Google Scholar] [CrossRef] [Green Version]
- Michałowska, M.; Rapiński, J. A review of tree species classification based on airborne LiDAR data and applied classifiers. Remote Sens. 2021, 13, 353. [Google Scholar] [CrossRef]
- Naidoo, L.; Cho, M.A.; Mathieu, R.; Asner, G. Classification of savanna tree species, in the Greater Kruger National Park region, by integrating hyperspectral and LiDAR data in a Random Forest data mining environment. ISPRS J. Photogramm. Remote Sens. 2012, 69, 167–179. [Google Scholar] [CrossRef]
- Potapov, P.; Li, X.; Hernandez-Serna, A.; Tyukavina, A.; Hansen, M.C.; Kommareddy, A.; Pickens, A.; Turubanova, S.; Tang, H.; Silva, C.E. Mapping global forest canopy height through integration of GEDI and Landsat data. Remote Sens. Environ. 2021, 253, 112165. [Google Scholar] [CrossRef]
- Liu, X.; Su, Y.; Hu, T.; Yang, Q.; Liu, B.; Deng, Y.; Tang, H.; Tang, Z.; Fang, J.; Guo, Q. Neural network guided interpolation for mapping canopy height of China’s forests by integrating GEDI and ICESat-2 data. Remote Sens. Environ. 2022, 269, 112844. [Google Scholar] [CrossRef]
- Li, M.; Li, C.; Jiang, H.; Fang, C.; Yang, J.; Zhu, Z.; Shi, L.; Liu, S.; Gong, P. Tracking bamboo dynamics in Zhejiang, China, using time-series of Landsat data from 1990 to 2014. Int. J. Remote Sens. 2016, 37, 1714–1729. [Google Scholar] [CrossRef]
- Gong, P.; Li, X.; Wang, J.; Bai, Y.; Chen, B.; Hu, T.; Liu, X.; Xu, B.; Yang, J.; Zhang, W. Annual maps of global artificial impervious area (GAIA) between 1985 and 2018. Remote Sens. Environ. 2020, 236, 111510. [Google Scholar] [CrossRef]
- Li, P.; Zhou, G.; Du, H.; Lu, D.; Mo, L.; Xu, X.; Shi, Y.; Zhou, Y. Current and potential carbon stocks in Moso bamboo forests in China. J. Environ. Manag. 2015, 156, 89–96. [Google Scholar] [CrossRef]
- Jarvis, A.; Reuter, H.I.; Nelson, A.; Guevara, E. Hole-Filled SRTM for the Globe Version 4. CGIAR-CSI SRTM 90m Database. Available online: http://srtm.csi.cgiar.org (accessed on 12 January 2023).
- Skakun, S.; Wevers, J.; Brockmann, C.; Doxani, G.; Aleksandrov, M.; Batič, M.; Frantz, D.; Gascon, F.; Gómez-Chova, L.; Hagolle, O. Cloud Mask Intercomparison eXercise (CMIX): An evaluation of cloud masking algorithms for Landsat 8 and Sentinel-2. Remote Sens. Environ. 2022, 274, 112990. [Google Scholar] [CrossRef]
- Huete, A.; Didan, K.; Miura, T.; Rodriguez, E.P.; Gao, X.; Ferreira, L.G. Overview of the radiometric and biophysical performance of the MODIS vegetation indices. Remote Sens. Environ. 2002, 83, 195–213. [Google Scholar] [CrossRef]
- Gitelson, A.A.; Viña, A.; Arkebauer, T.J.; Rundquist, D.C.; Keydan, G.; Leavitt, B. Remote estimation of leaf area index and green leaf biomass in maize canopies. Geophys. Res. Lett. 2003, 30. [Google Scholar] [CrossRef] [Green Version]
- Dash, J.; Curran, P. The MERIS terrestrial chlorophyll index. Int. J. Remote Sens. 2004, 25, 5403–5413. [Google Scholar] [CrossRef]
- Wang, J.; Xiao, X.; Qin, Y.; Dong, J.; Geissler, G.; Zhang, G.; Cejda, N.; Alikhani, B.; Doughty, R.B. Mapping the dynamics of eastern redcedar encroachment into grasslands during 1984–2010 through PALSAR and time series Landsat images. Remote Sens. Environ. 2017, 190, 233–246. [Google Scholar] [CrossRef] [Green Version]
- Lu, D.; Li, G.; Moran, E.; Dutra, L.; Batistella, M. The roles of textural images in improving land-cover classification in the Brazilian Amazon. Int. J. Remote Sens. 2014, 35, 8188–8207. [Google Scholar] [CrossRef]
- Zhang, M.; Gong, P.; Qi, S.; Liu, C.; Xiong, T. Mapping bamboo with regional phenological characteristics derived from dense Landsat time series using Google Earth Engine. Int. J. Remote Sens. 2019, 40, 9541–9555. [Google Scholar] [CrossRef]
- Zhang, M.; Keenan, T.F.; Luo, X.; Serra-Diaz, J.M.; Li, W.; King, T.; Cheng, Q.; Li, Z.; Andriamiarisoa, R.L.; Raherivelo, T.N.A.N. Elevated CO2 moderates the impact of climate change on future bamboo distribution in Madagascar. Sci. Total Environ. 2022, 810, 152235. [Google Scholar] [CrossRef] [PubMed]
- Kovacs, J.M.; Jiao, X.; Flores-de-Santiago, F.; Zhang, C.; Flores-Verdugo, F. Assessing relationships between Radarsat-2 C-band and structural parameters of a degraded mangrove forest. Int. J. Remote Sens. 2013, 34, 7002–7019. [Google Scholar] [CrossRef]
- Touzi, R.; Deschamps, A.; Rother, G. Phase of target scattering for wetland characterization using polarimetric C-band SAR. IEEE Trans. Geosci. Remote Sens. 2009, 47, 3241–3261. [Google Scholar] [CrossRef]
- Tan, S.; Zhang, X.; Wang, H.; Yu, L.; Du, Y.; Yin, J.; Wu, B. A CNN-Based Self-Supervised Synthetic Aperture Radar Image Denoising Approach. IEEE Trans. Geosci. Remote Sens. 2021, 60, 1–15. [Google Scholar] [CrossRef]






| Product | Variable | Time Interval | Website |
|---|---|---|---|
| Sentinel-2 Level 2A | RS reflectance | Monthly | https://earth.esa.int/web/sentinel/user-guides/sentinel-2-msi/product-types/level-2a (accessed on 12 January 2023) |
| Vegetation indices | Monthly | ||
| Phenology | Yearly | ||
| Morphology | Static | ||
| SRTM | Elevation | Static | https://srtm.csi.cgiar.org/ (accessed on 12 January 2023) |
| Slope | |||
| Aspect | |||
| NNGI | Tree height | Static | http://www.3decology.org/dataset-software/ (accessed on 12 January 2023) |
| Index | Equation |
|---|---|
| NDVI | |
| EVI | |
| GCVI | |
| MTCI | |
| LSWI |
| Predicted Label | |||||||||
|---|---|---|---|---|---|---|---|---|---|
| BF | DF | EF | CRO | AS | BAR | WS | UA | ||
| Field samples | BF | 65 | 4 | 2 | 0 | 0 | 0 | 0 | 0.92 |
| DF | 4 | 32 | 1 | 0 | 0 | 0 | 0 | 0.86 | |
| EF | 3 | 3 | 66 | 0 | 0 | 0 | 0 | 0.92 | |
| CRO | 0 | 1 | 0 | 63 | 2 | 0 | 0 | 0.95 | |
| AS | 0 | 0 | 2 | 4 | 20 | 2 | 0 | 0.71 | |
| BAR | 0 | 0 | 0 | 0 | 5 | 7 | 0 | 0.58 | |
| WS | 0 | 0 | 0 | 1 | 0 | 0 | 10 | 0.91 | |
| PA | 0.90 | 0.80 | 0.93 | 0.93 | 0.74 | 0.78 | 1.00 | OA = 0.89 | |
| F1-score | 0.91 | 0.83 | 0.92 | 0.94 | 0.73 | 0.67 | 0.95 | ||
| Window | Item | BF | DF | EF | CRO | AS | BAR | WT | OA |
|---|---|---|---|---|---|---|---|---|---|
| 1-month | Mean | 0.53 | 0.54 | 0.67 | 0.51 | 0.57 | 0.53 | 0.84 | 0.57 |
| Std | 0.15 | 0.12 | 0.16 | 0.2 | 0.13 | 0.16 | 0.71 | 0.13 | |
| 4-month | Mean | 0.77 | 0.77 | 0.81 | 0.8 | 0.77 | 0.72 | 0.91 | 0.77 |
| Std | 0.09 | 0.15 | 0.04 | 0.07 | 0.05 | 0.07 | 0.01 | 0.03 | |
| 6-month | Mean | 0.81 | 0.86 | 0.88 | 0.81 | 0.8 | 0.73 | 0.85 | 0.86 |
| Std | 0.04 | 0.07 | 0.02 | 0.03 | 0.03 | 0.09 | 0.02 | 0.02 | |
| 12-month | Mean | 0.86 | 0.91 | 0.92 | 0.88 | 0.81 | 0.72 | 0.90 | 0.87 |
| Std | 0.02 | 0 | 0.01 | 0.01 | 0.02 | 0.07 | 0.02 | 0.03 | |
| 24-month | Mean | 0.91 | 0.9 | 0.92 | 0.90 | 0.73 | 0.67 | 0.92 | 0.88 |
| Std | - | - | - | - | - | - | - | - |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2023 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Feng, X.; Tan, S.; Dong, Y.; Zhang, X.; Xu, J.; Zhong, L.; Yu, L. Mapping Large-Scale Bamboo Forest Based on Phenology and Morphology Features. Remote Sens. 2023, 15, 515. https://doi.org/10.3390/rs15020515
Feng X, Tan S, Dong Y, Zhang X, Xu J, Zhong L, Yu L. Mapping Large-Scale Bamboo Forest Based on Phenology and Morphology Features. Remote Sensing. 2023; 15(2):515. https://doi.org/10.3390/rs15020515
Chicago/Turabian StyleFeng, Xueliang, Shen Tan, Yun Dong, Xin Zhang, Jiaming Xu, Liheng Zhong, and Le Yu. 2023. "Mapping Large-Scale Bamboo Forest Based on Phenology and Morphology Features" Remote Sensing 15, no. 2: 515. https://doi.org/10.3390/rs15020515
APA StyleFeng, X., Tan, S., Dong, Y., Zhang, X., Xu, J., Zhong, L., & Yu, L. (2023). Mapping Large-Scale Bamboo Forest Based on Phenology and Morphology Features. Remote Sensing, 15(2), 515. https://doi.org/10.3390/rs15020515










