Enhancing Machine Learning Performance in Estimating CDOM Absorption Coefficient via Data Resampling
Abstract
:1. Introduction
2. Materials and Methods
2.1. Site Description and Data Acquisition
2.1.1. Study Area
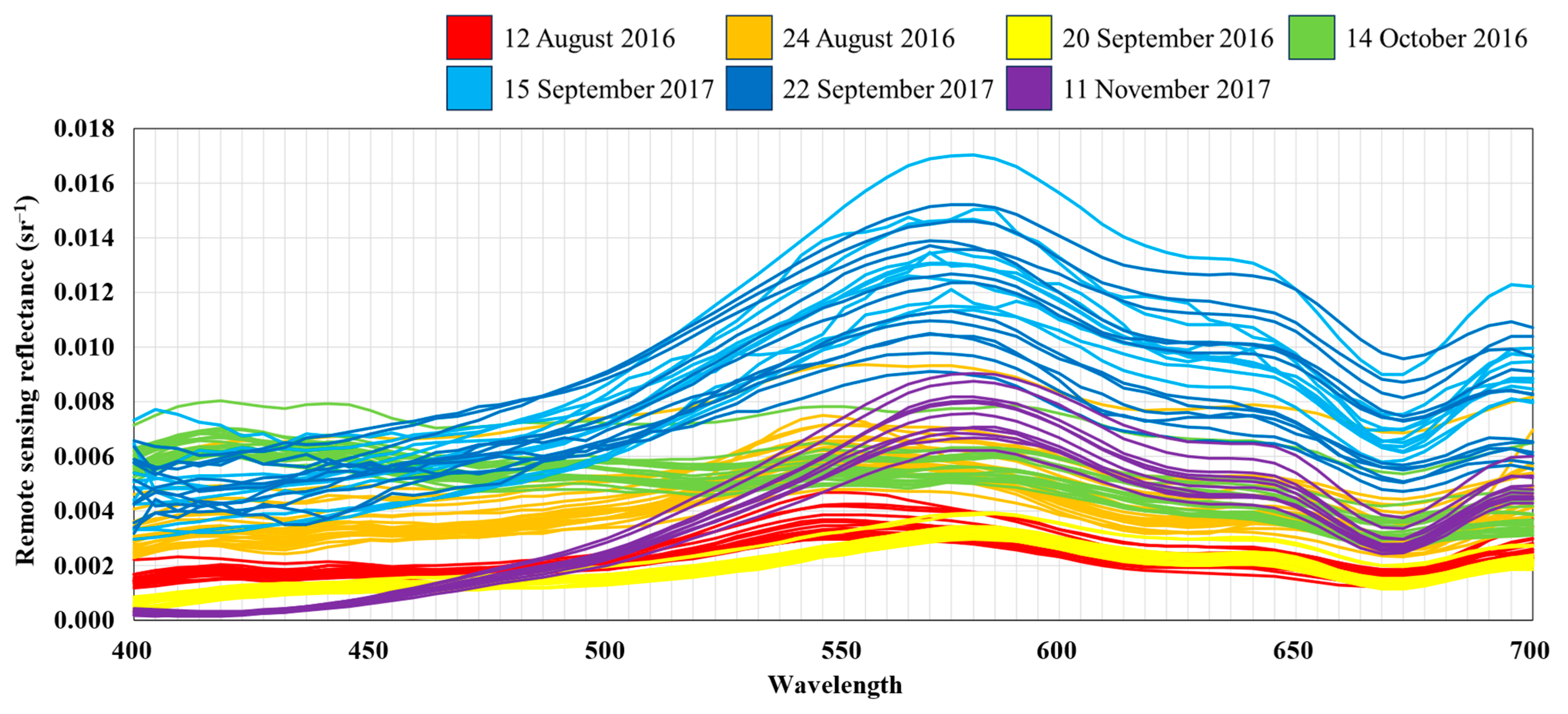
2.1.2. In Situ Reflectance Measurements and Airborne Hyperspectral Image
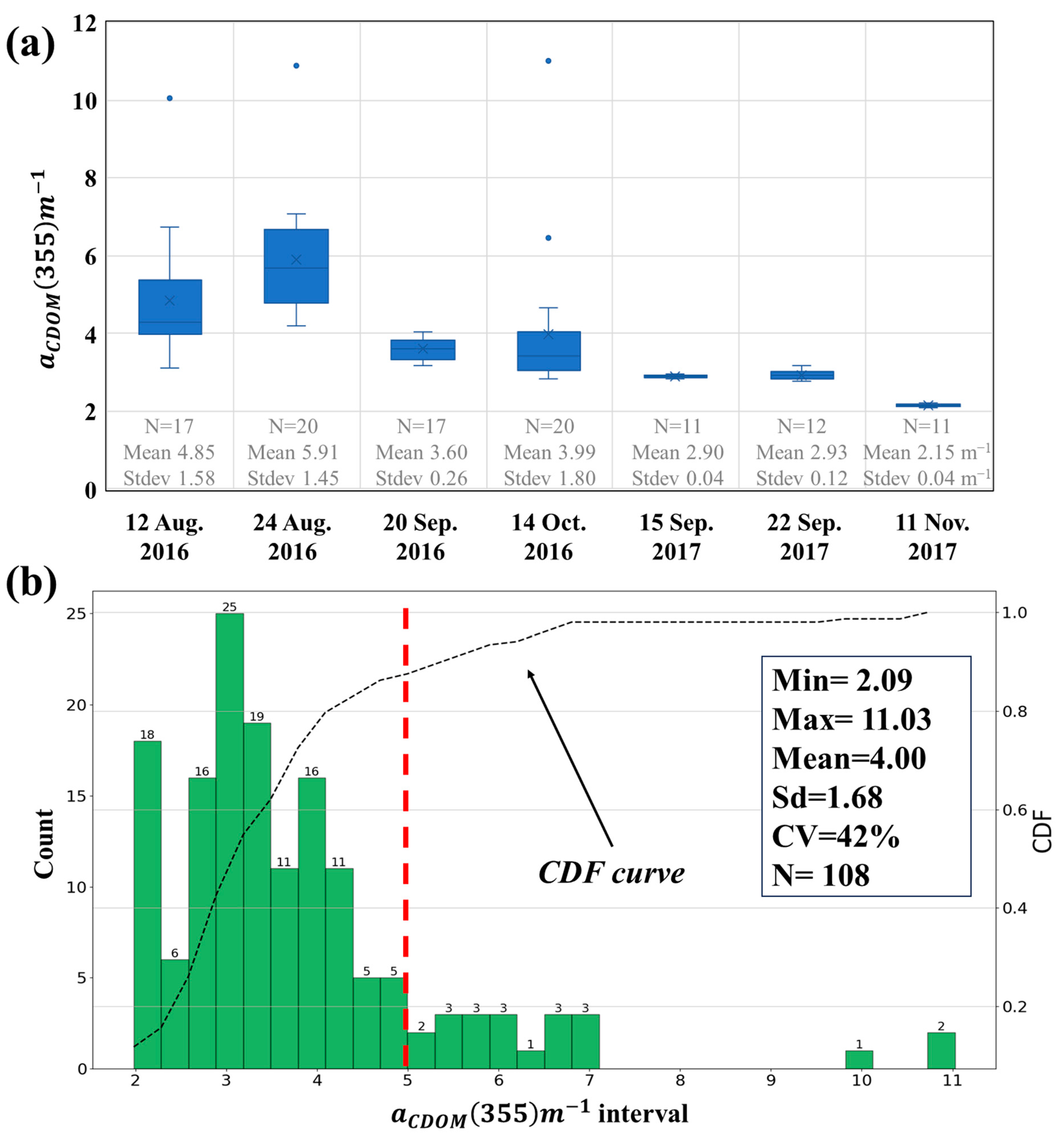
2.1.3. CDOM Absorption Coefficient
2.2. Feature Selection and Data Resampling Method
2.2.1. Feature Selection
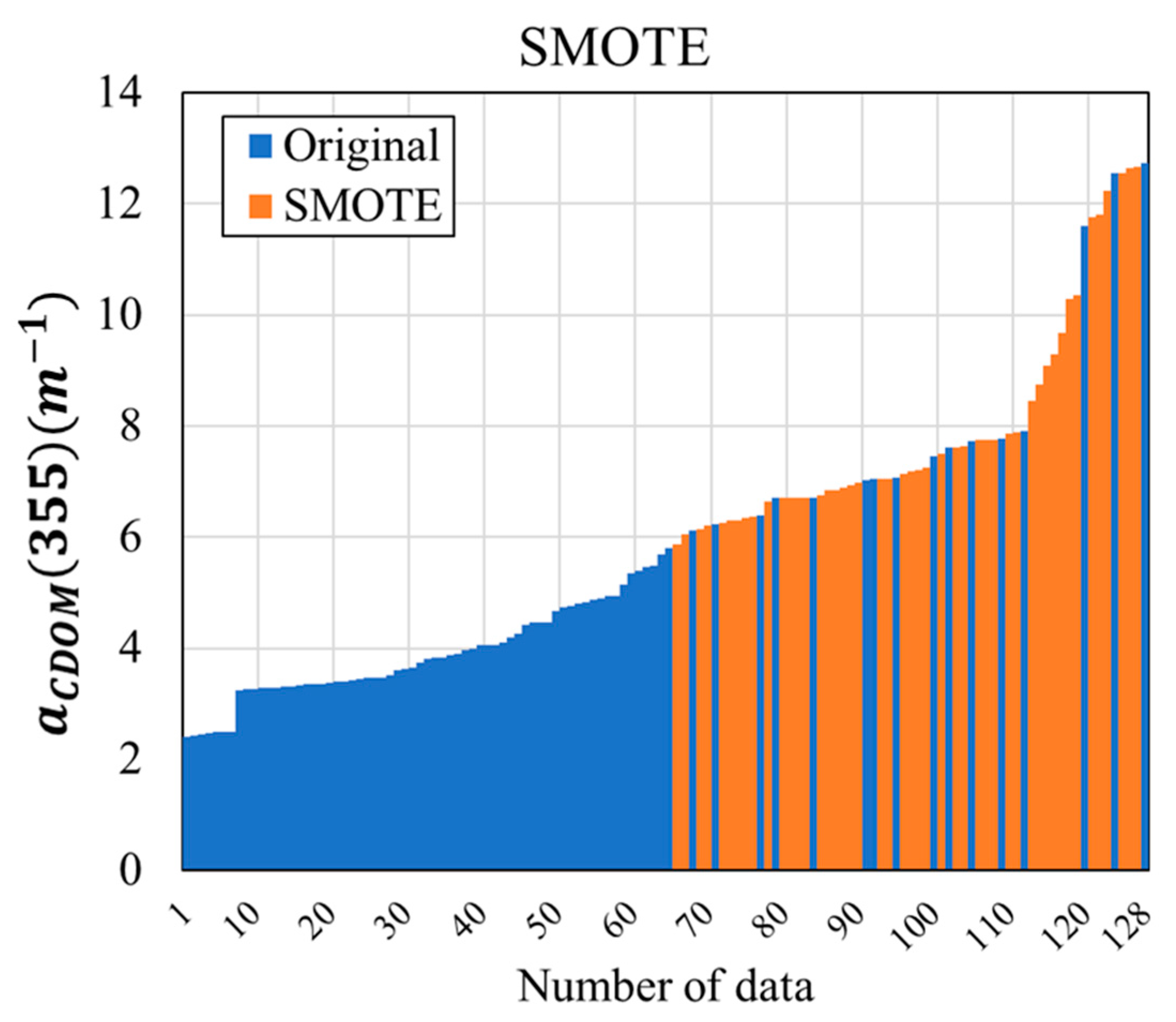
2.2.2. Data Resampling Method
2.3. Construction of Machine Learning Models and Evaluation of Model Performance
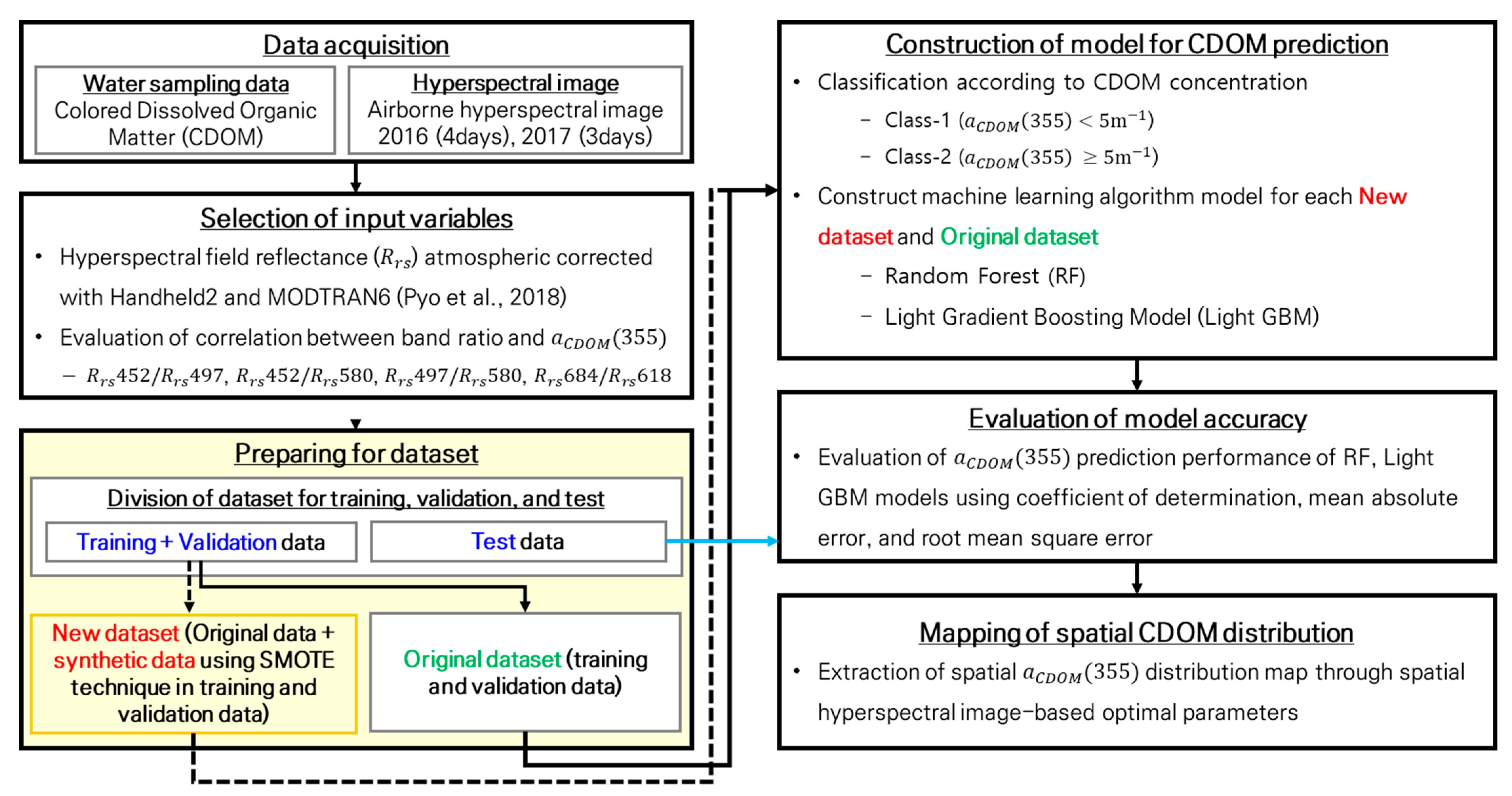
2.3.1. Model Process
2.3.2. Random Forest Algorithm
2.3.3. Light Gradient Boost Machine (Light GBM)
2.3.4. Model Accuracy
3. Results
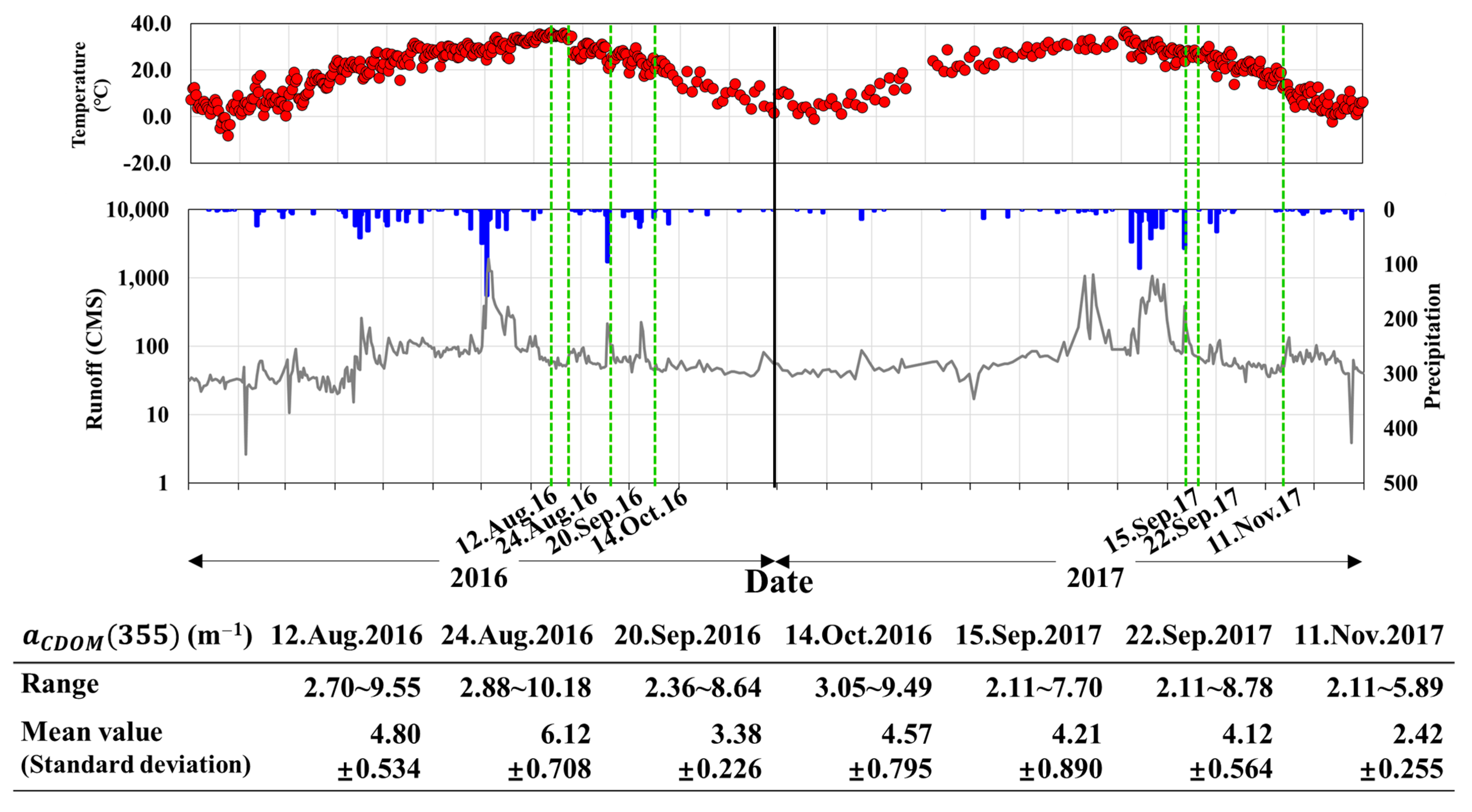
3.1. Descriptive Analysis of Chromophoric Dissolved Organic Matter (CDOM) in Reservoirs
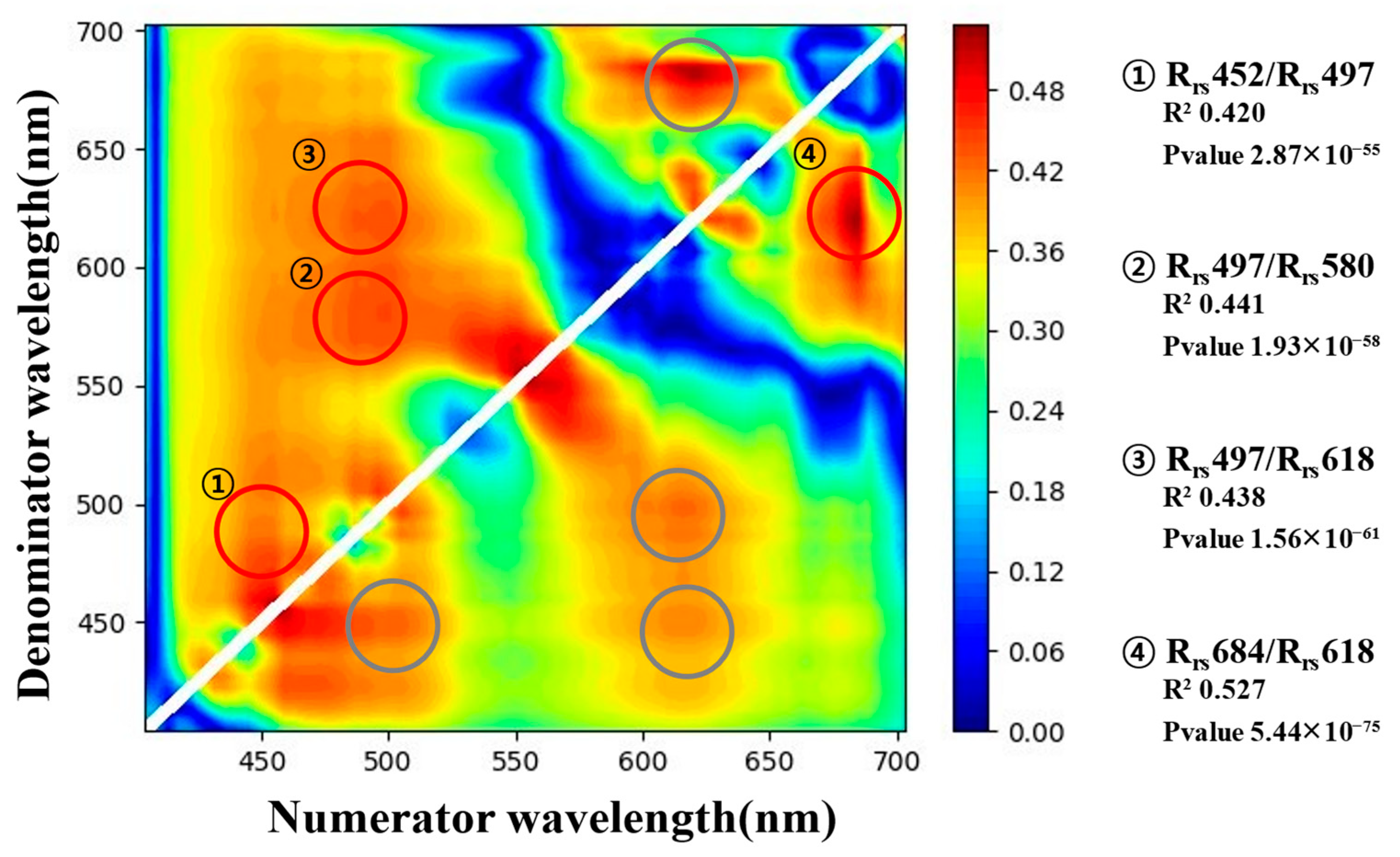
3.2. Results of Feature Selection
3.3. Comparison of Machine Learning Model Performance
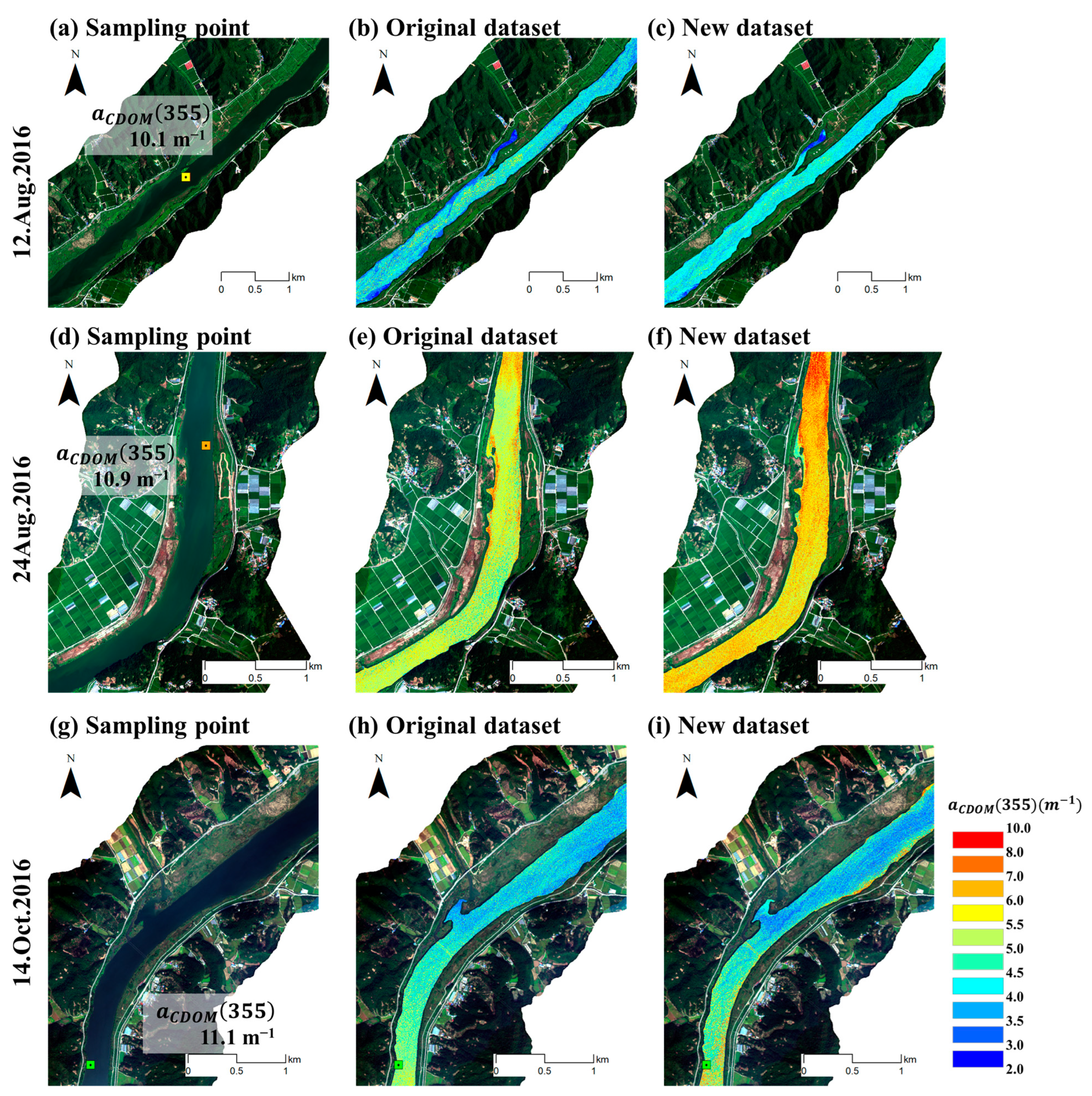
3.4. Analysis of CDOM High-Concentration Distribution Area
4. Discussion
4.1. Selection of Input Variables
4.2. Evaluation of Machine Learning Models and Application of Data Resampling
4.3. Spatial Distribution Results
5. Conclusions
- The selected input values that considered the overlap in the reflectance ratio R2 heatmap of the hyperspectral images were , , , and with R2 values of 0.420, 0.441, 0.438, and 0.527, respectively. The machine learning models were constructed using the four input variables with significant p-values.
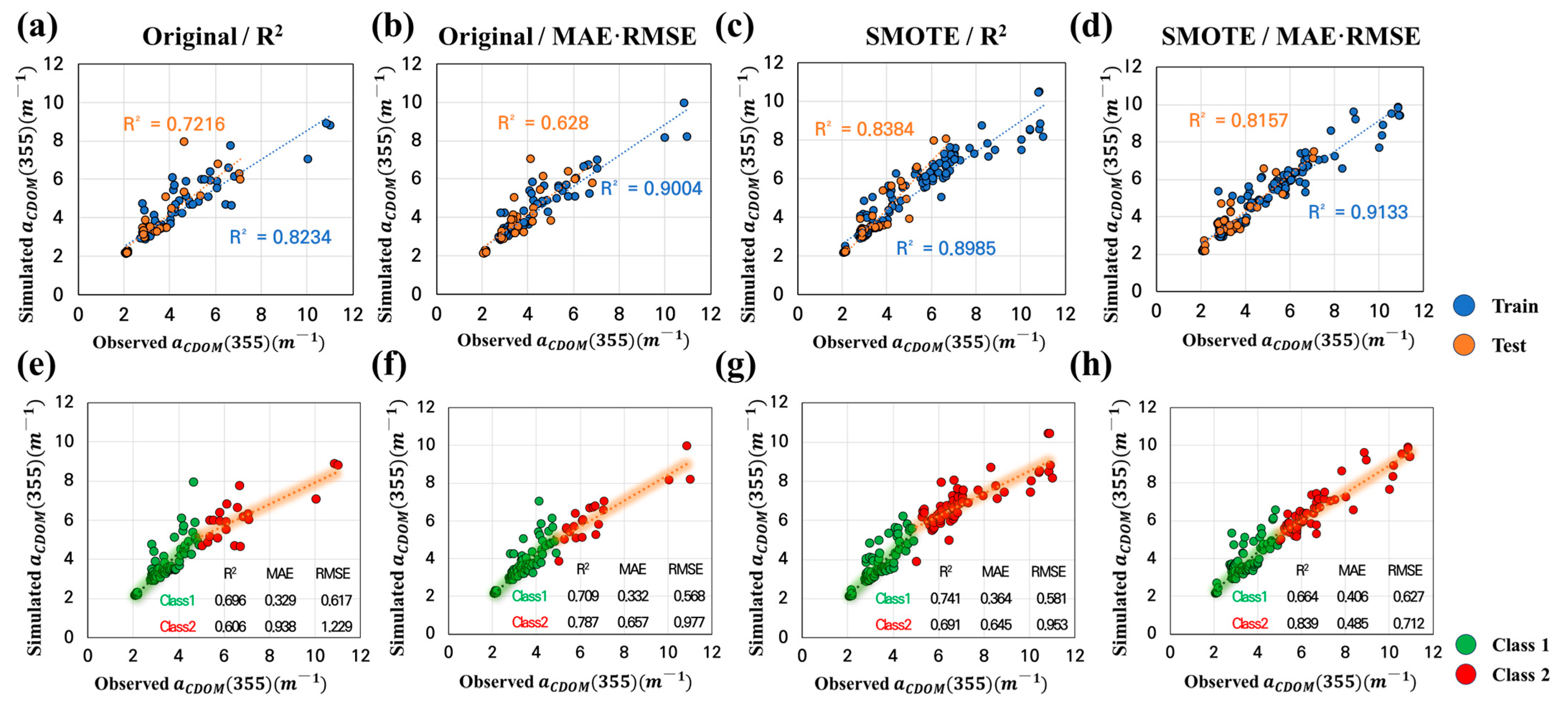
- To solve the imbalance problem, low-concentration (Class 1) and high-concentration (Class 2) sections were separated by 5 m−1 in the small CDOM dataset, and training and testing datasets for each class were extracted. The training data were divided into two subsets: the original dataset, which used only the training data, and the SMOTE dataset, in which SMOTE was applied to the training dataset. The machine learning models were constructed and evaluated for each dataset to compare the CDOM prediction performance of the original and SMOTE datasets.
- Both RF and Light GBM demonstrated considerable performance improvements in the best-case scenario when SMOTE was applied. The R2 values of RF were 0.881 and 0.816 in the training and test steps, whereas the R2 values of Light GBM were 0.993 and 0.772 in the training and test steps. The RF model showed better generalization performance than Light GBM.
- Spatial distribution was performed using the results of this study, and it was confirmed that the SMOTE dataset detected CDOM on high-concentration days more accurately than the original dataset.
Supplementary Materials
Author Contributions
Funding
Data Availability Statement
Acknowledgments
Conflicts of Interest
References
- Kirk, J.T.O. Light and Photosynthesis in Aquatic Ecosystems; Cambridge University Press: Cambridge, UK, 1994; ISBN 9788578110796. [Google Scholar]
- Zhao, Y.; Song, K.; Wen, Z.; Li, L.; Zang, S.; Shao, T.; Li, S.; Du, J. Seasonal Characterization of CDOM for Lakes in Semiarid Regions of Northeast China Using Excitation–Emission Matrix Fluorescence and Parallel Factor Analysis (EEM–PARAFAC). Biogeosciences 2016, 13, 1635–1645. [Google Scholar] [CrossRef]
- Kutser, T.; Pierson, D.C.; Kallio, K.Y.; Reinart, A.; Sobek, S. Mapping Lake CDOM by Satellite Remote Sensing. Remote Sens. Environ. 2005, 94, 535–540. [Google Scholar] [CrossRef]
- Coble, P.G. Marine Optical Biogeochemistry: The Chemistry of Ocean Color. Chem. Rev. 2007, 107, 402–418. [Google Scholar] [CrossRef]
- Ling, Z.; Sun, D.; Wang, S.; Qiu, Z.; Huan, Y.; Mao, Z.; He, Y. Remote Sensing Estimation of Colored Dissolved Organic Matter (CDOM) from GOCI Measurements in the Bohai Sea and Yellow Sea. Environ. Sci. Pollut. Res. 2020, 27, 6872–6885. [Google Scholar] [CrossRef] [PubMed]
- Menken, K.D.; Brezonik, P.L.; Bauer, M.E. Influence of Chlorophyll and Colored Dissolved Organic Matter (CDOM) on Lake Reflectance Spectra: Implications for Measuring Lake Properties by Remote Sensing. Lake Reserv. Manag. 2006, 22, 179–190. [Google Scholar] [CrossRef]
- Brezonik, P.L.; Olmanson, L.G.; Finlay, J.C.; Bauer, M.E. Factors Affecting the Measurement of CDOM by Remote Sensing of Optically Complex Inland Waters. Remote Sens. Environ. 2015, 157, 199–215. [Google Scholar] [CrossRef]
- Griffin, C.G.; Frey, K.E.; Rogan, J.; Holmes, R.M. Spatial and Interannual Variability of Dissolved Organic Matter in the Kolyma River, East Siberia, Observed Using Satellite Imagery. J. Geophys. Res. Biogeosciences 2011, 116, 1–12. [Google Scholar] [CrossRef]
- De Almeida, C.S.; Miccoli, L.S.; Andhini, N.F.; Aranha, S.; de Oliveira, L.C.; Artigo, C.E.; Em, A.A.R.; Em, A.A.R.; Bachman, L.; Chick, K.; et al. Remote Sensing of Ocean Colour in Coastal, and Other Optically-Complex, Waters; International Ocean Colour Coordinating Group (IOCCG): Dartmouth, NS, Canada, 2000; Volume 3. [Google Scholar]
- Zhang, H.; Yao, B.; Wang, S.; Wang, G. Remote Sensing Estimation of the Concentration and Sources of Coloured Dissolved Organic Matter Based on MODIS: A Case Study of Erhai Lake. Ecol. Indic. 2021, 131, 108180. [Google Scholar] [CrossRef]
- Jiang, G.; Ma, R.; Duan, H.; Loiselle, S.A.; Xu, J.; Liu, D. Remote Determination of Chromophoric Dissolved Organic Matter in Lakes, China. Int. J. Digit. Earth 2014, 7, 897–915. [Google Scholar] [CrossRef]
- Zhu, W.; Yu, Q. Inversion of Chromophoric Dissolved Organic Matter from EO-1 Hyperion Imagery for Turbid Estuarine and Coastal Waters. IEEE Trans. Geosci. Remote Sens. 2013, 51, 3286–3298. [Google Scholar] [CrossRef]
- Zhu, W.; Yu, Q.; Tian, Y.Q.; Becker, B.L.; Zheng, T.; Carrick, H.J. An Assessment of Remote Sensing Algorithms for Colored Dissolved Organic Matter in Complex Freshwater Environments. Remote Sens. Environ. 2014, 140, 766–778. [Google Scholar] [CrossRef]
- Ruescas, A.B.; Hieronymi, M.; Mateo-Garcia, G.; Koponen, S.; Kallio, K.; Camps-Valls, G. Machine Learning Regression Approaches for Colored Dissolved Organic Matter (CDOM) Retrieval with S2-MSI and S3-OLCI Simulated Data. Remote Sens. 2018, 10, 786. [Google Scholar] [CrossRef]
- Keller, S.; Maier, P.M.; Riese, F.M.; Norra, S.; Holbach, A.; Börsig, N.; Wilhelms, A.; Moldaenke, C.; Zaake, A.; Hinz, S. Hyperspectral Data and Machine Learning for Estimating CDOM, Chlorophyll a, Diatoms, Green Algae and Turbidity. Int. J. Environ. Res. Public Health 2018, 15, 1881. [Google Scholar] [CrossRef] [PubMed]
- Sun, X.; Zhang, Y.; Zhang, Y.; Shi, K.; Zhou, Y.; Li, N. Machine Learning Algorithms for Chromophoric Dissolved Organic Matter (Cdom) Estimation Based on Landsat 8 Images. Remote Sens. 2021, 13, 3560. [Google Scholar] [CrossRef]
- Chawla, N.V.; Japkowicz, N.; Kotcz, A. Editorial: Special issue on learning from imbalanced data sets. ACM SIGKDD Explor. Newsl. 2004, 6, 1–6. [Google Scholar] [CrossRef]
- Bourel, M.; Segura, A.M.; Crisci, C.; López, G.; Sampognaro, L.; Vidal, V.; Kruk, C.; Piccini, C.; Perera, G. Machine Learning Methods for Imbalanced Data Set for Prediction of Faecal Contamination in Beach Waters. Water Res. 2021, 202, 117450. [Google Scholar] [CrossRef] [PubMed]
- Kim, J.H.; Shin, J.K.; Lee, H.; Lee, D.H.; Kang, J.H.; Cho, K.H.; Lee, Y.G.; Chon, K.; Baek, S.S.; Park, Y. Improving the Performance of Machine Learning Models for Early Warning of Harmful Algal Blooms Using an Adaptive Synthetic Sampling Method. Water Res. 2021, 207, 117821. [Google Scholar] [CrossRef] [PubMed]
- Pyo, J.C.; Ligaray, M.; Kwon, Y.S.; Ahn, M.H.; Kim, K.; Lee, H.; Kang, T.; Cho, S.B.; Park, Y.; Cho, K.H. High-Spatial Resolution Monitoring of Phycocyanin and Chlorophyll-a Using Airborne Hyperspectral Imagery. Remote Sens. 2018, 10, 1180. [Google Scholar] [CrossRef]
- Bricaud, A.; Morel, A.; Prieur, L. Absorption by Dissolved Organic Matter of the Sea (Yellow Substance) in the UV and Visible Domains. Limnol. Oceanogr. 1981, 26, 43–53. [Google Scholar] [CrossRef]
- Li, P.; Chen, L.; Zhang, W.; Huang, Q. Spatiotemporal Distribution, Sources, and Photobleaching Imprint of Dissolved Organic Matter in the Yangtze Estuary and Its Adjacent Sea Using Fluorescence and Parallel Factor Analysis. PLoS ONE 2015, 10, e0130852. [Google Scholar] [CrossRef]
- Xu, J.; Fang, C.; Gao, D.; Zhang, H.; Gao, C.; Xu, Z.; Wang, Y. Optical Models for Remote Sensing of Chromophoric Dissolved Organic Matter (CDOM) Absorption in Poyang Lake. ISPRS J. Photogramm. Remote Sens. 2018, 142, 124–136. [Google Scholar] [CrossRef]
- Kim, J.; Jang, W.; Hwi Kim, J.; Lee, J.; Hwa Cho, K.; Lee, Y.G.; Chon, K.; Park, S.; Pyo, J.C.; Park, Y.; et al. Application of Airborne Hyperspectral Imagery to Retrieve Spatiotemporal CDOM Distribution Using Machine Learning in a Reservoir. Int. J. Appl. Earth Obs. Geoinf. 2022, 114, 103053. [Google Scholar] [CrossRef]
- Chawla, N.V.; Bowyer, K.W.; Hall, L.O.; Kegelmeyer, W.P. Snopes.Com: Two-Striped Telamonia Spider. J. Artif. Intell. Res. 2002, 16, 321–357. [Google Scholar] [CrossRef]
- Maldonado, S.; López, J.; Vairetti, C. An Alternative SMOTE Oversampling Strategy for High-Dimensional Datasets. Appl. Soft Comput. J. 2019, 76, 380–389. [Google Scholar] [CrossRef]
- Snieder, E.; Abogadil, K.; Khan, U.T. Resampling and Ensemble Techniques for Improving ANN-Based High-Flow Forecast Accuracy. Hydrol. Earth Syst. Sci. 2021, 25, 2543–2566. [Google Scholar] [CrossRef]
- Breiman, L. Random Forests. Mach. Learn. 2001, 45, 5–32. [Google Scholar] [CrossRef]
- Machado, M.R.; Karray, S.; De Sousa, I.T. LightGBM: An Effective Decision Tree Gradient Boosting Method to Predict Customer Loyalty in the Finance Industry. In Proceedings of the 2019 14th International Conference on Computer Science & Education (ICCSE), Toronto, ON, Canada, 19–21 August 2019; pp. 1111–1116. [Google Scholar] [CrossRef]
- Li, L.; Qiao, J.; Yu, G.; Wang, L.; Li, H.Y.; Liao, C.; Zhu, Z. Interpretable Tree-Based Ensemble Model for Predicting Beach Water Quality. Water Res. 2022, 211, 118078. [Google Scholar] [CrossRef] [PubMed]
- Al-Kharusi, E.S.; Tenenbaum, D.E.; Abdi, A.M.; Kutser, T.; Karlsson, J.; Bergström, A.K.; Berggren, M. Large-Scale Retrieval of Coloured Dissolved Organic Matter in Northern Lakes Using Sentinel-2 Data. Remote Sens. 2020, 12, 157. [Google Scholar] [CrossRef]
- Shao, T.; Song, K.; Du, J.; Zhao, Y.; Liu, Z.; Zhang, B. Retrieval of CDOM and DOC Using in Situ Hyperspectral Data: A Case Study for Potable Waters in Northeast China. J. Indian Soc. Remote Sens. 2016, 44, 77–89. [Google Scholar] [CrossRef]
- Kutser, T.; Casal Pascual, G.; Barbosa, C.; Paavel, B.; Ferreira, R.; Carvalho, L.; Toming, K. Mapping Inland Water Carbon Content with Landsat 8 Data. Int. J. Remote Sens. 2016, 37, 2950–2961. [Google Scholar] [CrossRef]
- Lee, Z.; Carder, K.L.; Arnone, R.A. Deriving Inherent Optical Properties from Water Color: A Multiband Quasi-Analytical Algorithm for Optically Deep Waters. Appl. Opt. 2002, 41, 5755. [Google Scholar] [CrossRef]
- Zhu, W.; Yu, Q.; Tian, Y.Q.; Chen, R.F.; Gardner, G.B. Estimation of Chromophoric Dissolved Organic Matter in the Mississippi and Atchafalaya River Plume Regions Using Above-Surface Hyperspectral Remote Sensing. J. Geophys. Res. 2011, 116, C02011. [Google Scholar] [CrossRef]
- Carder, K.L.; Chen, F.R.; Lee, Z.P.; Hawes, S.K.; Kamykowski, D. Semianalytic Moderate-Resolution Imaging Spectrometer Algorithms for Chlorophyll a and Absorption with Bio-Optical Domains Based on Nitrate-Depletion Temperatures. J. Geophys. Res. 1999, 104, 5403–5421. [Google Scholar] [CrossRef]
- Lee, Z.P. IOCCG IOCCG Report Number 05: Reports of the International Ocean-Colour Coordinating Group Remote Sensing of Inherent Optical Properties: Fundamentals, Tests of Algorithms, and Applications; IOCCG: Dartmouth, Canada, 2006; Volume 5, ISBN 9781896246567. [Google Scholar]
- Seidel, M.; Hutengs, C.; Oertel, F.; Schwefel, D.; Jung, A.; Vohland, M. Underwater Use of a Hyperspectral Camera to Estimate Optically Active Substances in Thewater Column of Fresh Water Lakes. Remote Sens. 2020, 12, 1745. [Google Scholar] [CrossRef]
- Hannadige, N.K.; Zhai, P.-W.; Gao, M.; Franz, B.A.; Hu, Y.; Knobelspiesse, K.; Jeremy Werdell, P.; Ibrahim, A.; Cairns, B.; Hasekamp, O.P. Atmospheric Correction over the Ocean for Hyperspectral Radiometers Using Multi-Angle Polarimetric Retrievals. Opt. Express 2021, 29, 4504. [Google Scholar] [CrossRef]
- Smith, R.C.; Baker, K.S. Optical Properties of the Clearest Natural Waters (200–800 Nm). Appl. Opt. 1981, 20, 177–184. [Google Scholar] [CrossRef] [PubMed]
- Ma, R.; Pan, D.; Duan, H.; Song, Q. Absorption and Scattering Properties of Water Body in Taihu Lake, China: Backscattering. Int. J. Remote Sens. 2009, 30, 2321–2335. [Google Scholar] [CrossRef]
- Hamel, L. Elements of Statistical Learning: Data Mining, Inference, and Prediction, 2nd ed.; Springer: New York, NY, USA, 2009. [Google Scholar]
- Cha, G.W.; Moon, H.J.; Kim, Y.M.; Hong, W.H.; Hwang, J.H.; Park, W.J.; Kim, Y.C. Development of a Prediction Model for Demolition Waste Generation Using a Random Forest Algorithm Based on Small Datasets. Int. J. Environ. Res. Public Health 2020, 17, 6997. [Google Scholar] [CrossRef] [PubMed]
- Meler, J.; Kowalczuk, P.; Ostrowska, M.; Ficek, D.; Zabłocka, M.; Zdun, A. Parameterization of the Light Absorption Properties of Chromophoric Dissolved Organic Matter in the Baltic Sea and Pomeranian Lakes. Ocean Sci. 2016, 12, 1013–1032. [Google Scholar] [CrossRef]
- Wang, C.; Deng, C.; Wang, S. Imbalance-XGBoost: Leveraging Weighted and Focal Losses for Binary Label-Imbalanced Classification with XGBoost. Pattern Recognit. Lett. 2020, 136, 190–197. [Google Scholar] [CrossRef]
- Chandra, W.; Suprihatin, B.; Resti, Y. Median-KNN Regressor-SMOTE-Tomek Links for Handling Missing and Imbalanced Data in Air Quality Prediction. Symmetry 2023, 15, 887. [Google Scholar] [CrossRef]
- Kim, J.H.; Lee, H.; Byeon, S.; Shin, J.; Lee, D.H.; Jang, J.; Chon, K.; Park, Y. Machine Learning-Based Early Warning Level Prediction for and Data Resampling. Toxics 2023, 11, 955. [Google Scholar] [CrossRef] [PubMed]
- Wen, Z.; Wang, Q.; Ma, Y.; Jacinthe, P.A.; Liu, G.; Li, S.; Shang, Y.; Tao, H.; Fang, C.; Lyu, L.; et al. Remote Estimates of Suspended Particulate Matter in Global Lakes Using Machine Learning Models. Int. Soil Water Conserv. Res. 2024, 12, 200–216. [Google Scholar] [CrossRef]
- Aurin, D.; Mannino, A.; Lary, D.J. Remote Sensing of CDOM, CDOM Spectral Slope, and Dissolved Organic Carbon in the Global Ocean. Appl. Sci. 2018, 8, 2687. [Google Scholar] [CrossRef]
- Jang, W.; Park, Y.; Pyo, J.; Park, S.; Kim, J.; Kim, J.H.; Cho, K.H.; Shin, J.K.; Kim, S. Optimal Band Selection for Airborne Hyperspectral Imagery to Retrieve a Wide Range of Cyanobacterial Pigment Concentration Using a Data-Driven Approach. Remote Sens. 2022, 14, 1754. [Google Scholar] [CrossRef]
- Berk, A.; Conforti, P.; Kennett, R.; Perkins, T.; Hawes, F.; van den Bosch, J. Modtran® 6: A major upgrade of the modtran® radiative transfer code. In Proceedings of the Hyperspectral Image and Signal Processing: Evolution in Remote Sensing (WHISPERS), Lausanne, Switzerland, 24–27 June 2014; pp. 1–4. [Google Scholar] [CrossRef]
- Duan, S.-B.; Li, Z.-L.; Tang, B.-H.; Wu, H.; Ma, L.; Zhao, E.; Li, C. Land surface reflectance retrieval from hyperspectral data collected by an unmanned aerial vehicle over the baotou test site. PLoS ONE 2013, 8, e66972. [Google Scholar] [CrossRef]

| Model | Method | Train R2 | Test R2 | Train MAE | Test MAE | Train RMSE | Test RMSE |
| Random Forest | Original | 0.645 | 0.500 | 0.645 | 0.716 | 1.076 | 1.012 |
| (±0.116) | (±0.132) | (±0.129) | (±0.141) | (±0.182) | (±0.223) | ||
| SMOTE | 0.798 | 0.476 | 0.620 | 0.880 | 0.984 | 1.338 | |
| (±0.127) | (±0.148) | (±0.219) | (±0.202) | (±0.300) | (±0.325) | ||
| Light Gradient Boosting Machine | Original | 0.618 | 0.564 | 0.757 | 0.697 | 1.252 | 0.882 |
| (±0.077) | (±0.135) | (±0.086) | (±0.096) | (±0.108) | (±0.134) | ||
| SMOTE | 0.844 | 0.456 | 0.569 | 0.907 | 0.893 | 1.357 | |
| (±0.088) | (±0.161) | (±0.220) | (±0.203) | (±0.332) | (±0.341) |
| Model | Method | Model Accuracy | Train R2 | Test R2 | Train MAE | Test MAE | Train RMSE | Test RMSE |
| Random Forest | Original | R2 | 0.823 | 0.722 | 0.433 | 0.493 | 0.756 | 0.802 |
| MAE/RMSE | 0.900 | 0.628 | 0.341 | 0.556 | 0.604 | 0.830 | ||
| SMOTE | R2 | 0.898 | 0.838 | 0.471 | 0.566 | 0.765 | 0.777 | |
| MAE/RMSE | 0.881 | 0.816 | 0.468 | 0.495 | 0.793 | 0.682 | ||
| Light Gradient Boosting Machine | Original | R2 | 0.945 | 0.583 | 0.590 | 0.691 | 0.906 | 0.867 |
| MAE | 0.738 | 0.628 | 0.341 | 0.556 | 0.604 | 0.830 | ||
| RMSE | 0.813 | 0.571 | 0.459 | 0.599 | 0.881 | 0.881 | ||
| SMOTE | R2/MAE/RMSE | 0.993 | 0.772 | 0.142 | 0.531 | 0.225 | 0.837 |
Disclaimer/Publisher’s Note: The statements, opinions and data contained in all publications are solely those of the individual author(s) and contributor(s) and not of MDPI and/or the editor(s). MDPI and/or the editor(s) disclaim responsibility for any injury to people or property resulting from any ideas, methods, instructions or products referred to in the content. |
© 2024 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Kim, J.; Kim, J.H.; Jang, W.; Pyo, J.; Lee, H.; Byeon, S.; Lee, H.; Park, Y.; Kim, S. Enhancing Machine Learning Performance in Estimating CDOM Absorption Coefficient via Data Resampling. Remote Sens. 2024, 16, 2313. https://doi.org/10.3390/rs16132313
Kim J, Kim JH, Jang W, Pyo J, Lee H, Byeon S, Lee H, Park Y, Kim S. Enhancing Machine Learning Performance in Estimating CDOM Absorption Coefficient via Data Resampling. Remote Sensing. 2024; 16(13):2313. https://doi.org/10.3390/rs16132313
Chicago/Turabian StyleKim, Jinuk, Jin Hwi Kim, Wonjin Jang, JongCheol Pyo, Hyuk Lee, Seohyun Byeon, Hankyu Lee, Yongeun Park, and Seongjoon Kim. 2024. "Enhancing Machine Learning Performance in Estimating CDOM Absorption Coefficient via Data Resampling" Remote Sensing 16, no. 13: 2313. https://doi.org/10.3390/rs16132313





