1. Introduction
Knowing the tree species composition of a forest enables the estimation of the forest’s economic value and produces valuable information for studying forest ecosystems. Today, forest inventory in Scandinavia is based on an area-based approach [
1,
2] where laser scanning (point density about 1 pts/m
2) and aerial images (resolution typically 0.5 m) are used as inventory data. However, using these approaches, species-specific diameter distributions have poor prediction accuracy and improvement in tree species detection is needed.
Forest data using remote sensing methods has mostly been concentrating on forest stand (i.e., the ecologically homogeneous and spatially continuous part of a forest) and plot level data. However, stand-level forest variables are typically an average or sum from the set of trees composing the stand. In calculating forest inventory variables such as the volume and biomass of the growing stock, tree-level models are typically used nowadays [
3,
4,
5]. Very high resolution remote sensing data allows moving from the stand level to the level of individual trees, which has certain benefits, for example in precision forestry, forest management planning, biomass estimation and modeling forest growth [
6].
Increasing the level of detail of the forest can also improve detailed modeling of forests, which can be used to predict forest growth and improve satellite-based remote sensing of forests by more accurately modeling the radiative transfer within the forest canopy.
Forest and tree species classification using multi- or hyperspectral imaging or laser scanning has been widely studied [
7,
8,
9]. However, the data has mainly been captured from manned aircrafts or satellites, wherefore studies have been focusing more on the forest or plot level. The challenge in passive imaging has been the dependence on sunlight and the high impact of the changing and different illumination conditions on the radiometry of the data [
9,
10,
11,
12,
13]. Shadowing and brightening of individual tree crowns cause the pixels of a single tree crown to scale from really dark pixels to really bright pixels. A few methods have been developed and suggested to reduce the effect of the changing illumination in the forest canopy [
14], but none of them have been extensively tested with high resolution spectral imaging data from individual trees. In addition, illumination changes can be beneficial in classification tasks, since they potentially provide species-specific information of the tree structure [
15,
16].
Airborne Laser Scanning (ALS), both discrete and full-waveform, has been used to classify trees in boreal forests [
17,
18,
19,
20,
21]. The most demanding task using passive imaging systems has always been discriminating pines from spruces due to their spectral similarity. However, structural data from laser scanning has shown it can capture the structural differences of these species, such as the vertical extent of the leafy canopy [
18,
22,
23].
Individual trees have been detected using passive data [
24], mainly using image segmentation using textural features [
25,
26], but the development of dense image matching methods and computing power has enabled the production of high resolution photogrammetric point clouds. Their ability to produce structural data from a forest has been presented in the literature [
27,
28,
29,
30]. Photogrammetric point clouds produce great top-of-the-canopy information that enables the computation of Canopy Height Models (CHM) [
29] and thus detection of individual trees [
31]. However, the notable limitation of photogrammetric point clouds compared to laser scanning is that passive imaging does not have good penetration ability [
30], especially with aerial data from manned aircrafts. Thus, laser scanning that can more deeply penetrate forest canopies has been the main method of providing information on forest structure [
1,
2], especially at individual tree level [
32]. Photogrammetric point clouds derived from data captured using manned aircrafts have been used, but the spatial resolution is not usually accurate enough to produce good detection results.
Small unmanned aerial vehicles (UAVs) have been rapidly incorporated in various remote sensing applications, including forestry [
33,
34,
35]. The use of UAVs in aerial imaging has enabled measurements with higher spatial resolution, improving the resolution of photogrammetric point clouds and the acquisition of three-dimensional (3D) structural data from the forest [
36,
37]. Some studies have been using data from UAVs in individual tree height determination with good results [
38]. Multispectral data acquired from UAVs has been used in tree species classification, but the data has been limited to RGB images and one near-infrared (NIR) channel using NIR-modified cameras [
39,
40,
41]. The development of low-weight hyperspectral imaging sensors is likely to increase the use of spectral data collected from UAVs in forestry applications [
34].
Recently, the development of small hyperspectral imaging sensors has enabled high spectral and spatial resolution measurements from UAVs. Several pushbroom hyperspectral sensors have been implemented in UAVs [
42,
43,
44,
45,
46]. The novel hyperspectral cameras operating in the frame format principle offer interesting possibilities for UAV remote sensing by stable imaging geometry and by giving an opportunity to make 3D hyperspectral measurements [
47,
48,
49]. Näsi et al. [
50] presented the first study with 3D hyperspectral UAV imaging in individual tree-level analysis of bark beetle damage in spruce forests. To the best of the authors’ knowledge, the classification of individual trees using hyperspectral imagery from UAVs has not yet been studied.
Novel hyperspectral imaging technology based on a variable air gap Fabry–Pérot interferometer (FPI) operating in the visible to near-infrared spectral range (500–900 nm; VNIR) [
47,
48,
49] was used in this study. The FPI technology makes it possible to manufacture a lightweight, frame format hyperspectral imager operating on the time-sequential principle. The FPI camera can be easily mounted on a small UAV together with an RGB camera, which enables the simultaneous hyperspectral imaging with high spatial resolution photogrammetric point cloud creation [
50].
The objective of this study is to test the use of high resolution photogrammetric point clouds together with hyperspectral UAV imaging in individual tree detection and classification in boreal forests. In particular, the data processing challenges in a real forest environment will be studied and the importance of different spectral and structural features in tree species classification will be assessed. Previous studies using UAV data have not utilized both spectral and structural data in tree species classification, nor have they provided reliable complete workflows to detect and classify individual trees from large forested scenes. The materials and methods are presented in
Section 2. The results are given in
Section 3 and discussed
Section 4. Conclusions are provided in
Section 5.
2. Materials and Methods
2.1. Study Area and Reference Tree Data Collection
The study area was the Vesijako research forest area in the municipality of Padasjoki in southern Finland (approximately 61°24′N and 25°02′E). The area has been used as a research forest by the Natural Resources Institute of Finland (and its predecessor, the Finnish Forest Research Institute). Eleven experimental test sites from stands dominated by pine (Pinus sylvestris), spruce (Picea abies), birch (Betula pendula) and larch (Larix sibirica) were used in this study. The test sites represented development stages from young to middle-aged and mature stands (no seedling stands or clear cut areas). Within each test site, there were 1–16 experimental plots treated with differing silvicultural schemes and cutting systems (altogether 56 experimental plots). The size of the experimental plots was 1000–2000 m2. Within the experimental plots, all trees with breast-height diameter of at least 50 mm were measured as tally trees in 2012–2013. For each tally tree, the following variables were recorded: relative location within the plot, tree species, and diameter at breast height. Height was measured for a number of sample trees in each plot and estimated for all tally trees using height models calibrated by sample tree measurements. The geographic location of the experimental plot corner points was measured with a Global Positioning System (GPS) device, and the locations were processed with local base station data, with an average error of approximately 1 m.
For this study, a total of 300 fixed-radius (9 m) circular sample plots were placed on the experimental plots in the eleven test sites (4–8 circular plots per each experimental plot, depending on the size and shape of the experimental plots). The plot variables of the circular sample plots were calculated on the basis of the tree maps of the experimental plots. The statistics of the main forest variables in the field measurement data are presented in
Table 1 and
Table 2. Some of the sample plots have an exceptionally high amount of growing stock compared to values typical for this geographic area, which can be seen in the maximum values and standard deviation of the volumes in the field observations of this study area. The areas with the highest amount of growing stock are dominated by pine and larch.
Figure 1 shows the locations of the study areas.
The reference trees collected in the field measurements were visually compared to the collected UAV orthomosaics in order to check their geometric correspondence at the individual tree level (orthomosaic calculation is described in
Section 2.3). Some misalignment could be observed, which was due to challenges in tree positioning in the ground conditions, due to the georeferencing quality of the UAV orthomosaics, as well as due to different characteristics at the object view and the aerial view. The misaligned reference trees were manually aligned with the trees in the UAV orthomosaics or removed if the corresponding tree could not be identified reliably from the UAV orthomosaics. This was performed using Quantum GIS (QGIS) by overlaying the field data over the RGB and FPI mosaics.
2.2. Remote Sensing Data Capture
Altogether, 11 test sites were captured using a small UAV in eight separate flights on 25–26 June 2014, in the Vesijako test site (
Table 3 and
Table 4). For georeferencing purposes, three to nine Ground Control Points (GCPs) with cross-shaped signals with an arm length of 3 m and width of 30 cm were installed in each test area. Reflectance panels of size 1 m × 1 m and with a nominal reflectivity of 0.03, 0.1 and 0.5 were installed in the area for reflectance transformation purposes [
51]. The reference reflectance values were measured in a laboratory with an estimated accuracy of 2%–5% using the FIGIFIGO goniospectrometer [
52].
The UAV remote system belonging to the Finnish Geospatial Research Institute (FGI) was used in the data capture. The UAV platform frame was a Tarot 960 hexacopter with Tarot 5008 (340 KV) brushless electric motors. The autopilot was a Pixhawk equipped with Arducopter 3.15 firmware. The system’s payload capacity is about 3 kg and the flight time is 10–30 min, depending on payload, battery, conditions, and flight style. A novel hyperspectral camera based on a tuneable Fabry–Pérot interferemoter (FPI) [
47,
48,
49] was used to capture the spectral data. The FPI camera captures frame-format hyperspectral images in a time-sequential mode. Due to the sequential exposure of the individual bands (0.075 s between adjacent exposures, 1.8 s during the entire data cube with 24 exposures), each band of the data cube has a slightly different position and orientation, which has to be taken into account in the post-processing phase. The image size was 1024 × 648 pixels, and the pixel size was 11 μm. The FPI camera has a focal length of 10.9 mm; the field of view (FOV) is ±18° in the flight direction, ±27° in the cross-flight direction, and ±31° at the format corner. The camera system has an irradiance sensor (based on the Intersil ISL29004 photodetector) to measure the wideband irradiance during each exposure [
53]. The UAV was also equipped with the Ocean Optics spectrometer and irradiance sensor to monitor illumination conditions; due to some technical problems, the data quality was not suitable for the radiometric correction. A GPS receiver is used to record the exact time of the first exposure of each data cube. Spectral settings can be selected according to the requirements. In this study, altogether, 33 bands were used with the full width of the half maximum (FWHM) of 11–31 nm. The settings used in this study are given in
Table 5. In order to capture high spatial resolution data, the UAV was also equipped with an ordinary RGB compact digital camera, the Samsung NX1000. The camera has a 23.5 × 15.7 mm complementary metal-oxide semiconductor (CMOS) sensor with 20.3 megapixels and a 16 mm lens. The UAV system is presented in
Figure 2.
The flying height was 83–94 m from the ground level, providing an average Ground Sampling Distance (GSD) of 8.6 cm for the FPI images and 2.3 cm for the RGB images on the ground level. The flight height was 62–73 m from the tree tops (calculated for an average tree height of 21 m). Thus, the average GSDs were 6.5 cm and 1.8 cm at tree tops for the FPI and RGB data sets, respectively. The flight speed was about 4 m/s. The FPI image blocks had average forward and side overlaps of 67% and 61%, respectively, at the nominal ground level and 58% and 50%, respectively, at the tree top level. For the RGB blocks, the average forward and side overlaps were 78% and 73%, respectively, at the ground level. At the level of treetops, the average forward and side overlaps were 72% and 65%, respectively. The overlaps of RGB blocks were suitable for the orientation processing and point cloud generation. The overlapping of FPI images were quite low in the forested scene with large height differences, thus the combined processing with RGB images was necessary to enable the highest quality geometric reconstruction.
Imaging conditions were quite windless, but illumination varied a lot in different flights. Illumination conditions were cloudy and quite uniform during flights v01, v02, v0607 and v0910. Test site v08 was captured under sunny conditions, and during flights v0304, v05 and v11 the illumination conditions varied between sunny to cloudy. Irradiance recordings by the Intersil ISL29004 irradiance sensor during the flights are presented in
Figure 3. The recordings were quite uniform during the flights captured in cloudy conditions. The variability in irradiance measurements during flights v0304, v05, v08 and v11 were partially due to changing weather and partially due to tilting of the irradiance sensor in different flight directions, thus obtaining different levels of irradiation.
The national ALS data by the National Land Survey of Finland (NLS) was used to provide the ground level, and it was also used as the geometric reference for evaluating the geometric quality of photogrammetric processing [
54]. The minimum point density of the NLS’s ALS data is half a point per square metre, and the elevation accuracy of the points in well-defined surfaces is 15 cm. The horizontal accuracy of the data is 60 cm. The ALS data used in this study was collected on 12 May 2012.
2.3. UAV Data Processing
Rigorous processing was required in order to derive quantitative information from the imagery. The processing of FPI camera images is similar to any frame format camera images that cover the area of interest with a large number of images. The major difference is the processing of the nonaligned spectral bands due to the time-sequential imaging principle (
Section 2.2) for which a registration procedure has been developed. The image data processing chain for tree parameter estimation included the following steps:
Applying radiometric laboratory calibration corrections to the FPI images.
Determination of the geometric imaging model, including interior and exterior orientations of the images.
Using dense image matching to create a Digital Surface Model (DSM).
Registration of the spectral bands of FPI images.
Determination of a radiometric imaging model to transform the digital numbers (DNs) data to reflectance.
Calculating the hyperspectral and RGB image mosaics.
Subsequent remote sensing analysis
The image preprocessing and photogrammetric processing (steps 1–3) were carried out using 3.5 GHz quad-core PC with 88 GB RAM and a GeForce GTX 980 graphics processing unit (GPU). For the band registration, radiometric processing and mosaic calculation (steps 4–6), several regular office PCs were used in parallel. Remote sensing analysis was performed using 3.5 GHz quad-core PC with 24 GB RAM.
In the following sections, the geometric (2, 3, 4) and radiometric (1, 5, 6) processing steps and estimation process (7) used in this investigation are described.
2.3.1. Geometric Processing
Geometric processing included the determination of the orientations of the images and determination of the 3D object model. The Agisoft PhotoScan Professional commercial software (AgiSoft LLC, St. Petersburg, Russia) and the FGI’s in-house C++ software (spectral band registration) were used for the geometric processing.
PhotoScan performs photo-based 3D reconstruction based on feature detection and dense matching, and it is widely used in processing UAV images [
50,
55]. In the orientation processing, the quality was set to “high”, which means that the full resolution images were used. The settings for the number of key points per image were 40,000 and for the final number of tie points per image 1000; an automated camera calibration was performed simultaneously with image orientation (self-calibration). The initial processing provided image orientations and sparse point clouds in the internal coordinate system of the software. For the data, an automatic outlier removal was performed on the basis of the residuals (re-projection error) (10% of the points with the largest errors were removed), as well as standard deviations of the tie point 3D coordinates (reconstruction uncertainty) (10% of the points with the largest uncertainty were removed). Some outlier points were also removed manually from the sparse point cloud (points on the sky and below the ground). The image orientations were transformed to the ETRS-TM35FIN coordinate system using the GCPs in the area. The outputs of the process were the camera calibrations (Interior Orientation Parameters—IOP), the image exterior orientations in the object coordinate system (Exterior Orientation Parameters—EOP), and the 3D coordinates of the tie points. Orientations of the FPI hypercubes were determined in a simultaneous processing with the RGB images in the PhotoScan. Three FPI image bands (reference bands) were included simultaneously in the processing: band 4: L
0 = 520.8 nm, dt = 1.125 s; band 12: L
0 = 590.4 nm, dt = 1.725 s; band 16: L
0 = 630.7 nm, dt = 0.15 s. (L0 is the central wavelength and dt is the time difference to the first exposure of the data cube.) The orientations of those FPI image bands that were not included in the PhotoScan processing were determined using the space resection method developed at the FGI. This method used the tie points calculated during the PhotoScan processing as GCPs and determined the corresponding image coordinates by correlation matching of each unoriented band to the reference band 4 and finally calculated the space resection. This processing produced EOPs for all the bands in all hypercubes.
For accurate dense point cloud generation, the orientations of the RGB datasets were also determined separately without the FPI images. Dense point clouds with 5 cm point interval were then generated using two-times downsampled RGB images. A mild filtering was used to eliminate outliers, which allowed high height differences for the data set.
2.3.2. Radiometric Processing
Our preferred approach was to analyze different test areas simultaneously. Thus, it was necessary to scale the radiometric values in each data set to a similar reference scale. Calibration of the DNs to reflectance factors was the preferred approach. We installed the reflectance reference panels close to the takeoff place in each test site in order to carry out the reflectance transformation using the empirical line method. Unfortunately, the targets were inside the forest and surrounded by tall trees, thus the illumination conditions in reference targets did not correspond to the illumination conditions on top of the canopy. Because of this, they did not provide accurate reflectance calibration by the empirical line method. A further challenge was that the illumination conditions were variable during many of the flights, providing large relative differences in radiometric values within the blocks and between different blocks (
Figure 3).
A modified approach was used to eliminate radiometric differences of images. We applied the radiometric block adjustment and in-flight irradiance data to normalize the differences within and between the blocks [
49,
53,
56]. We selected test site v06 as the reference test site and calculated the empirical line calibration using this area (a
abs_ref, b
abs_ref). The area for the reflectance panels was relatively open in block v06, and the surrounding vegetation did not shade the panels. We selected a reference image in each block
i (
) and normalized the rest of the images
j in the block to this image using the relative scaling factor a
rel_vi_j. The reference image was selected so that its illumination conditions represented well the conditions of the majority of the data set. The a priori values for the relative correction parameters a
rel_vi_j were derived from the Intersil ISL29004 irradiance measurements:
where irradiance
vi_j and irradiance
vi_ref are the irradiance measurements during the acquisition of image
j and reference image
ref. The a
rel_vi_j values were adjusted using the radiometric block adjustment method [
49,
53,
56]. The relative scaling factor (s
vi_ref) was calculated between each block and the reference block v06 using the Intersil irradiance values of reference images and scaled using the exposure times of the reference block and the block
i (t
exp_ref, t
exp_vi). Median filtering was used in the irradiance values to eliminate the instability of the irradiance measurements:
Thus, the final equation for the transformation from DNs to reflectance was
We did not apply correction for the bidirectional reflectance (BRDF) effects in the data set. BRDF effects were nonexistent in the image blocks captured under cloudy conditions. For the data sets captured in sunny conditions, the maximum view angles to the object of different blocks were 2.6°–4.8° in the flight direction and 3.9°–9.4° in the cross-flight direction at the top of the canopy; thus, the BRDF effects could be assumed to be insignificant. The radiometric model parameters were calculated separately for each band.
2.3.3. Mosaic Calculation
Hyperspectral orthophoto mosaics were calculated with 10 cm GSD from the FPI images using the FGI’s in-house mosaicking software. Mosaics were calculated for each band separately and then combined to form a uniform hyperspectral data cube over each block area. The radiometric processing described above (absolute calibration and relative normalizations within and between image blocks) were used for the FPI images. The RGB mosaics were calculated using the PhotoScan mosaicking module with a 5 cm GSD.
2.4. Workflow for Individual Tree Detection and Classification
The final derived products from the UAV data that were used in this study are:
Photogrammetric point cloud with a 10 cm point interval computed from the RGB data;
RGB orthomosaic with 5 cm GSD (used for visual evaluation for the reference data and classification of individual trees);
FPI orthomosaic with 33 spectral bands and 10 cm GSD.
Figure 4 presents the workflow for training and validating the classifier using the reference data and detecting the individual trees and classifying them using the trained classifier. The reference data collected from the field is used to create the classification model. The classification model utilizing both spectral and structural features was tested. The spectral features were computed from the FPI mosaics using Matlab (Matlab R2015b, The MathWorks, Inc., Natick, MA, USA) and structural features from the photogrammetric point clouds using LAStools software (LAStools, version 160703, rapidlasso GmbH). Feature selection was tested to improve classification performance and to evaluate the significance of various features. The feature selection and classification training and testing were done using Weka software (Weka 3.8, University of Waikato). The individual trees were detected from the photogrammetric point clouds using FUSION software (FUSION version 3.50, US Department of Agriculture, Forest Service, Pacific Northwest Research Station). The classification features for the detected trees were derived using the same methods as with the reference data. In the end, the classification model created using the reference data was used to predict the classes of the detected trees. A detailed description of the steps in this workflow is given in the following sections.
2.4.1. Spectral Features
The FPI hyperspectral data cube consisted of 33 different spectral bands. The spectral features for classification were derived from the FPI orthomosaic by selecting pixels within a circular area around the tree center using a one-meter circle radius. Several different spectral features were computed, which are summarized in
Table 6. Most of the features are mean and median spectra calculated from differently selected pixel sets. Two of the features, namely MeanSpectraNormalized and ContinuumRemovedSpectra, are spectral features that normalize the spectra which aims at reducing the effects of shadowing and illumination differences. The feature MeanSpectraNormalized normalizes the spectra by dividing each spectral band by the sum of all bands. The ContinuumRemovedSpectra is a feature where the spectra is normalized by dividing the spectrum by a convex hull (i.e., continuum) over the spectrum [
57].
2.4.2. Structural Features
The 3D structural features were computed from the RGB point clouds with 0.1 m GSD and using algorithms provided by LAStools software. The computed structural parameters are explained in
Table 7. The detailed workflow for computing the structural features is described in
Appendix A.
In order to avoid the creation of features that are biased to classify trees based on their height and not the point-height distribution, all tree metrics that are directly proportional to the tree height were normalized by dividing the feature value by the maximum height of the tree. Without this normalization, the Larch trees in our study area, for example, could be correctly classified only based on their height since the Larch samples were all from the same test site with a low variation in tree heights. This would decrease the generalization of the classification. Due to the same reason, the maximum height was not used as a classification feature.
2.4.3. Feature Selection
The total number of spectral and structural features was 347. In order to find good features with a high prediction ability, feature selection and its effect on the classification were also investigated.
The feature selection was performed using Weka 3.8 software. The derived features included many features that had a high possibility to be highly correlated between each other, such as different spectral features at the same spectral band; feature selection methods that take into account the correlation between the individual features were used. The chosen method was Correlation-based Feature Selection (CFS) [
60] implemented for evaluating a subset of features (CfsSubsetEvaluation in Weka). This method strives for subsets that correlate highly with the class value and have a low internal correlation between single features. The method evaluates the worth of a subset of attributes by considering the individual predictive ability of each feature along with the degree of redundancy between them. This is beneficial especially with spectral data from vegetation where spectral features are usually characterized better by a combination of different spectral bands rather than using information from only one spectral band. The use of vegetation indices is also based on this. Vegetation indices (VIs) have been used especially with sensors having a limited number of spectral bands (less than 10). The FPI camera provides 33 spectral band data and a good classification method should extract the same significant information from this data that could possibly be obtained by calculating VIs.
The CfsSubsetEvaluation method can be used with different search methods for feature subset evaluation. The feature selection method was performed with three different search methods:
GeneticSearch: Performs a search using the simple genetic algorithm [
61].
BestFirst: Searches the space of attribute subsets by greedy hill climbing augmented with a backtracking facility.
GreedyStepWise: Performs a greedy forward or backward search through the space of attribute subsets.
The three feature selection methods were performed using 10-fold cross validation. The features selected at least eight times out of the ten runs were chosen as the best features by each method. The final feature set included features that were among the best features using any two of the three feature selection methods. Feature selection was performed with and without normalizing spectral features (i.e., MeanSpectraNormalized and ContinuumRemovedSpectra) to evaluate their benefits compared to unnormalizing spectral features.
2.4.4. Classification Methods
The classification was performed using Weka software. Several different types of classification algorithms were tested in the classification, including:
The default parameters of the software were used in the classifiers. The parameters that were used with each classification method are described in
Appendix B.
2.4.5. Classification Performance Evaluation
Since the reference data was slightly imbalanced (See
Section 3.2), several evaluation metrics that describe classification performance and generality were computed. The general accuracy and reliability of the classification model is evaluated with overall accuracy and Cohen’s Kappa. Species-specific classification performance was evaluated with precision, recall, F-score, and confusion matrices.
Overall accuracy is the total number of correctly classified samples (i.e., sum of true positives for all classes) divided by the total number of samples. Recall (i.e., User Accuracy) is the proportion of trees which were classified to a specific class among all trees that truly belong to that class. Recall is also known as the completeness.
where
tp is the number of samples predicted positive that are actually positive (True positive), and
fn is the number of samples predicted negative that are actually positive (False negative).
Precision (i.e., exactness or producer accuracy) is the proportion of the examples that truly belong to a specific class among all those classified as that specific class.
where
fp is the number of samples predicted positive that are actually negative (False positive).
F-score (or F-measure) is the harmonic mean of recall and precision and thus it expresses the balance between the recall and precision.
The Cohen’s Kappa statistic measures the agreement of prediction with the true class. It is a metric that compares an observed accuracy with an expected accuracy (i.e., it takes into account the random chance of classifying correctly).
where
k is the number of classes,
nti is the number of samples truly in class
i,
nci is the number of samples classified to class
i and
N is the total number of samples in all classes.
The validation of the classification performance was done using leave-one-out cross-validation (LOOCV). This method is a special case of k-fold cross-validation where the number of folds is equal to the number of samples. Thus, each sample is used once for testing while other samples are used for training. The final classification model used for prediction is a model trained using all samples.
The classification performance using different feature sets was also evaluated for importance of spectral and structural features. This evaluation was only done using relatively fast classification methods, since using the full feature set (for example, with the MLP and LOOCV methods) is extremely time-consuming.
2.4.6. Individual Tree Detection
The individual trees were detected from the photogrammetric point clouds with 0.1 m GSD. The tree detection was done using the FUSION software. The summarized workflow for the tree detection goes as follows:
Convert ASCII-xyz file to LAS format
Point cloud thinning
Point cloud to Digital Surface Model (DSM) using Triangulated Irregular Network (TIN) model
DSM to Canopy Height Model (CHM) utilizing Digitral Terrain Model (DTM) computed from NLS ALS data.
Tree detection from the CHM using a local maxima-based method developed in [
66,
67].
The final output was a CSV file with the following information: tree location, tree height and crown width, and minimum and maximum height.
The parameters at 2–5 were optimized by visually evaluating the resulting tree maps over the orthomosaics. These parameters relate to how much spatial smoothing and sampling is used in producing the CHM and detecting trees from it. These are parameters that depend on the forest type (i.e., typical crown shapes and canopy density) and photogrammetric point cloud smoothness. The best compromise between false positive and true negative was the goal of the visual evaluation and parameter optimization. The detailed description of this workflow is given in
Appendix C.
2.4.7. Evaluation of Individual Tree Detection and Classification Performance
To demonstrate the ability of UAV-based photogrammetric point clouds and hyperspectral imaging to detect and classify individual trees, the best performing classifier was used to classify all trees detected from the test sites.
The accuracy of the tree detection and classification was estimated by comparing the reference data with the detected and classified trees in each test site. The tree detection accuracy was performed by searching for whether there is a tree detected within a specific radius to each reference tree. Different search radiuses (1 m, 1.5 m and 2 m) were tested to find out if there is a difference between the tree crown centers determined from images mosaics and crown centers detected from CHMs.
The classification accuracy and reliability of detected trees was estimated visually using the resulting classified tree maps and RGB orthomosaic and by computing statistics of the probability of the prediction given by the trained classification model.
3. Results
3.1. UAV Data Processing
Geometric processing of all the data sets was successful with the PhotoScan software. Statistics of the orientation processing are given in
Table 8. The numbers of images in the integrated blocks were 176–758. The reprojection errors were mostly 0.6–0.9 pixels. For block v07, the reprojection errors were higher, up to 2.5 pixels for an unknown reason. Possible explanations could be inaccuracies in the GCPs or the poor determinability of the IOPs and EOPs due to the small size of this block. However, the output block quality appeared to be good when comparing the point cloud generated from this block to the ALS reference point cloud.
The image-matching using the RGB images provided dense photogrammetric point clouds with 500–700 points per m
2, and with an approximate 5 cm point distance. The comparison between the photogrammetric and ALS-interpolated surfaces was made visually using profile plotting and difference surfaces. This method was used to provide a rough quality assessment of the processing. A more accurate assessment was not possible because specific height reference data was not implemented in the target area. For each separate area, ten profiles with all points from the photogrammetric DSM and ALS point cloud in ±1 m width along the profile were computed in South–North and East–West directions. Examples of profiles in test site v05 are given in
Figure 5, and an example of a photogrammetric point cloud of test site v05 is presented in
Figure 6. The profile plots revealed that the photogrammetric surfaces generally followed the structure that was visible in the ALS point cloud: individual trees seemed well-aligned and minor differences in canopy top level and ground level were observable. As anticipated, the surface generated by photogrammetric techniques did not reach the ground level as often as ALS. It was noteworthy that in gaps where the photogrammetric surface reached the ground, the ground-level estimate was mostly below the ALS ground. Subtracting photogrammetric DSM from the interpolated ALS reference DSM created a difference surface. A slight mismatch in the xy-direction resulted in large differences in the DSM difference; thus, the comparison worked best with test sites with flat surfaces such as fields or roads. There was a slight tilt in the photogrammetric surface in some test sites (block v06 ±3 m, block v09) or curvature in which the middle of the area was lower and the edges higher than the ALS reference (block v05 ±1 m, v07, v11). The most probable causes for the slight systematic deformations were uncertainties in IOPs and EOPs and potential inaccuracies in GCPs. Furthermore, in many cases, the point intersection angles when matching points in small openings were very narrow, which reduced the accuracy of the derived points. However, the quality of georeferencing could be considered good and appropriate for this investigation.
The results of the block adjustments were utilized in the registration of the all the bands to the reference band. The registration process was successful, and only minor misalignments could be observed between the bands in some cases.
A visual assessment of mosaics after radiometric processing indicated that the radiometry uniformity was sufficient in most of the cases (
Figure 7) and suitable reflectance values were obtained from individual image blocks. The most challenging test sites were v0304, v05, and v11, which all had a variable illumination. The radiometric normalization method did not provide sufficient uniformity for the test site v0304, so it was left out of the analysis. In the other two test sites (v05 and v11), the processing eliminated the major radiometric differences. The resulting spectra are discussed in
Section 3.3.
3.2. Reference Data Processing
Due to inaccuracies in the field data collection and the RGB and FPI mosaics, there were notable misalignments between the field data tree locations and the trees visually observed from the mosaics as discussed in
Section 2.1. The misalignment was worst in the areas with high tree density. In some areas, the misalignment was systematic and the data could be aligned just by moving all the reference trees in that specific area to some direction. However, in some areas, the misalignment was not systematic, therefore every tree had to be moved one-by-one to the correct position if the correct position and species of the tree could be identified from the orthomosaic. Some test sites included several tree species at different canopy heights. In such areas, only the highest trees could be identified.
If reference data had to be moved, particular attention was given to moving the reference data to the matching species. In most cases, it was possible to identify the tree species from the RGB orthomosaic. However, at some areas with high tree density and multiple tree species, it was not possible to identify the correct trees, and thus the reference data had to be omitted. Reference data for the test site v0304 could not be aligned properly, and thus it was omitted completely from the analysis.
Although it was possible to fix the locations of the misaligned reference trees to match the correct tree species in the mosaics, it was not always possible to be certain that the reference tree was exactly the correct tree in the orthomosaics. This means that other reference data (such as height and DBH) might be incorrect, and thus could not be utilized in this study. Over half of the original data (approximately 8800 trees) had to be omitted from the analysis, since it could not be aligned to correct tree species reliably. The final reference data included 4151 trees from nine different test sites. The proportion of different tree species and their distribution to different test sites are summarized in
Table 9.
3.3. Feature Selection
The most significant features according to the feature selection methods are summarized in
Table 10. The first seven features of the full feature set with normalizing spectral features (i.e., MeanSpectraNormalized and ContinuumRemovedSpectra) were always included in the final feature set regardless of the method used. The last fifteen features were among the best predicting features using two of the three methods. If the normalizing spectral features were omitted from the analysis, the features included 14 features, from which the first three were selected by each feature selection method.
Most of the significant features were spectral features. Examples of the mean and median spectra and the effect of normalization can be seen in
Figure 8,
Figure 9 and
Figure 10. When considering the average spectra of species calculated from all the test sites, the differences between the spruce and pine were the smallest. The differences in the birch and larch were larger than the other two (
Figure 8). The mean and median spectra are almost equal which indicates that the species-specific spectral values are quite uniformly distributed and there are not any significant outliers. When comparing the spectral signatures of the species in different test sites, the differences were less than 25% in most cases (
Figure 9). These differences were caused by the uncertainties in the relative calibration procedure, but they were partially also due to the natural variability within each species. The spruce spectrum of the test site v11 is clearly brighter than with the other test sites. The normalization slightly reduced the differences but notable differences were still present after normalization. The test site v11 had changes in the illumination during the data capture and the radiometric correction was not able to remove all the illumination changes which affected the radiometric quality of the final data.
When the full feature set was used, most of the significant spectral features were normalized spectral features. Normalizing the spectra reduces the effects of shadowing and illumination differences, which is probably the reason why they performed best with this data, since some of the data had been collected in different flights and time of day with varying illumination conditions. Illumination and shadowing differences are reduced, especially when the illumination conditions are specular, but it also reduces the possible scale differences with data collected on different occasions in diffuse illumination conditions. The relative spectral differences were reduced, especially at the near infra-red wavelengths where changes in view angle and tree structure have the strongest impact. At the same time, the differences at visible wavelengths are still present, and thus most of the best features are at visible wavelengths, but few NIR features are also present.
If the normalizing spectral features were omitted from the analysis, the final feature set included more NIR features and only features in the red band (i.e., 657 nm) from the visible spectral region. Without the normalization, the relative differences in the different species are higher in the NIR region and smaller in the visible wavelength region, and thus the NIR features without normalization were the best predictors. In addition, a few more 3D structural features were included in the final feature set with the feature set without normalizing spectral features.
The most significant 3D structural features were mostly those features at the canopy top layers, which was an expected result since passive optical imaging does not have good penetration ability. The minimum height normalized by the maximum height performed well, as it provides a good estimate of the canopy extent downwards, which provides species-specific information (for example, between pines and spruces), since the canopy of spruces extends lower than pines and birches. However, it must be noted that with photogrammetrically-produced point clouds, the penetration ability is also highly dependent on the density of the forest (gaps between trees), which is not always a species-specific property.
3.4. Classification
The classification was performed using various feature configurations, including all features, all features without normalizing spectral features and mean spectra only. The evaluation of different classification methods for feature-selected datasets and mean spectra only are summarized in
Table 11.
The best performing classification algorithms were MLP and RandomForest, which provided 94.9% and 95.2% overall accuracies for feature-selected dataset with all features, respectively. None of the classification algorithms performed exceptionally poorly in the classification, but the NaiveBayes method did not provide good results (87.1%). The NaiveBayes method was probably most affected by the imbalance data, which resulted in poorer results. The classification results with feature selection and using a feature set without the normalizing spectral features produced slightly worse classification results compared to those achieved with the normalizing features. Without the normalizing spectral features, Random Forest provided the best result with 93% overall accuracy.
The classification was also tested using only the mean spectra. The results indicate that using only the mean spectra, good classification results can be achieved. The accuracy of classification using only the mean spectra and MLP method was 95.2%. The accuracy of the other classification methods differed by approximately 1% from those achieved with the full feature set with feature selection, except the NaiveBayes method which produced only 70.7% accuracy.
The evaluation of classification performance with different feature sets using k-NN and RandomForest classifiers are presented in
Table 12 and
Table 13. The best performance was achieved when using all 347 features. The accuracies were 95.5% for k-NN and 95.1% for RandomForest. When all features were used, the feature selection did not improve the classification accuracy, but neither did it drastically worsen the results. Feature reduction based on the feature selection significantly improved the learning time of the classification, especially with more complex methods such as MLP. Good performance was also achieved by using only the spectral features (94.7% and 94.9% for Random Forest and k-NN, respectively). Random Forest and k-NN (k = 3) achieved 94.8% and 94.9% accuracies, respectively, when all the features were used with the feature set without normalizing spectral features, which was less than 1% worse than with the full feature set. The worst results were obtained when only the structural features were used in classification. The Kappa values are slightly smaller than the accuracy, which indicates a small effect of the imbalanced data.
The species-specific results for the k-NN, RandomForest, and MLP methods with a feature-selected full feature set are presented in confusion matrices in
Table 14,
Table 15,
Table 16 and
Table 17. As expected, the most errors in classification occurred among the classification of pines and spruces. The most frequent error was spruces classified as pines, which might be a result of the imbalance in the data (most of the training data are pines). Birches could be classified with a high recall and precision (both over 95%) since they have a strong spectral difference compared to coniferous species, especially in the NIR channels (
Figure 8). When using normalized features, the best balance between recall and precision is obtained with the RandomForest and MLP which both had an F-score of 0.93. The confusion matrix for RandomForest using a feature set without normalizing spectral features (
Table 17) and feature selection indicates that most errors in this case occur due to the poor true positive classification rate of Larch trees.
3.5. Individual Tree Detection and Species Prediction
The individual tree detection was evaluated visually and by using the reference data. A visual examination of the located trees and orthomosaics indicated that most of the trees in the test sites could be found. The most notable areas where the position of the detected trees and the trees visually observed from the orthomosaic differed were areas where the point clouds were incomplete due to matching failures or occlusions (for example, due to strong shadows or insufficient overlaps), and thus had local inaccuracies in the 3D point cloud and CHM. The most notable undetected trees were shorter trees at lower levels of the canopy, which could be seen from the orthomosaics but could not be detected from the point clouds due to the limited ability of passive sensors to penetrate the canopy. Some of the errors might have been induced by the fact that the reference trees were manually placed with the help of orthomosaics. The tree top peak in the point clouds might have been at a different location than the tree center visually detected from the orthomosaics.
The tree detection in the test sites was tested using the reference data with three different search radiuses (
Table 18). Increasing the search radius had a significant impact on the detection rate in some of the test sites, which indicates that there was a notable difference between the tree crown centers determined from image mosaics and automatic detection from CHM. The best results with 64%–96% tree identification rates were obtained with the 2 m search radius. However, due to the lack of reference data for testing, the accuracy estimation results of the tree detection are only indicative.
Visual inspection of the classification of the individual trees indicated that most of the trees were classified correctly. Most errors occurred between pines and spruces and in areas that had a lot of shadowing (for example in area v08, which was measured under completely sunny skies). The species-specific and area-specific prediction probabilities presented in
Table 19 indicate that the trained MLP classifier could classify most of the detected trees with a high probability (90%–97%). The best prediction could be provided for pines (97%), and thus also in test sites with many pines. The probability for the prediction of larch (90%) was the lowest, which also causes the v11 test site to have lowest mean probabilities since it has many larch trees.
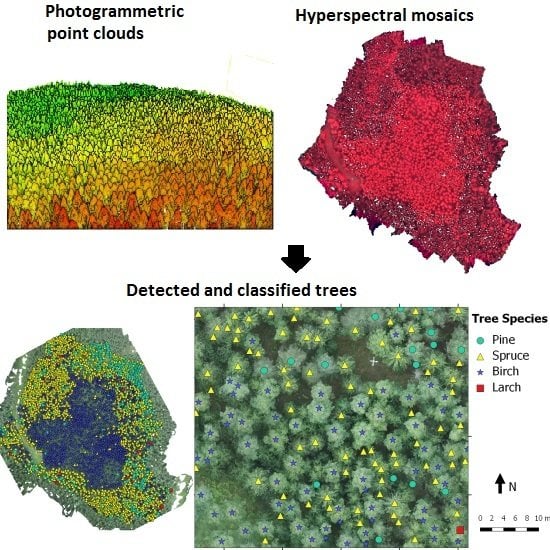
Figure 11 presents an example of test site v05 with detected and classified trees.
3.6. Processing Times
In the following, we present estimates of the computing times needed to process the datasets. The given time-estimates are computing times for the research system and non-optimized computers. In addition, the process included some interactive steps, such as measuring GCPs and reflectance panels in images, as well as interactive quality control; these steps are expected to impact minimally on the computation times when automated in the future.
Most of the processing time was spent on computing the image orientation and calculating the photogrammetric point clouds which took at the minimum 3.5 h and at the maximum 11.5 h for test sites v02 and v0304, respectively. The band-wise exterior orientation calculation took approximately 5 s for the individual band and image, resulting in processing times of 1.5 and 6 h for the smallest and largest blocks, respectively. The radiometric block adjustment and mosaic calculation took approximately 3–6 min per hypercube depending on the parameters and size of the area, thus resulting in 2 and 13 h for the smallest and largest blocks, respectively. These numbers are affected by many factors, such as the quality of approximate values, selected quality levels and complexity of the area.
The spectral and structural feature extraction took approximately 2.3 h and 10 h for all test sites, respectively. The used feature selection methods took approximately 5 min. The classification model learning and evaluation had much variability. The fastest classification method using the feature selected dataset was the k-NN method which could be trained and evaluated using the LOOCV method in less than one minute. For RandomForest and MLP methods, it took 3.6 h and 12.3 h, respectively. The complete individual tree detection workflow (i.e., from point cloud to tree locations) took approximately 40 min for all test sites and the tree species prediction of the detected trees using the trained MLP model took approximately 5 s for all trees.
4. Discussion
This investigation developed a small UAV hyperspectral imaging and a photogrammetry-based remote sensing method for individual tree detection and classification in a boreal forest. The method was assessed using, altogether, eleven forest test sites with 4151 reference trees.
4.1. Aspects of Data Processing
Datasets were captured in deep forest scene, and conditions during the data capture were typical for the northern climate zone during summer with variable cloudiness and rain showers. Variable conditions are challenging for analysis methods based on passive spectral information. The conventional radiometric correction methods based on reflectance panels or radiative transfer modeling [
42,
43,
44,
45,
46] are not suitable in such conditions. The novel radiometric processing approach based on radiometric block adjustment and onboard irradiance measurements provided promising results. The reflectance panels could not be utilized in several test sites in this study because they were too deep in the forest and shadowed by the trees. In many operational applications, the installation of reflectance panels is likely to be challenging; thus, it is important to develop methods that are independent of those. Important future improvements will include the enhancement of the irradiance spectrometer to be intolerant to platform tilting as well as improving the system calibration. The UAV system used in this study was equipped with the irradiance spectrometer but the data quality was not sufficient for performing accurate enough reflectance correction. Future enhancements are expected to improve the accuracy and level of automation and ultimately removing the need for ground reflectance panels. Approaches using object characteristics could also be suitable. For example, one possible method would be to use reflectance information of known natural targets in the scenes (such as trees) to perform radiometric normalization between different areas whose data has been collected in different flights. An important topic for future studies will be how different illumination conditions (direct sun illumination and different shadow depth) should be accounted for in the data analysis in a forested environment and which parts of it should be compensated for. For example, Korpela et al. [
15] classified spectral tree features based on the illumination condition derived from the 3D object model.
The processing and analysis were highly automated. The major interactive processes included the deployment of geometric and radiometric in situ targets and the field reference data capture. We expect that with future improvements in direct georeferencing performance and system radiometric calibration, the data processing chain could be fully automated. Further considerations are needed to automate the tree species reference data. For example, spectral libraries [
68] could provide a good reference if the data were properly calibrated. With this data set, we also used interactive analysis in some phases of the classification, but there are possibilities to automate these steps.
A lot of different software including commercial, open and in-house was chained and much of the software was not optimized for the material used in this study or with respect to speed. The computing times in photogrammetric and radiometric processing were approximately 8 h to 32 h depending on the size of the block; the combined feature extraction, selection and classification took approximately 25 h with the MLP method, which was the slowest method. All the operations can be highly automated, optimized and parallelized after sufficient information about the measurement problem is available, thus faster processing can be implemented in the future. In the future studies, we will also investigate more challenging forests with a larger variability in tree species.
4.2. Consideration of Identification and Classification Results
Spectral and 3D point cloud features were used in the classification. The feature selection results indicated that spectral features were more significant in discriminating different tree species than the structural features derived from the photogrammetric point clouds. Based on the feature selection and classification results, the normalizing spectral features proved to be slightly better than non-normalizing spectral features.
The best classification results were obtained with RandomForest and MLP which both gave 95% overall accuracies and F-scores of 0.93. No previous studies were found where both spectral and structural data measured from UAV had been used to classify individual trees. Previous UAV-based studies have been based on only multispectral data captured using NIR-modified cameras. Lisein et al. [
41] performed classification using RandomForest and achieved an overall classification error of 16% (out of bag error) with five different tree species. Michez et al. [
39] achieved a classification accuracy of 84% with five different tree species but the results were closer to pixel-based classification where a single tree was not represented as a whole.
In previous studies which have not been based on UAV data, Dalponte et al. [
69] have used high resolution airborne hyperspectral imagery to classify individual trees with 93.5% overall accuracy with three classes, namely pine, spruce and broadleaves. In addition, Lee et al. [
70] have tested combined ALS data and airborne hyperspectral imagery in tree species classification (six mainly deciduous species) in Northern European conditions, achieving users’ accuracies between 36.4% and 100% (total average = 61.1%) at the individual tree level. Vaughn et al. [
71] used ALS data in separating five tree species (two conifers and three deciduous) in the Pacific Northwest, U.S.A, achieving users’ accuracies between 81.5% and 95% (total average = 85.4%). An accuracy of 88%–90% was achieved by Korpela et al. [
72] in classifying Scots pine, Norway spruce, and birch using airborne LiDAR data.
Success rates in identifying individual trees based on the photogrammetric surface models with 5 cm point density were 26%–88%, 46%–95%, or 64%–96% for the search radius of 1 m, 1.5 m, or 2 m, respectively, used in evaluating the identification of the tree. Here, the geometric consistency of ground truth and the DSM used in the assessment of results plays an important role. The results are good compared to previous studies by Kaartinen et al. [
73] and Wang et al. [
74] where similar individual tree detection rates have been obtained for dominant trees (i.e., tallest trees in the neighborhood) using ALS data. Individual tree detection from photogrammetric point clouds created from UAV data have been tested, for example, by Sperclich et al. [
75] who achieved 90% detection rates for 2 m search radius, but the experiment concentrated only on a single forest plot and to the best of the authors’ knowledge, more extensive studies have not been conducted.
The local maxima-based method was successful for the boreal forest where the trees have crowns with a sharp shape, but it is expected that further tuning is needed to obtain successful results in forests with less separable crowns. Here, the spectral features might provide additional information.
Tree density has a strong impact on the detection accuracy of individual trees in forests using point cloud data [
73,
74]. However, the tree density estimates for our study area are based on tree densities for individual plots within each test site. These were calculated using the original reference data without manual reference data modification. The number of reference plots in each test site varied, and the tree densities in the plots in the same test site also had significant variations. Thus, the tree densities of the plots did not represent the entire test sites properly and this information could not be used reliably to evaluate the effect of tree densities to tree detection rates in the test sites.
Various classifiers used for tree species classification include, e.g., discriminant analysis, maximum likelihood, spectral angle mapper, support vector machine, radial basis function, neural networks, and nearest neighbor classifiers [
76]. Korpela et al. [
72] have tested discriminant analysis, k-nearest neighbor, and RandomForest estimators in tree species classification using airborne LiDAR data; the RandomForest method generally gave the best classification results. In this study, the Multilayer Perceptron (MLP) and RandomForest provided the best classification results. MLP has not been used in tree species classification as widely as RandomForest. A study by Feret and Asner [
77] used MLP in tree species discrimination in tropics but received poor results.
The reference data was slightly imbalanced, as pines formed the majority of the data. This had some small effects on classification training and performance evaluation. However, the effects were not perceived to be significant. It must be noted that this study used a constant one-meter radius circle to segment tree crown pixels. This might have an effect on the results, and in the future its effect should be studied to test whether automatic segmentation of individual crowns from images or CHMs could provide better results. Automatic tree crown segmentation is especially important with forests with broader and variable sized tree crowns. Spectral data only from the most nadir views of the photogrammetric image block were utilized in the classification; in further studies, the impact of using multiple views should also be studied.
5. Conclusions
This study investigated the ability of unmanned airborne vehicle (UAV) based photogrammetry and hyperspectral imaging in individual tree detection and classification in boreal forests. Altogether, eleven small forest test sites with 4151 reference trees were included in the analysis. A novel frame format hyperspectral imager based on a tunable Fabry–Pérot interferometer (FPI) was used in this study. The results were very promising, showing that small UAV-based remote sensing has the potential to provide accurate results in automatic tree detection and species classification even when data is captured under poor conditions, in variable illumination conditions. The spectral features derived from the hyperspectral images produced good results in tree species classification. The photogrammetric 3D point cloud features did not improve the species classification results in this study, but the individual tree detection from photogrammetric point clouds produced good results.
To the best of the authors’ knowledge, this was the first investigation studying tree species classification in a boreal forest utilizing small UAV-based hyperspectral frame imaging and photogrammetric point clouds. The classification study was comprehensive; it compared various classifiers and different features in the image classification task. The data set used was extensive and represented well the practical, challenging situations of UAV-based remote sensing tasks in boreal forests. Furthermore, we presented significant results of the potential of the novel radiometric block adjustment approach when integrated with UAV-based irradiance data in practical conditions.
Our study provided suggestions for a number of improvements in the entire remote sensing system, from the sensor technology to the final analysis process. The installation of reflectance panels and ground control points proved to be challenging in the densely-forested scene. Hardware improvements to the positioning and orientation system, the onboard irradiance measurement system, and better radiometric calibrations of the camera and spectrometer could enable better accuracy and automation in geometric and radiometric processing. These enhancements could even remove the need for ground reference targets. In addition, by using spectral libraries as reference data, the classifier learning process could be automated, reducing or completely removing the need for field-based training data collection. The hyperspectral frame format imaging also could provide many more features to the classification process than used in this study. For example, frame imaging technology allows the extraction of multiple features for each pixel instead of using a single spectral feature for each pixel. The extracted spectral features were based on all the spectral bands of the used hyperspectral imager and feature selection was utilized to identify the most significant spectral features for the classification. Comparisons to the approaches using derived spectral features, such as vegetation indices, will be investigated in the future. Further improvements to the point cloud processing workflow should include automation of the parameter selection for the canopy height model generation and tree detection based on the point cloud characteristics. These novel methods are expected to provide a powerful tool for automating various environmental close-range remote sensing tasks in the very near future; the technology is already operational and we expect that the performance will improve further with improved processing methods and sensor integration.