α-Linolenic Acid-Rich Diet Influences Microbiota Composition and Villus Morphology of the Mouse Small Intestine
Abstract
1. Introduction
2. Materials and Methods
2.1. Animals
2.2. Histological Analysis of the Small Intestine
2.2.1. Proliferation Assay
2.2.2. Periodic Acid-Schiff (PAS) Staining
2.3. GC-MS/MS Quantification of Fatty Acids in Intestinal Tissue and Diet
2.4. Microbial Composition of the Small Intestine
2.5. Metagenome Prediction and Characterization
2.6. Statistical Analysis of Fatty Acid Profile and Small Intestinal Morphology
3. Results
3.1. Fatty Acid Profile of Different Diets
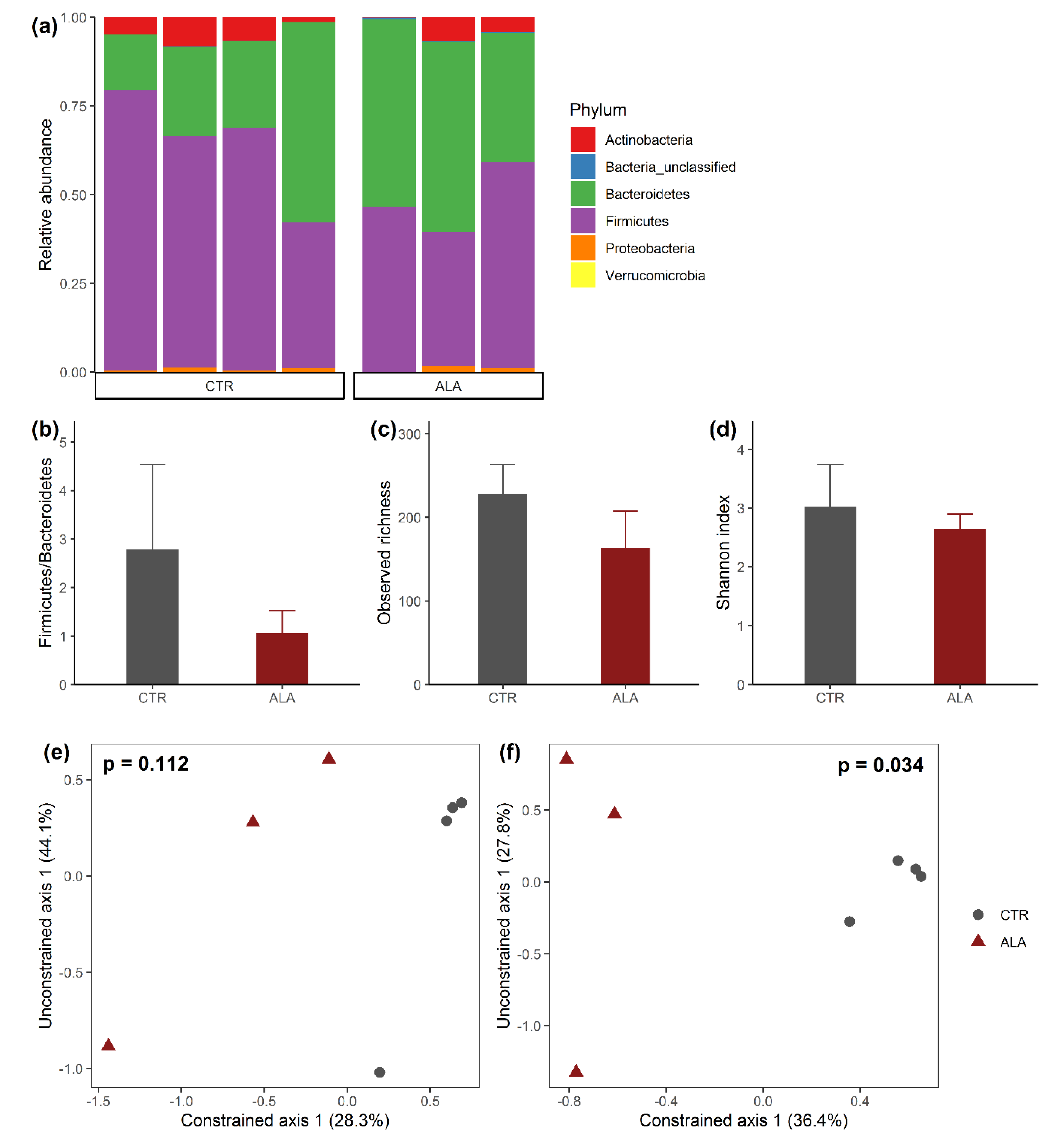
3.2. ALA-Rich Diet Alters the Composition of the Microbiota in the Mid Small Intestine of Adult Mice
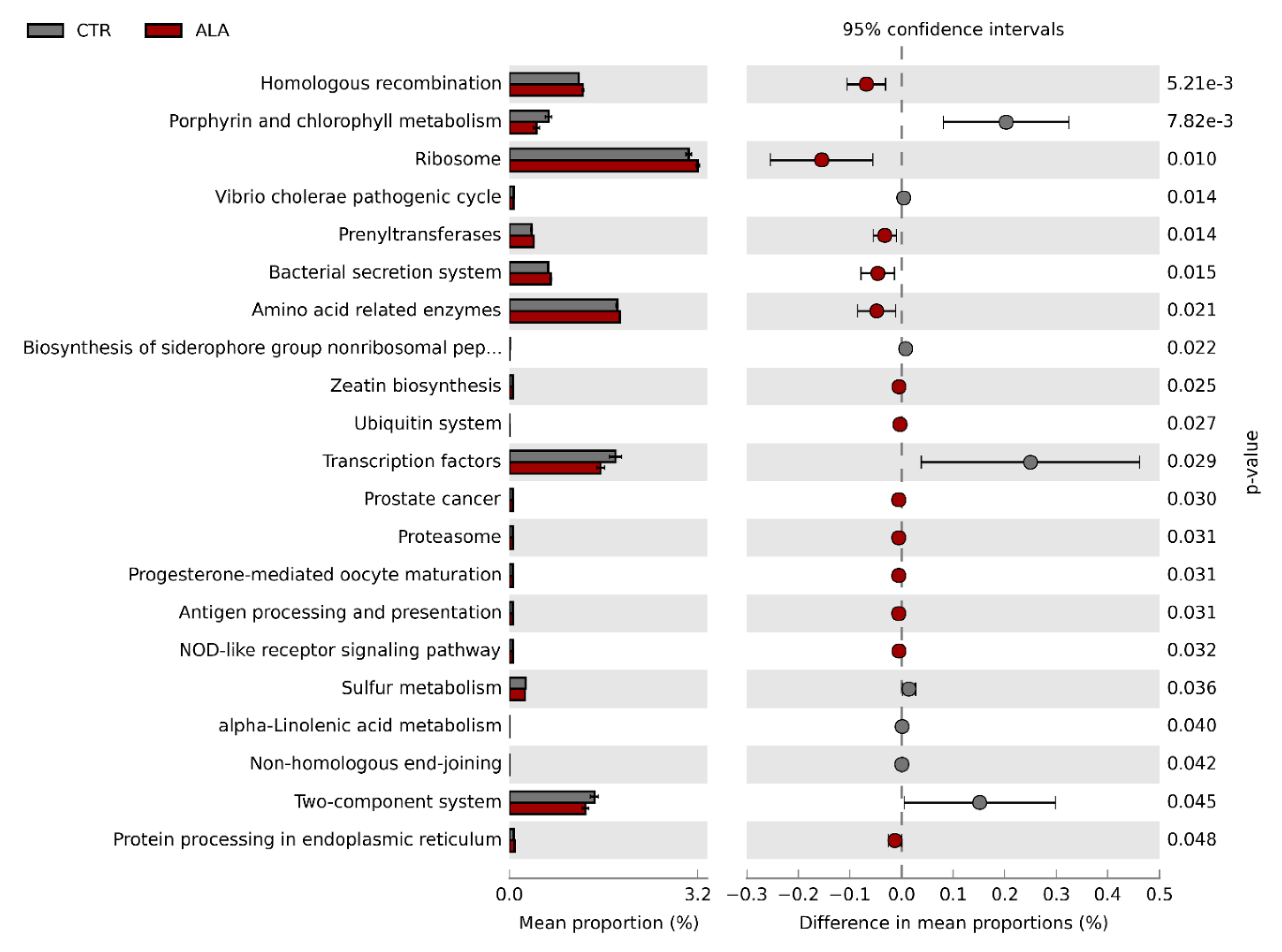
3.3. ALA-Rich Diet Might Impact the Metagenome Profile of Commensal Microbiota in the Small Intestine
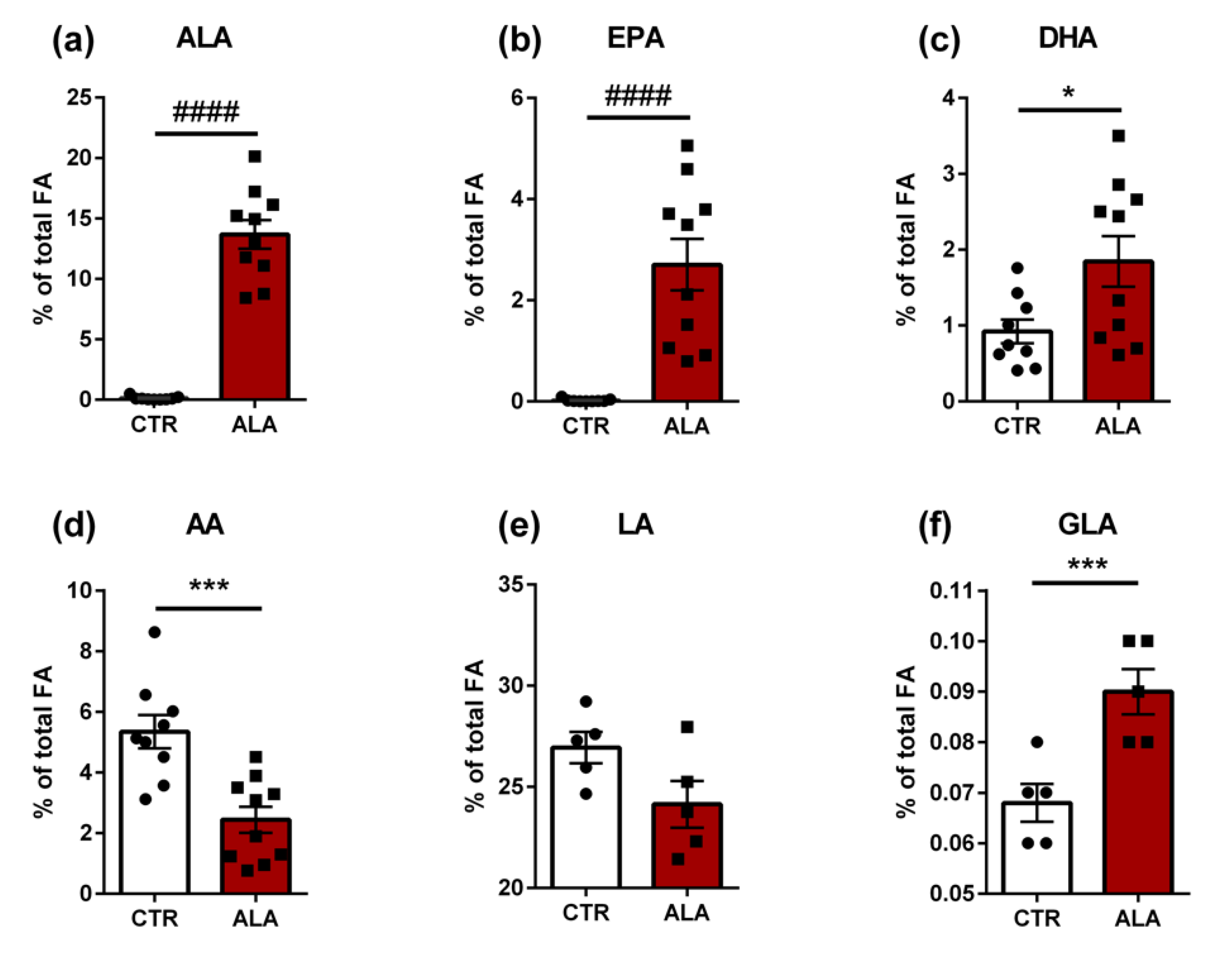
3.4. ALA-Rich Diet Induces Changes in the Fatty Acid Composition of the Jejunum
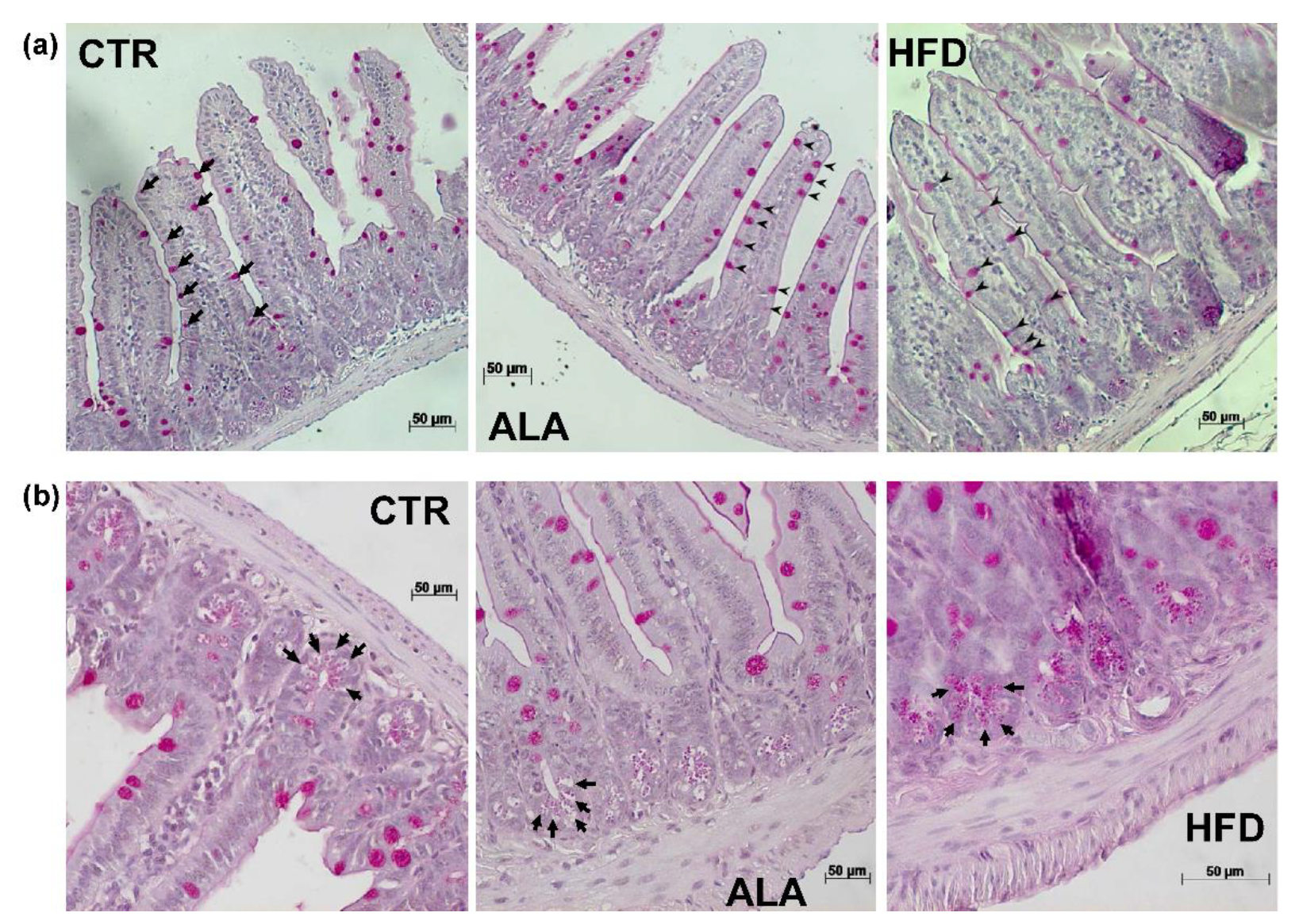
3.5. ALA-Rich Diet Shapes Small Intestinal Morphology
4. Discussion
Limitation of the Analysis
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
References
- Caligiuri, S.P.B.; Parikh, M.; Stamenkovic, A.; Pierce, G.N.; Aukema, H.M. Dietary modulation of oxylipins in cardiovascular disease and aging. Am. J. Physiol. Heart Circ. Physiol. 2017, 313, H903–H918. [Google Scholar] [CrossRef] [PubMed]
- Carroll, A.E.; Hwang, D.H. Decreased formation of porstaglandins derived from arachidonic acid by dietary linolenate in rats. Am. J. Clin. Nutr. 1980, 33, 590–597. [Google Scholar] [CrossRef]
- Horii, T.; Satouchi, K.; Kobayashi, Y.; Saito, K.; Watanabe, S.; Yoshida, Y.; Okuyama, H. Effect of dietary alpha-linolenate on platelet-activating factor production in rat peritoneal polymorphonuclear leukocytes. J. Immunol. 1991, 147, 1607. [Google Scholar]
- Thies, F.; Miles, E.A.; Nebe-von-Caron, G.; Powell, J.R.; Hurst, T.L.; Newsholme, E.A.; Calder, P.C. Influence of dietary supplementation with long-chain n−3 or n−6 polyunsaturated fatty acids on blood inflammatory cell populations and functions and on plasma soluble adhesion molecules in healthy adults. Lipids 2001, 36, 1183–1193. [Google Scholar] [CrossRef] [PubMed]
- Caligiuri, S.P.B.; Aukema, H.M.; Ravandi, A.; Guzman, R.; Dibrov, E.; Pierce, G.N. Flaxseed consumption reduces blood pressure in patients with hypertension by altering circulating oxylipins via an α-linolenic acid-induced inhibition of soluble epoxide hydrolase. Hypertension 2014, 64, 53–59. [Google Scholar] [CrossRef]
- Caligiuri, S.P.B.; Rodriguez-Leyva, D.; Aukema, H.M.; Ravandi, A.; Weighell, W.; Guzman, R.; Pierce, G.N. Dietary flaxseed reduces central aortic blood pressure without cardiac involvement but through changes in plasma oxylipins. Hypertension 2016, 68, 1031–1038. [Google Scholar] [CrossRef]
- Kulkarni, P.S.; Eakins, K.E. The enzymatic conversion of prostaglandin endoperoxides to thromboxane-A2-like activity by human iris microsomes. Prostaglandins 1977, 14, 601–605. [Google Scholar] [CrossRef]
- Ibrahim, A.; Aziz, M.; Hassan, A.; Mbodji, K.; Collasse, E.; Coëffier, M.; Bounoure, F.; Savoye, G.; Déchelotte, P.; Marion-Letellier, R. Dietary α-linolenic acid–rich formula reduces adhesion molecules in rats with experimental colitis. Nutrition 2012, 28, 799–802. [Google Scholar] [CrossRef]
- Reifen, R.; Karlinsky, A.; Stark, A.H.; Berkovich, Z.; Nyska, A. α-Linolenic acid (ALA) is an anti-inflammatory agent in inflammatory bowel disease. J. Nutr. Biochem. 2015, 26, 1632–1640. [Google Scholar] [CrossRef]
- Trebble, T.M.; Pearl, D.S.; Eiden, M.; Masoodi, M.; Brümmer, B.J.; Gullick, D.; Mills, G.; Brown, J.F.; Shute, J.K.; Mckeever, T.M.; et al. Altered colonic mucosal availability of n-3 and n-6 polyunsaturated fatty acids in ulcerative colitis and the relationship to disease activity. J. Crohns Colitis 2014, 8, 70–79. [Google Scholar] [CrossRef]
- Tyagi, A.; Kumar, U.; Reddy, S.; Santosh, V.S.; Mohammed, S.B.; Ehtesham, N.Z.; Ibrahim, A. Attenuation of colonic inflammation by partial replacement of dietary linoleic acid with α-linolenic acid in a rat model of inflammatory bowel disease. Br. J. Nutr. 2012, 108, 1612–1622. [Google Scholar] [CrossRef] [PubMed]
- Narisawa, T.; Takahashi, M.; Kotanagi, H.; Kusaka, H.; Yamazaki, Y.; Koyama, H.; Fukaura, Y.; Nishizawa, Y.; Kotsugai, M.; Isoda, Y. Inhibitory effect of dietary perilla oil rich in the n-3 polyunsaturated fatty acid alpha-linolenic acid on colon carcinogenesis in rats. Jpn. J. Cancer Res. Gann 1991, 82, 1089–1096. [Google Scholar] [CrossRef] [PubMed]
- Tian, Y.; Wang, H.; Yuan, F.; Li, N.; Huang, Q.; He, L.; Wang, L.; Liu, Z. Perilla oil has similar protective effects of fish oil on high-fat diet-induced nonalcoholic fatty liver disease and gut dysbiosis. BioMed Res. Int. 2016, 2016, 11. [Google Scholar] [CrossRef] [PubMed]
- Patterson, E.; O’Doherty, R.M.; Murphy, E.F.; Wall, R.; O’Sullivan, O.; Nilaweera, K.; Fitzgerald, G.F.; Cotter, P.D.; Ross, R.P.; Stanton, C. Impact of dietary fatty acids on metabolic activity and host intestinal microbiota composition in C57BL/6J mice. Br. J. Nutr. 2014, 111, 1905–1917. [Google Scholar] [CrossRef]
- Salsinha, A.S.; Pimentel, L.L.; Fontes, A.L.; Gomes, A.M.; Rodríguez-Alcalá, L.M. Microbial production of conjugated linoleic acid and conjugated linolenic acid relies on a multienzymatic system. Microbiol. Mol. Biol. Rev. 2018, 82. [Google Scholar] [CrossRef]
- Hennessy, A.A.; Ross, R.P.; Devery, R.; Stanton, C. The health promoting properties of the conjugated isomers of α-linolenic acid. Lipids 2011, 46, 105–119. [Google Scholar] [CrossRef]
- Yuan, G.-F.; Chen, X.-E.; Li, D. Conjugated linolenic acids and their bioactivities: A review. Food Funct. 2014, 5, 1360–1368. [Google Scholar] [CrossRef]
- Fontes, A.L.; Pimentel, L.L.; Simões, C.D.; Gomes, A.M.P.; Rodríguez-Alcalá, L.M. Evidences and perspectives in the utilization of CLNA isomers as bioactive compounds in foods. Crit. Rev. Food Sci. Nutr. 2017, 57, 2611–2622. [Google Scholar] [CrossRef]
- Gorissen, L.; Raes, K.; Weckx, S.; Dannenberger, D.; Leroy, F.; De Vuyst, L.; De Smet, S. Production of conjugated linoleic acid and conjugated linolenic acid isomers by Bifidobacterium species. Appl. Microbiol. Biotechnol. 2010, 87, 2257–2266. [Google Scholar] [CrossRef]
- Hennessy, A.A.; Barrett, E.; Paul Ross, R.; Fitzgerald, G.F.; Devery, R.; Stanton, C. The production of conjugated α-linolenic, γ-linolenic and stearidonic acids by strains of Bifidobacteria and Propionibacteria. Lipids 2012, 47, 313–327. [Google Scholar] [CrossRef]
- Kishino, S.; Ogawa, J.; Ando, A.; Shimizu, S. Conjugated α-linolenic acid production from α-linolenic acid by Lactobacillus plantarum AKU 1009a. Eur. J. Lipid Sci. Technol. 2003, 105, 572–577. [Google Scholar] [CrossRef]
- Ogawa, J.; Kishino, S.; Ando, A.; Sugimoto, S.; Mihara, K.; Shimizu, S. Production of conjugated fatty acids by lactic acid bacteria. J. Biosci. Bioeng. 2005, 100, 355–364. [Google Scholar] [CrossRef] [PubMed]
- Druart, C.; Bindels, L.B.; Schmaltz, R.; Neyrinck, A.M.; Cani, P.D.; Walter, J.; Ramer-Tait, A.E.; Delzenne, N.M. Ability of the gut microbiota to produce PUFA-derived bacterial metabolites: Proof of concept in germ-free versus conventionalized mice. Mol. Nutr. Food Res. 2015, 59, 1603–1613. [Google Scholar] [CrossRef] [PubMed]
- Ohue-Kitano, R.; Yasuoka, Y.; Goto, T.; Kitamura, N.; Park, S.-B.; Kishino, S.; Kimura, I.; Kasubuchi, M.; Takahashi, H.; Li, Y.; et al. α-Linolenic acid–derived metabolites from gut lactic acid bacteria induce differentiation of anti-inflammatory M2 macrophages through G protein-coupled receptor 40. FASEB J. 2017, 32, 304–318. [Google Scholar] [CrossRef]
- Benoit, B.; Kayal, F.; Bruno, J.; Estienne, M.; Plaisancié, P.; Debard, C.; Ducroc, R. Saturated and unsaturated fatty acids differently modulate colonic goblet cells in vitro and in rat pups. J. Nutr. 2015, 145, 1754–1762. [Google Scholar] [CrossRef] [PubMed]
- Konieczka, P.; Barszcz, M.; Choct, M.; Smulikowska, S. The interactive effect of dietary n-6: n-3 fatty acid ratio and vitamin E level on tissue lipid peroxidation, DNA damage in intestinal epithelial cells, and gut morphology in chickens of different ages. Poult. Sci. 2018, 97, 149–158. [Google Scholar] [CrossRef] [PubMed]
- Schober, Y.; Wahl, H.G.; Renz, H.; Nockher, W.A. Determination of red blood cell fatty acid profiles: Rapid and high-confident analysis by chemical ionization-gas chromatography-tandem mass spectrometry. J. Chromatogr. B 2017, 1040, 1–7. [Google Scholar] [CrossRef]
- Walters, W.; Hyde, E.R.; Berg-Lyons, D.; Ackermann, G.; Humphrey, G.; Parada, A.; Gilbert, J.A.; Jansson, J.K.; Caporaso, J.G.; Fuhrman, J.A.; et al. Improved bacterial 16S rRNA gene (V4 and V4-5) and fungal internal transcribed spacer marker gene primers for microbial community surveys. MSystems 2015, 1, e00009–e00015. [Google Scholar] [CrossRef]
- Parada, A.E.; Needham, D.M.; Fuhrman, J.A. Every base matters: Assessing small subunit rRNA primers for marine microbiomes with mock communities, time series and global field samples. Environ. Microbiol. 2016, 18, 1403–1414. [Google Scholar] [CrossRef]
- Apprill, A.; McNally, S.; Parsons, R.; Weber, L. Minor revision to V4 region SSU rRNA 806R gene primer greatly increases detection of SAR11 bacterioplankton. Aquat. Microb. Ecol. 2015, 75, 129–137. [Google Scholar] [CrossRef]
- Schloss, P.D.; Westcott, S.L.; Ryabin, T.; Hall, J.R.; Hartmann, M.; Hollister, E.B.; Lesniewski, R.A.; Oakley, B.B.; Parks, D.H.; Robinson, C.J.; et al. Introducing mothur: Open-source, platform-independent, community-supported software for describing and comparing microbial communities. Appl. Environ. Microbiol. 2009, 75, 7537–7541. [Google Scholar] [CrossRef] [PubMed]
- Kozich, J.J.; Westcott, S.L.; Baxter, N.T.; Highlander, S.K.; Schloss, P.D. Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform. Appl. Environ. Microbiol. 2013, 79, 5112–5120. [Google Scholar] [CrossRef] [PubMed]
- Rognes, T.; Flouri, T.; Nichols, B.; Quince, C.; Mahé, F. VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016, 4, e2584. [Google Scholar] [CrossRef] [PubMed]
- Fuchs, B.M.; Quast, C.; Pruesse, E.; Peplies, J.; Knittel, K.; Ludwig, W.; Glöckner, F.O. SILVA: A comprehensive online resource for quality checked and aligned ribosomal RNA sequence data compatible with ARB. Nucleic Acids Res. 2007, 35, 7188–7196. [Google Scholar] [CrossRef]
- Kulam-Syed-Mohideen, A.S.; Chai, B.; McGarrell, D.M.; Cardenas, E.; Garrity, G.M.; Fish, J.; Tiedje, J.M.; Wang, Q.; Farris, R.J.; Marsh, T.; et al. The Ribosomal Database Project: Improved alignments and new tools for rRNA analysis. Nucleic Acids Res. 2008, 37, D141–D145. [Google Scholar] [CrossRef]
- McMurdie, P.J.; Holmes, S. phyloseq: An R package for reproducible interactive analysis and graphics of microbiome census data. PLoS ONE 2013, 8, e61217. [Google Scholar] [CrossRef] [PubMed]
- Anderson, M.J.; Willis, T.J. Canonical analysis of principal coordinates: A useful method of constrained ordination for ecology. Ecology 2003, 84, 511–525. [Google Scholar] [CrossRef]
- Todorov, H.; Fournier, D.; Gerber, S. Principal components analysis: Theory and application to gene expression data analysis. Genom. Comput. Biol. 2018, 4, e100041. [Google Scholar] [CrossRef]
- Love, M.I.; Huber, W.; Anders, S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014, 15, 550. [Google Scholar] [CrossRef]
- Langille, M.G.I.; Zaneveld, J.; Caporaso, J.G.; McDonald, D.; Knights, D.; Reyes, J.A.; Clemente, J.C.; Burkepile, D.E.; Vega Thurber, R.L.; Knight, R.; et al. Predictive functional profiling of microbial communities using 16S rRNA marker gene sequences. Nat. Biotechnol. 2013, 31, 814. [Google Scholar] [CrossRef]
- McDonald, D.; Price, M.N.; Goodrich, J.; Nawrocki, E.P.; DeSantis, T.Z.; Probst, A.; Andersen, G.L.; Knight, R.; Hugenholtz, P. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 2012, 6, 610–618. [Google Scholar] [CrossRef] [PubMed]
- Parks, D.H.; Tyson, G.W.; Hugenholtz, P.; Beiko, R.G. STAMP: Statistical analysis of taxonomic and functional profiles. Bioinformatics 2014, 30, 3123–3124. [Google Scholar] [CrossRef]
- Shin, H.-S.; Kim, S.-W. Lipid composition of perilla seed. J. Am. Oil Chem. Soc. 1994, 71, 619–622. [Google Scholar] [CrossRef]
- Ding, Y.; Mokgolodi, N.C.; Hu, Y.; Shi, L.; Ma, C.; Liu, Y.-J. Characterization of fatty acid composition from five perilla seed oils in China and its relationship to annual growth temperature. J. Med. Plants Res. 2012, 6, 1645–1651. [Google Scholar]
- Peirett, P. Fatty acid content and chemical composition of vegetative parts of Perilla (Perilla frutescens) after different growth lengths. J. Med. Plants Res. 2011, 5, 72–78. [Google Scholar] [CrossRef]
- Gwari, G.; Lohani, H.; Haider, S.Z.; Bhandari, U.; Chauhan, N.; Rawat, D.S. Fatty acid and nutrient composition of Perilla (Perilla frutescens L.) accessions collected from Uttarakhand. Int. J. Phytopharm. 2014, 5, 379–382. [Google Scholar]
- Kiouptsi, K.; Jäckel, S.; Pontarollo, G.; Grill, A.; Kuijpers, M.J.E.; Wilms, E.; Weber, C.; Sommer, F.; Nagy, M.; Neideck, C.; et al. The Microbiota Promotes Arterial Thrombosis in Low-Density Lipoprotein Receptor-Deficient Mice. MBio 2019, 10, e02298-19. [Google Scholar] [CrossRef]
- Hildebrandt, M.A.; Hoffmann, C.; Sherrill-Mix, S.A.; Keilbaugh, S.A.; Hamady, M.; Chen, Y.-Y.; Knight, R.; Ahima, R.S.; Bushman, F.; Wu, G.D. High-fat diet determines the composition of the murine gut microbiome independently of obesity. Gastroenterology 2009, 137, 1716–1724. [Google Scholar] [CrossRef]
- Mujico, J.R.; Baccan, G.C.; Gheorghe, A.; Díaz, L.E.; Marcos, A. Changes in gut microbiota due to supplemented fatty acids in diet-induced obese mice. Br. J. Nutr. 2013, 110, 711–720. [Google Scholar] [CrossRef]
- Ley, R.E.; Bäckhed, F.; Turnbaugh, P.; Lozupone, C.A.; Knight, R.D.; Gordon, J.I. Obesity alters gut microbial ecology. Proc. Natl. Acad. Sci. USA 2005, 102, 11070–11075. [Google Scholar] [CrossRef]
- Ley, R.E.; Turnbaugh, P.J.; Klein, S.; Gordon, J.I. Human gut microbes associated with obesity. Nature 2006, 444, 1022. [Google Scholar] [CrossRef] [PubMed]
- Turnbaugh, P.J.; Ley, R.E.; Mahowald, M.A.; Magrini, V.; Mardis, E.R.; Gordon, J.I. An obesity-associated gut microbiome with increased capacity for energy harvest. Nature 2006, 444, 1027. [Google Scholar] [CrossRef] [PubMed]
- Balfegó, M.; Canivell, S.; Hanzu, F.A.; Sala-Vila, A.; Martínez-Medina, M.; Murillo, S.; Mur, T.; Ruano, E.G.; Linares, F.; Porras, N.; et al. Effects of sardine-enriched diet on metabolic control, inflammation and gut microbiota in drug-naïve patients with type 2 diabetes: A pilot randomized trial. Lipids Health Dis. 2016, 15, 78. [Google Scholar] [CrossRef] [PubMed]
- Biavati, B.; Vescovo, M.; Torriani, S.; Bottazzi, V. Bifidobacteria: History, ecology, physiology and applications. Ann. Microbiol. 2000, 50, 117–131. [Google Scholar]
- Druart, C.; Neyrinck, A.M.; Vlaeminck, B.; Fievez, V.; Cani, P.D.; Delzenne, N.M. Role of the lower and upper intestine in the production and absorption of gut microbiota-derived PUFA metabolites. PLoS ONE 2014, 9, e87560. [Google Scholar] [CrossRef] [PubMed]
- Lopez, M.S.; Tan, I.S.; Yan, D.; Kang, J.; McCreary, M.; Modrusan, Z.; Austin, C.D.; Xu, M.; Brown, E.J. Host-derived fatty acids activate type VII secretion in Staphylococcus aureus. Proc. Natl. Acad. Sci. USA 2017, 114, 11223–11228. [Google Scholar] [CrossRef]
- Kenny, J.G.; Ward, D.; Josefsson, E.; Jonsson, I.-M.; Hinds, J.; Rees, H.H.; Lindsay, J.A.; Tarkowski, A.; Horsburgh, M.J. The Staphylococcus aureus response to unsaturated long chain free fatty acids: Survival mechanisms and virulence implications. PLoS ONE 2009, 4, e4344. [Google Scholar] [CrossRef]
- Katan, M.B.; Zock, P.L.; Brouwer, I.A. Dietary α-linolenic acid is associated with reduced risk of fatal coronary heart disease, but increased prostate cancer risk: A meta-analysis. J. Nutr. 2004, 134, 919–922. [Google Scholar] [CrossRef]
- Ramon, J.M.; Bou, R.; Romea, S.; Alkiza, M.E.; Jacas, M.; Ribes, J.; Oromi, J. Dietary fat intake and prostate cancer risk: A case–control study in Spain. Cancer Causes Control 2000, 11, 679–685. [Google Scholar] [CrossRef]
- De Stéfani, E.; Deneo-Pellegrini, H.; Boffetta, P.; Ronco, A.; Mendilaharsu, M. α-linolenic acid and risk of prostate cancer: A case-control study in Uruguay. Cancer Epidemiol. Prev. Biomark. 2000, 9, 335–338. [Google Scholar]
- Dessì, M.; Noce, A.; Bertucci, P.; Manca di Villahermosa, S.; Zenobi, R.; Castagnola, V.; Addessi, E.; Di Daniele, N. Atherosclerosis, dyslipidemia, and inflammation: The significant role of polyunsaturated fatty acids. ISRN Inflamm. 2013, 2013, 191823. [Google Scholar] [CrossRef] [PubMed]
- Burdge, G.C. Metabolism of α-linolenic acid in humans. Prostaglandins Leukot. Essent. Fat. Acids 2006, 75, 161–168. [Google Scholar] [CrossRef] [PubMed]
- Punchard, N.A.; Green, A.T.; Mullins, J.G.; Thompson, R.P.H. Analysis of the intestinal absorption of essential fatty acids in vivo in the rat. Prostaglandins Leukot. Essent. Fat. Acids 2000, 62, 27–33. [Google Scholar] [CrossRef]
- Nagasawa, A.; Suzuki, J.; Hase, T.; Murase, T.; Wakisaka, T.; Tokimitsu, I. Dietary α-linolenic acid–rich diacylglycerols reduce body weight gain accompanying the stimulation of intestinal β-oxidation and related gene expressions in C57BL/KsJ-db/db mice. J. Nutr. 2002, 132, 3018–3022. [Google Scholar] [CrossRef]
- Wang, S.; Wu, D.; Matthan, N.R.; Lamon-Fava, S.; Lecker, J.L.; Lichtenstein, A.H. Reduction in dietary omega-6 polyunsaturated fatty acids: Eicosapentaenoic acid plus docosahexaenoic acid ratio minimizes atherosclerotic lesion formation and inflammatory response in the LDL receptor null mouse. Atherosclerosis 2009, 204, 147–155. [Google Scholar] [CrossRef]
- Jenkins, A.P.; Thompson, R.P. Does the fatty acid profile of dietary fat influence its trophic effect on the small intestinal mucosa? Gut 1993, 34, 358–364. [Google Scholar] [CrossRef]
- Abrams, G.D.; Baurer, H.; Sprinz, H. Influence of the normal flora on mucosal morphology and cellular renewal in the ileum. A comparison of germ-free and conventional mice. Lab. Investig. 1963, 12, 355–364. [Google Scholar]
- Zheng, X.; Tsuchiya, K.; Okamoto, R.; Iwasaki, M.; Kano, Y.; Sakamoto, N.; Nakamura, T.; Watanabe, M. Suppression of hath1 gene expression directly regulated by hes1 via notch signaling is associated with goblet cell depletion in ulcerative colitis. Inflamm. Bowel Dis. 2011, 17, 2251–2260. [Google Scholar] [CrossRef]
- Docter, D.; Westmeier, D.; Markiewicz, M.; Stolte, S.; Knauer, S.K.; Stauber, R.H. The nanoparticle biomolecule corona: Lessons learned—Challenge accepted. Chem. Soc. Rev. 2015, 44, 6094–6121. [Google Scholar] [CrossRef]
- Westmeier, D.; Hahlbrock, A.; Reinhardt, C.; Frohlich-Nowoisky, J.; Wessler, S.; Vallet, C.; Poschl, U.; Knauer, S.K.; Stauber, R.H. Nanomaterial-microbe cross-talk: Physicochemical principles and (patho)biological consequences. Chem. Soc. Rev. 2018, 47, 5312–5337. [Google Scholar] [CrossRef]
- Siemer, S.; Hahlbrock, A.; Vallet, C.; McClements, D.J.; Balszuweit, J.; Voskuhl, J.; Docter, D.; Wessler, S.; Knauer, S.K.; Westmeier, D.; et al. Nanosized food additives impact beneficial and pathogenic bacteria in the human gut: A simulated gastrointestinal study. NPJ Sci. Food 2018, 2, 22. [Google Scholar] [CrossRef] [PubMed]
- Stauber, R.H.; Siemer, S.; Becker, S.; Ding, G.B.; Strieth, S.; Knauer, S.K. Small Meets Smaller: Effects of Nanomaterials on Microbial Biology, Pathology, and Ecology. ACS Nano 2018, 12, 6351–6359. [Google Scholar] [CrossRef] [PubMed]
- Siemer, S.; Westmeier, D.; Vallet, C.; Steinmann, J.; Buer, J.; Stauber, R.H.; Knauer, S.K. Breaking resistance to nanoantibiotics by overriding corona-dependent inhibition using a pH-switch. Mater. Today 2018, 26, 19–29. [Google Scholar] [CrossRef]
- Weiss, S.; Xu, Z.Z.; Peddada, S.; Amir, A.; Bittinger, K.; Gonzalez, A.; Lozupone, C.; Zaneveld, J.R.; Vázquez-Baeza, Y.; Birmingham, A.; et al. Normalization and microbial differential abundance strategies depend upon data characteristics. Microbiome 2017, 5, 27. [Google Scholar] [CrossRef] [PubMed]



| Ingredient | Relative amount in control chow (% of diet) | Relative amount in ALA-Rich diet (% of diet) |
| Proteins | 17.61% | ~14.1% |
| Fat | 5.1% | ~24% |
| Fiber | 4.05% | ~3.24% |
| Disaccharides | 11.1% | ~8.88% |
| Polysaccharides | 47.2% | ~37.76% |
| Fatty acid | Relative amount (% of diet) | Relative amount (% of diet) |
| Palmitic acid C-16:0 | 0.36% | 4.1% |
| Stearic acid C-18:0 | 0.35% | 5.55% |
| Oleic acid C-18:1 cis | 0.93% | 1.61% |
| Linoleic acid C-18:2 cis | 3.3%% | 2.96% |
| α-Linolenic acid C-18:3 n3 | 0.03% | 9.27% |
| γ-Linolenic acid C18:3 n6 | 0.0002% | 0.05% |
| Arachidic acid C-20:0 | 0.04% | 0.21% |
| Eicosanoic acid C-20:1 | 0.01% | 0.04% |
| Behenic acid C-22:0 | 0.04% | 0.04% |
| Erucic acid C-22:1 | 0.02% | 0.14% |
| Lignoceric acid C-24:0 | 0.01% | 0.02% |
| Metabolizable energy | 3518 kcal/kg | ~4582 kcal/kg |
| Ingredient | Relative amount (% of diet) |
| Proteins | 17.3% |
| Carbohydrates | 48.5% |
| Fat | 21.2% |
| Fatty acid | Relative amount (% of diet) |
| Saturated fat | 13.1% |
| C-16:1 | 0.323% |
| Oleic acid C-18:1 cis | 4.43% |
| C-18:1 isomers | 0.85% |
| Linoleic acid C-18:2 cis | 0.49% |
| C-18:2 isomers | 0.28% |
| α-Linolenic acid C18:3 n3 | 0.15% |
| Metabolizable energy | 4500 kcal/kg |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Todorov, H.; Kollar, B.; Bayer, F.; Brandão, I.; Mann, A.; Mohr, J.; Pontarollo, G.; Formes, H.; Stauber, R.; Kittner, J.M.; et al. α-Linolenic Acid-Rich Diet Influences Microbiota Composition and Villus Morphology of the Mouse Small Intestine. Nutrients 2020, 12, 732. https://doi.org/10.3390/nu12030732
Todorov H, Kollar B, Bayer F, Brandão I, Mann A, Mohr J, Pontarollo G, Formes H, Stauber R, Kittner JM, et al. α-Linolenic Acid-Rich Diet Influences Microbiota Composition and Villus Morphology of the Mouse Small Intestine. Nutrients. 2020; 12(3):732. https://doi.org/10.3390/nu12030732
Chicago/Turabian StyleTodorov, Hristo, Bettina Kollar, Franziska Bayer, Inês Brandão, Amrit Mann, Julia Mohr, Giulia Pontarollo, Henning Formes, Roland Stauber, Jens M. Kittner, and et al. 2020. "α-Linolenic Acid-Rich Diet Influences Microbiota Composition and Villus Morphology of the Mouse Small Intestine" Nutrients 12, no. 3: 732. https://doi.org/10.3390/nu12030732
APA StyleTodorov, H., Kollar, B., Bayer, F., Brandão, I., Mann, A., Mohr, J., Pontarollo, G., Formes, H., Stauber, R., Kittner, J. M., Endres, K., Watzer, B., Nockher, W. A., Sommer, F., Gerber, S., & Reinhardt, C. (2020). α-Linolenic Acid-Rich Diet Influences Microbiota Composition and Villus Morphology of the Mouse Small Intestine. Nutrients, 12(3), 732. https://doi.org/10.3390/nu12030732








