Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition
Abstract
:1. Introduction
2. Materials and Methods
2.1. Animals and Diet
2.2. Plasma Biochemical Profile
2.3. Fasting Blood Glucose, Intraperitoneal Glucose Tolerance Test, and Homeostatic Index of Insulin Resistance
2.4. mRNA-Seq Experiment
2.5. Histopathology Analysis
2.6. Microbial Community Analysis
2.7. Statistical Analysis
3. Results
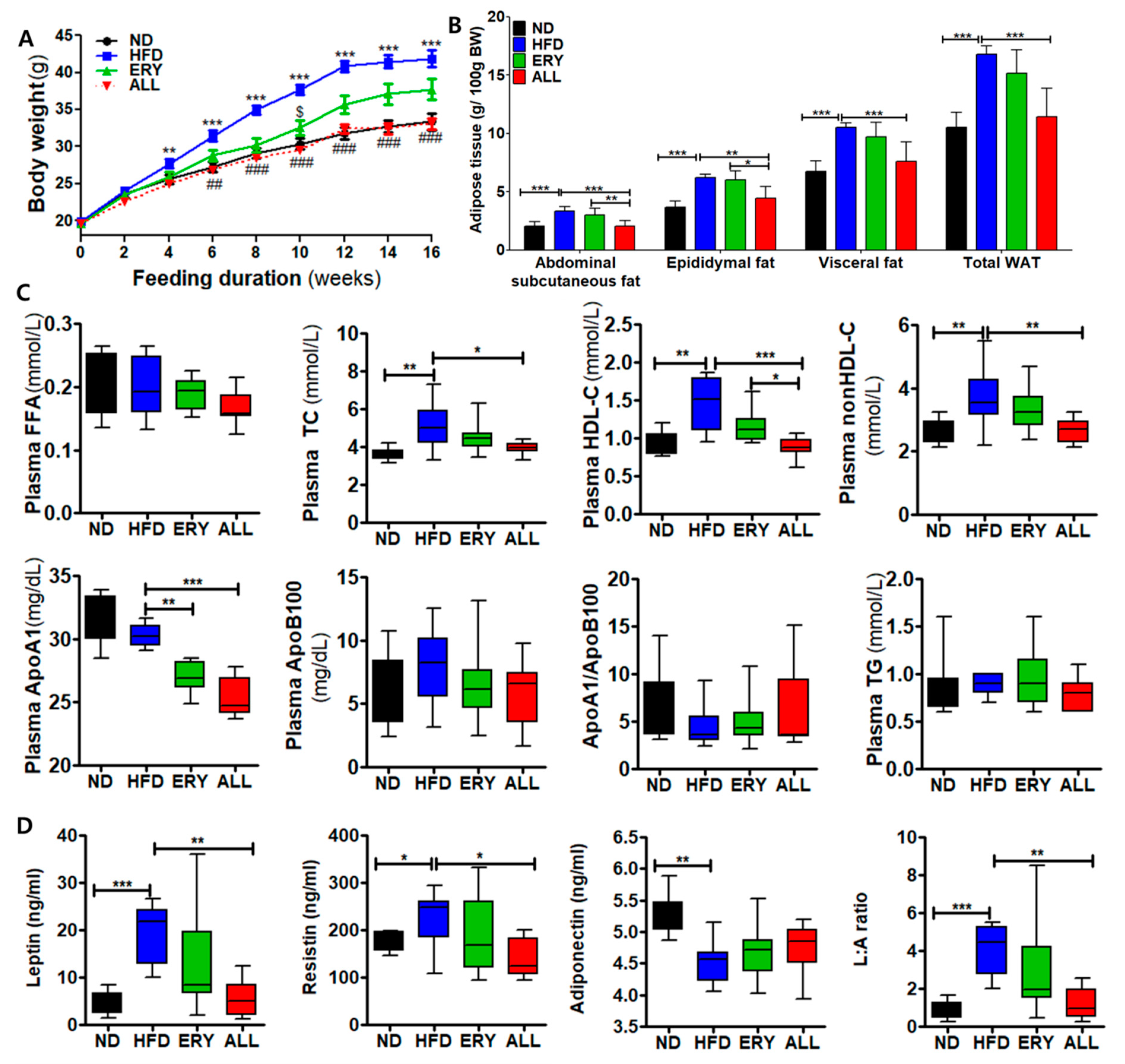
3.1. Allulose Lowered Body Fat and Plasma Lipid Levels in Diet-Induced Obese (DIO) Mice
3.2. Allulose Lowered Insulin Resistance and Glucose Tolerance by Modulating the Activities of Hepatic Glucose-Regulating Enzymes in DIO Mice
3.3. Allulose Reduced Hepatic Lipid Accumulation and Fibrosis by Modulating the Inflammatory Response in DIO Mice
3.4. Allulose Altered the Transcriptional Responses in the Liver Tissue of DIO Mice
3.5. Allulose Influenced the Microbiome Composition in DIO Mouse Feces
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Conflicts of Interest
References
- Takahara, M.; Shimomura, I. Metabolic syndrome and lifestyle modification. Rev. Endocr. Metab. Disord. 2014, 15, 317–327. [Google Scholar] [CrossRef] [PubMed]
- Redinger, R.N. Fat storage and the biology of energy expenditure. Transl. Res. 2009, 154, 52–60. [Google Scholar] [CrossRef] [PubMed]
- Strissel, K.J.; Stancheva, Z.; Miyoshi, H.; Perfield, J.W.; DeFuria, J.; Jick, Z.; Greenberg, A.S.; Obin, M.S. Adipocyte death, adipose tissue remodeling, and obesity complications. Diabetes 2007, 56, 2910–2918. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Whitmore, C. Type 2 diabetes and obesity in adults. Br. J. Nurs. 2010, 19, 880–886. [Google Scholar] [CrossRef]
- Sekirov, I.; Russell, S.L.; Antunes, L.C.M.; Finlay, B.B. Gut microbiota in health and disease. Physiol. Rev. 2010, 90, 859–904. [Google Scholar] [CrossRef] [Green Version]
- Everard, A.; Cani, P.D. Diabetes, obesity and gut microbiota. Best Pract. Res. Clin. Gastroenterol. 2013, 27, 73–83. [Google Scholar] [CrossRef] [Green Version]
- He, Y.; Wu, W.; Zheng, H.-M.; Li, P.; McDonald, D.; Sheng, H.-F.; Chen, M.-X.; Chen, Z.-H.; Ji, G.-Y.; Mujagond, P. Regional variation limits applications of healthy gut microbiome reference ranges and disease models. Nat. Med. 2018, 24, 1532–1535. [Google Scholar] [CrossRef]
- Lontchi-Yimagou, E.; Sobngwi, E.; Matsha, T.E.; Kengne, A.P. Diabetes mellitus and inflammation. Curr. Diabetes Rep. 2013, 13, 435–444. [Google Scholar] [CrossRef]
- Randeria, S.N.; Thomson, G.J.; Nell, T.A.; Roberts, T.; Pretorius, E. Inflammatory cytokines in type 2 diabetes mellitus as facilitators of hypercoagulation and abnormal clot formation. Cardiovasc. Diabetol. 2019, 18, 72. [Google Scholar] [CrossRef] [Green Version]
- Lee, E.; Jung, S.-R.; Lee, S.-Y.; Lee, N.-K.; Paik, H.-D.; Lim, S.-I. Lactobacillus plantarum strain Ln4 attenuates diet-induced obesity, insulin resistance, and changes in hepatic mRNA levels associated with glucose and lipid metabolism. Nutrients 2018, 10, 643. [Google Scholar] [CrossRef] [Green Version]
- Zhu, C.; Song, K.; Shen, Z.; Quan, Y.; Tan, B.; Luo, W.; Wu, S.; Tang, K.; Yang, Z.; Wang, X. Roseburia intestinalis inhibits interleukin-17 excretion and promotes regulatory T cells differentiation in colitis. Mol. Med. Rep. 2018, 17, 7567–7574. [Google Scholar] [CrossRef] [PubMed]
- Wang, X.; Ota, N.; Manzanillo, P.; Kates, L.; Zavala-Solorio, J.; Eidenschenk, C.; Zhang, J.; Lesch, J.; Lee, W.P.; Ross, J. Interleukin-22 alleviates metabolic disorders and restores mucosal immunity in diabetes. Nature 2014, 514, 237–241. [Google Scholar] [CrossRef] [PubMed]
- Liu, W.-C.; Yang, M.-C.; Wu, Y.-Y.; Chen, P.-H.; Hsu, C.-M.; Chen, L.-W. Lactobacillus plantarum reverse diabetes-induced Fmo3 and ICAM expression in mice through enteric dysbiosis-related c-Jun NH2-terminal kinase pathways. PLoS ONE 2018, 13, e0196511. [Google Scholar] [CrossRef] [PubMed]
- Komaroff, A.L. The microbiome and risk for obesity and diabetes. JAMA 2017, 317, 355–356. [Google Scholar] [CrossRef] [PubMed]
- Forslund, K.; Hildebrand, F.; Nielsen, T.; Falony, G.; Le Chatelier, E.; Sunagawa, S.; Prifti, E.; Vieira-Silva, S.; Gudmundsdottir, V.; Pedersen, H.K. Disentangling type 2 diabetes and metformin treatment signatures in the human gut microbiota. Nature 2015, 528, 262–266. [Google Scholar] [CrossRef] [PubMed]
- Bray, G.A.; Nielsen, S.J.; Popkin, B.M. Consumption of high-fructose corn syrup in beverages may play a role in the epidemic of obesity. Am. J. Clin. Nutr. 2004, 79, 537–543. [Google Scholar] [CrossRef]
- Ludwig, D.S.; Peterson, K.E.; Gortmaker, S.L. Relation between consumption of sugar-sweetened drinks and childhood obesity: A prospective, observational analysis. Lancet 2001, 357, 505–508. [Google Scholar] [CrossRef]
- Malik, V.S.; Popkin, B.M.; Bray, G.A.; Després, J.-P.; Willett, W.C.; Hu, F.B. Sugar-sweetened beverages and risk of metabolic syndrome and type 2 diabetes: A meta-analysis. Diabetes Care 2010, 33, 2477–2483. [Google Scholar] [CrossRef] [Green Version]
- Ludwig, D.S.; Ebbeling, C.B. The carbohydrate-insulin model of obesity: Beyond “calories in, calories out”. JAMA Intern. Med. 2018, 178, 1098–1103. [Google Scholar] [CrossRef]
- Tandel, K.R. Sugar substitutes: Health controversy over perceived benefits. J. Pharmacol. Pharmacother. 2011, 2, 236. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kroger, M.; Meister, K.; Kava, R. Low-calorie sweeteners and other sugar substitutes: A review of the safety issues. Compr. Rev. Food Sci. Food Saf. 2006, 5, 35–47. [Google Scholar] [CrossRef]
- Han, Y.; Han, H.J.; Kim, A.H.; Choi, J.Y.; Cho, S.J.; Park, Y.B.; Jung, U.J.; Choi, M.S. d-Allulose supplementation normalized the body weight and fat-pad mass in diet-induced obese mice via the regulation of lipid metabolism under isocaloric fed condition. Mol. Nutr. Food Res. 2016, 60, 1695–1706. [Google Scholar] [CrossRef]
- Hossain, A.; Yamaguchi, F.; Matsuo, T.; Tsukamoto, I.; Toyoda, Y.; Ogawa, M.; Nagata, Y.; Tokuda, M. Rare sugar D-allulose: Potential role and therapeutic monitoring in maintaining obesity and type 2 diabetes mellitus. Pharmacol. Ther. 2015, 155, 49–59. [Google Scholar] [CrossRef]
- Iwasaki, Y.; Sendo, M.; Dezaki, K.; Hira, T.; Sato, T.; Nakata, M.; Goswami, C.; Aoki, R.; Arai, T.; Kumari, P. GLP-1 release and vagal afferent activation mediate the beneficial metabolic and chronotherapeutic effects of D-allulose. Nat. Commun. 2018, 9, 1–17. [Google Scholar] [CrossRef] [PubMed]
- Noronha, J.C.; Braunstein, C.R.; Glenn, A.J.; Khan, T.A.; Viguiliouk, E.; Noseworthy, R.; Blanco Mejia, S.; Kendall, C.W.; Wolever, T.M.; Leiter, L.A. The effect of small doses of fructose and allulose on postprandial glucose metabolism in type 2 diabetes: A double-blind, randomized, controlled, acute feeding, equivalence trial. Diabetes Obes. Metab. 2018, 20, 2361–2370. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Kwon, E.-Y.; Yu, M.K.; Lee, S.J.; Kim, H.-J.; Kim, S.-B.; Kim, Y.H.; Choi, M.-S. A preliminary study for evaluating the dose-dependent effect of D-allulose for fat mass reduction in adult humans: A randomized, double-blind, placebo-controlled trial. Nutrients 2018, 10, 160. [Google Scholar] [CrossRef] [Green Version]
- Matthews, D.; Hosker, J.; Rudenski, A.; Naylor, B.; Treacher, D.; Turner, R. Homeostasis model assessment: Insulin resistance and β-cell function from fasting plasma glucose and insulin concentrations in man. Diabetologia 1985, 28, 412–419. [Google Scholar] [CrossRef] [Green Version]
- Glasgow, V.L.; Foreman, G. U-search: A program to teach end user searching at an academic health sciences library. In The End User Search in the Health Science; Taylor & Francis Online: Oxfordshire, UK, 2013; p. 137. [Google Scholar]
- DeSantis, T.Z.; Hugenholtz, P.; Larsen, N.; Rojas, M.; Brodie, E.L.; Keller, K.; Huber, T.; Dalevi, D.; Hu, P.; Andersen, G.L. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 2006, 72, 5069–5072. [Google Scholar] [CrossRef] [Green Version]
- Benjamini, Y.; Hochberg, Y. Controlling the false discovery rate: A practical and powerful approach to multiple testing. J. R. Stat. Soc. 1995, 57, 289–300. [Google Scholar] [CrossRef]
- Park, P.-J.; Kim, C.-W.; Cho, S.-Y.; Rha, C.-S.; Seo, D.-B.; Lee, S.-J. Aqueous spray-dried green tea extract regulates body weight and epididymal fat accumulation in mice. Korean J. Food Sci. Technol. 2010, 42, 103–108. [Google Scholar]
- White, U.A.; Tchoukalova, Y.D. Sex dimorphism and depot differences in adipose tissue function. Biochim. Et Biophys. Acta 2014, 1842, 377–392. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Fabbrini, E.; Sullivan, S.; Klein, S. Obesity and nonalcoholic fatty liver disease: Biochemical, metabolic, and clinical implications. Hepatology 2010, 51, 679–689. [Google Scholar] [CrossRef] [PubMed]
- Browning, J.D.; Szczepaniak, L.S.; Dobbins, R.; Nuremberg, P.; Horton, J.D.; Cohen, J.C.; Grundy, S.M.; Hobbs, H.H. Prevalence of hepatic steatosis in an urban population in the United States: Impact of ethnicity. Hepatology 2004, 40, 1387–1395. [Google Scholar] [CrossRef] [PubMed]
- Yu, X.X.; Pandey, S.K.; Booten, S.L.; Murray, S.F.; Monia, B.P.; Bhanot, S. Reduced adiposity and improved insulin sensitivity in obese mice with antisense suppression of 4E-BP2 expression. Am. J. Physiol. 2008, 294, E530–E539. [Google Scholar] [CrossRef] [Green Version]
- Hatting, M.; Tavares, C.D.; Sharabi, K.; Rines, A.K.; Puigserver, P. Insulin regulation of gluconeogenesis. Ann. New York Acad. Sci. 2018, 1411, 21. [Google Scholar] [CrossRef]
- Scheen, A. Pathophysiology of type 2 diabetes. Acta Clin. Belg. 2003, 58, 335–341. [Google Scholar] [CrossRef]
- Ritchie, S.; Connell, J. The link between abdominal obesity, metabolic syndrome and cardiovascular disease. Nutr. Metab. Cardiovasc. Dis. 2007, 17, 319–326. [Google Scholar] [CrossRef]
- Kahn, S. The relative contributions of insulin resistance and beta-cell dysfunction to the pathophysiology of type 2 diabetes. Diabetologia 2003, 46, 3–19. [Google Scholar] [CrossRef] [Green Version]
- Hotamisligil, G.S. Inflammation and metabolic disorders. Nature 2006, 444, 860–867. [Google Scholar] [CrossRef]
- Shah, A.; Mehta, N.; Reilly, M.P. Adipose inflammation, insulin resistance, and cardiovascular disease. J. Parenter. Enter. Nutr. 2008, 32, 638–644. [Google Scholar] [CrossRef] [Green Version]
- Galbo, T.; Perry, R.J.; Jurczak, M.J.; Camporez, J.-P.G.; Alves, T.C.; Kahn, M.; Guigni, B.A.; Serr, J.; Zhang, D.; Bhanot, S. Saturated and unsaturated fat induce hepatic insulin resistance independently of TLR-4 signaling and ceramide synthesis in vivo. Proc. Natl. Acad. Sci. USA 2013, 110, 12780–12785. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Han, Y.; Yoon, J.; Choi, M.S. Tracing the Anti-Inflammatory Mechanism/Triggers of D-Allulose: A Profile Study of Microbiome Composition and mRNA Expression in Diet-Induced Obese Mice. Mol. Nutr. Food Res. 2020, 64, 1900982. [Google Scholar] [CrossRef] [PubMed]
- Li, Q.; Chang, Y.; Zhang, K.; Chen, H.; Tao, S.; Zhang, Z. implication of the gut microbiome composition of type 2 diabetic patients from northern china. Sci. Rep. 2020, 10, 1–8. [Google Scholar] [CrossRef]
- Sanz, Y.; Rastmanesh, R.; Agostonic, C. Understanding the role of gut microbes and probiotics in obesity: How far are we? Pharmacol. Res. 2013, 69, 144–155. [Google Scholar] [CrossRef] [PubMed]
- Han, Y.; Park, H.; Choi, B.-R.; Ji, Y.; Kwon, E.-Y.; Choi, M.-S. Alteration of Microbiome Profile by D-Allulose in Amelioration of High-Fat-Diet-Induced Obesity in Mice. Nutrients 2020, 12, 352. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Horie, M.; Miura, T.; Hirakata, S.; Hosoyama, A.; Sugino, S.; Umeno, A.; Murotomi, K.; Yoshida, Y.; Koike, T. Comparative analysis of the intestinal flora in type 2 diabetes and nondiabetic mice. Exp. Anim. 2017, 66, 405–416. [Google Scholar] [CrossRef] [Green Version]
- Zhang, F.; Wang, M.; Yang, J.; Xu, Q.; Liang, C.; Chen, B.; Zhang, J.; Yang, Y.; Wang, H.; Shang, Y. Response of gut microbiota in type 2 diabetes to hypoglycemic agents. Endocrine 2019, 66, 485–493. [Google Scholar] [CrossRef] [PubMed]
- De La Cuesta-Zuluaga, J.; Mueller, N.T.; Corrales-Agudelo, V.; Velásquez-Mejía, E.P.; Carmona, J.A.; Abad, J.M.; Escobar, J.S. Metformin is associated with higher relative abundance of mucin-degrading Akkermansia muciniphila and several short-chain fatty acid–producing microbiota in the gut. Diabetes Care 2017, 40, 54–62. [Google Scholar] [CrossRef] [PubMed] [Green Version]




| ND | HFD | ERY | ALL | |
|---|---|---|---|---|
| Initial BW (g) | 19.96 ± 1.13 | 19.94 ± 1.11 | 19.61 ± 1.12 | 19.66 ± 1.41 |
| Final BW (g) | 33.34 ± 1.49 | 41.85 ± 3.19 *** | 37.72 ± 3.98 | 33.11 ± 2.13 ### |
| Total BWG (g/16 wk) | 13.38 ± 1.81 | 21.91 ± 2.99 *** | 18.11 ± 3.50 | 13.45 ± 2.64 ### |
| Food Intake (g/day) | 3.65 ± 0.10 | 3.06 ± 0.15 | 2.62 ± 0.12 $ | 3.20 ± 0.16 |
| Energy Intake (kcal/day) | 14.24 ± 0.39 | 14.03 ± 0.68 | 11.48 ± 0.53 $ | 14.03 ± 0.73 |
| FER | 0.03 ± 0.01 | 0.06 ± 0.00 *** | 0.06 ± 0.00 | 0.04 ± 0.00 ##, @@ |
© 2020 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (http://creativecommons.org/licenses/by/4.0/).
Share and Cite
Han, Y.; Kwon, E.-Y.; Choi, M.-S. Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition. Nutrients 2020, 12, 2113. https://doi.org/10.3390/nu12072113
Han Y, Kwon E-Y, Choi M-S. Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition. Nutrients. 2020; 12(7):2113. https://doi.org/10.3390/nu12072113
Chicago/Turabian StyleHan, Youngji, Eun-Young Kwon, and Myung-Sook Choi. 2020. "Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition" Nutrients 12, no. 7: 2113. https://doi.org/10.3390/nu12072113
APA StyleHan, Y., Kwon, E. -Y., & Choi, M. -S. (2020). Anti-Diabetic Effects of Allulose in Diet-Induced Obese Mice via Regulation of mRNA Expression and Alteration of the Microbiome Composition. Nutrients, 12(7), 2113. https://doi.org/10.3390/nu12072113





