Clinically Relevant Genes and Proteins Modulated by Tocotrienols in Human Colon Cancer Cell Lines: Systematic Scoping Review
Abstract
:1. Introduction
2. Materials and Methods
2.1. Search Strategy
2.2. Search Method
2.3. Criteria to Select Studies
2.4. Functional Bioinformatics Analysis
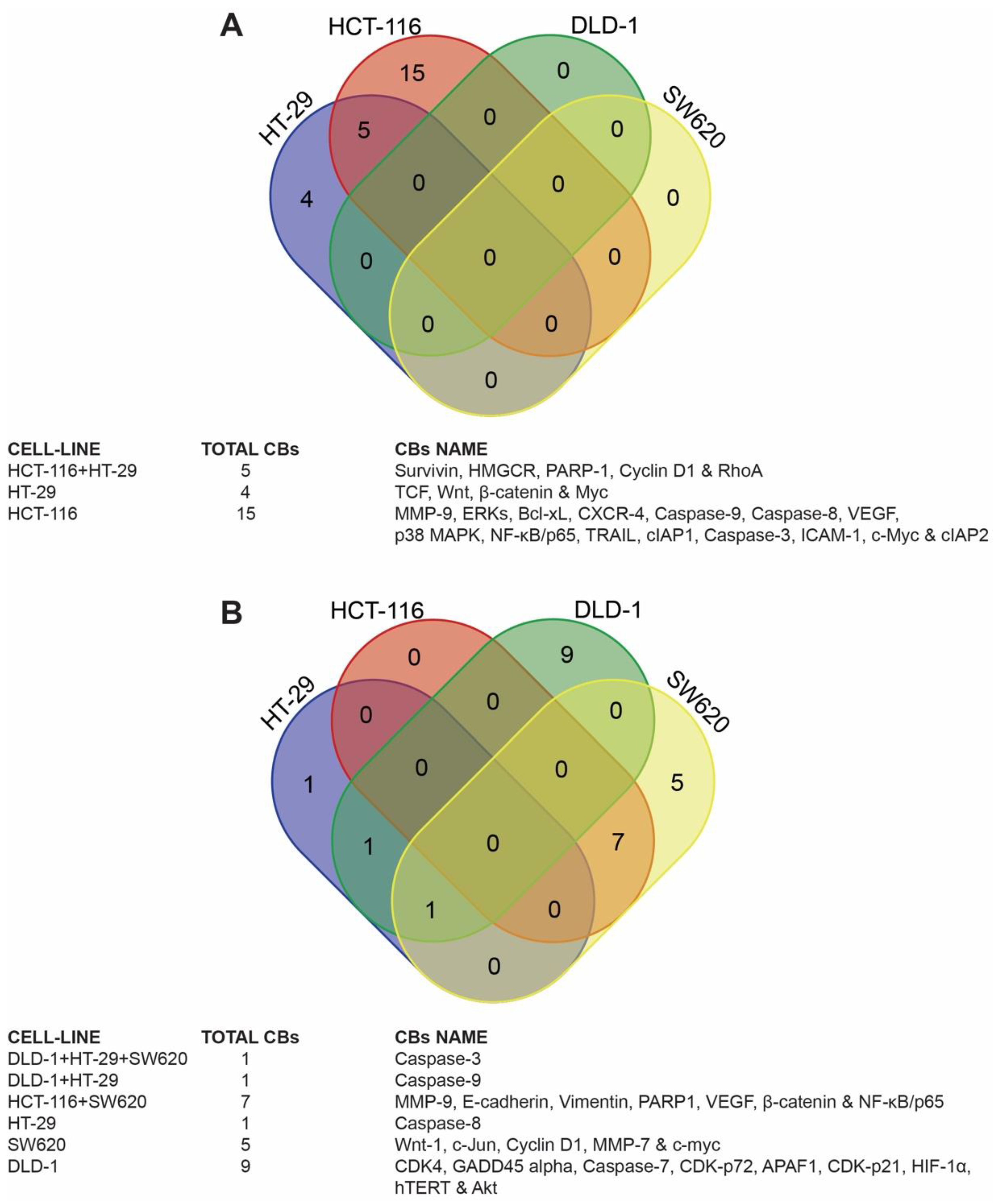
2.4.1. Venn Diagram Analysis
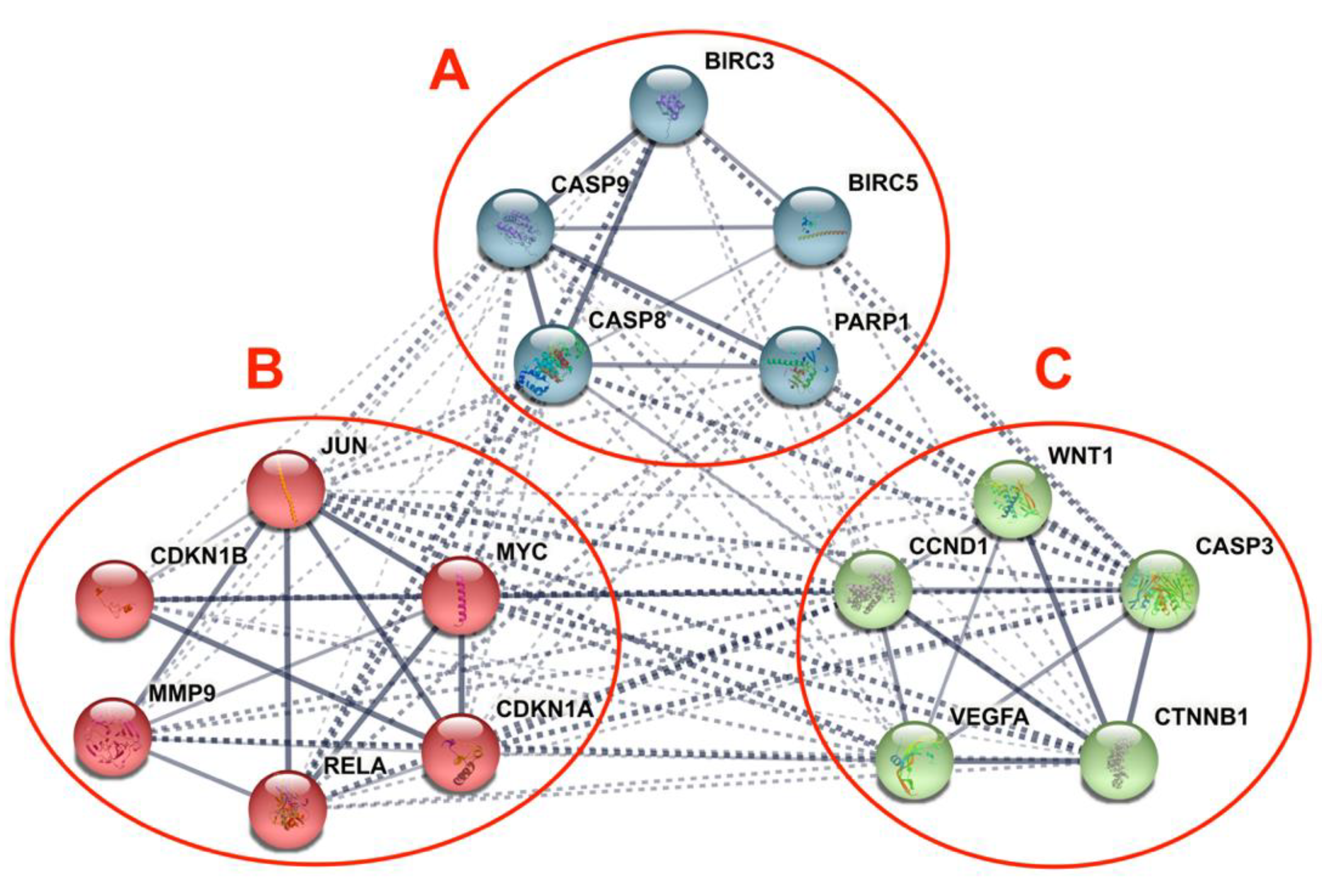
2.4.2. STRING Interaction Analysis
2.4.3. Cytoscape Molecular Network Analysis
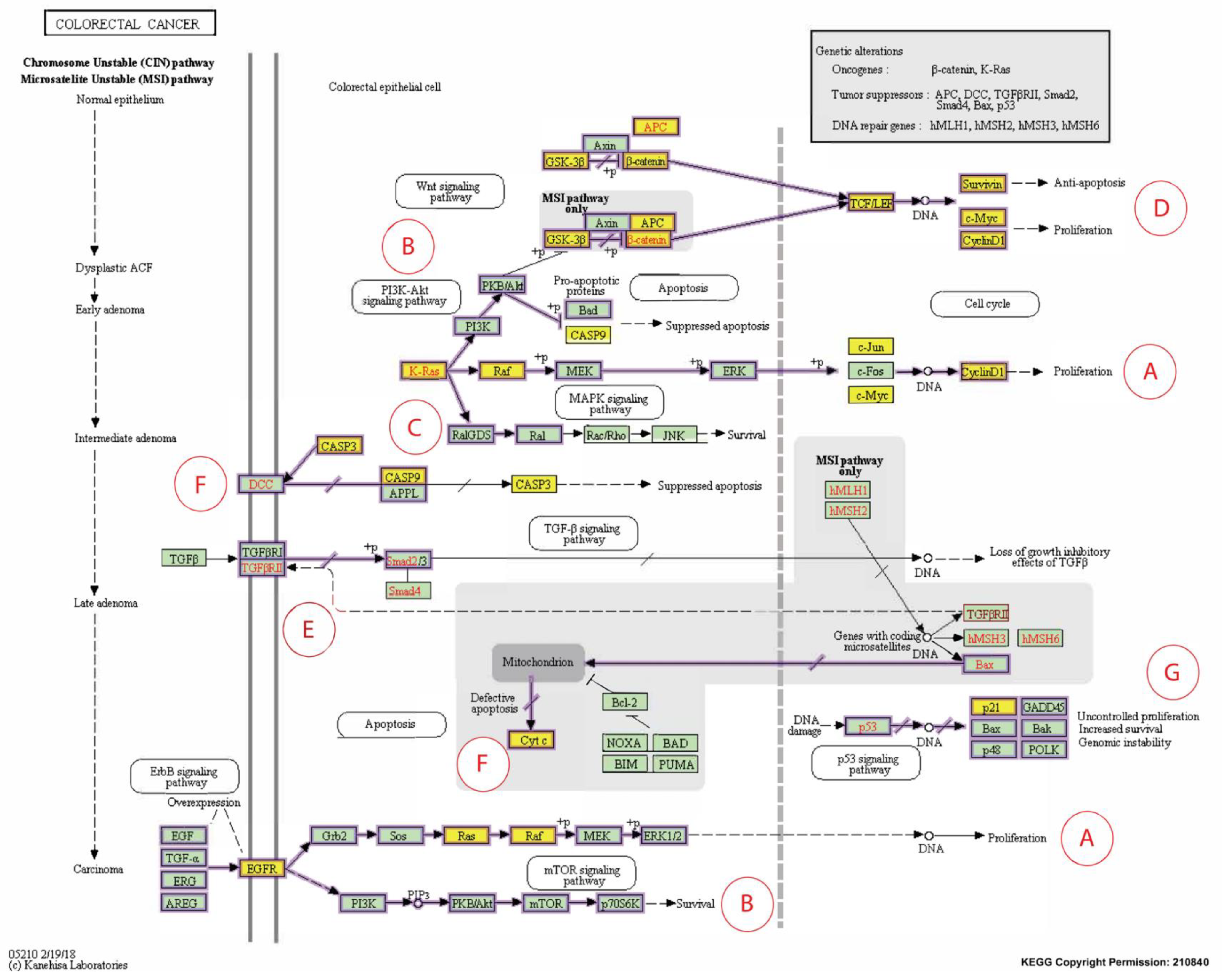
2.4.4. KEGG Pathway Analysis
3. Results
3.1. Selection of Articles
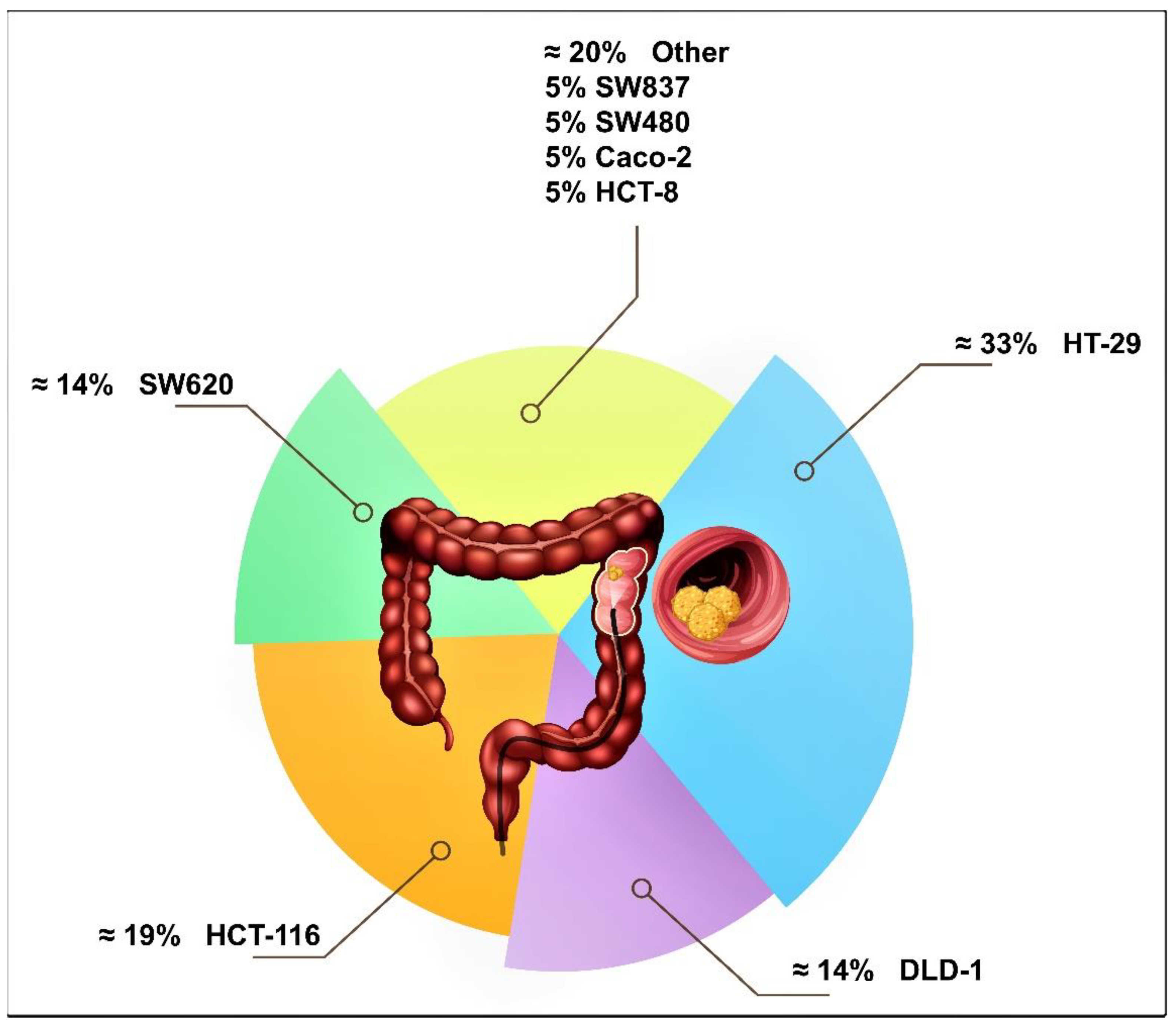
3.2. Human Colon Cancer Cell Lines Studied
3.3. Identification of Candidate Biomarkers in the Human Colon Cancer Cell Lines
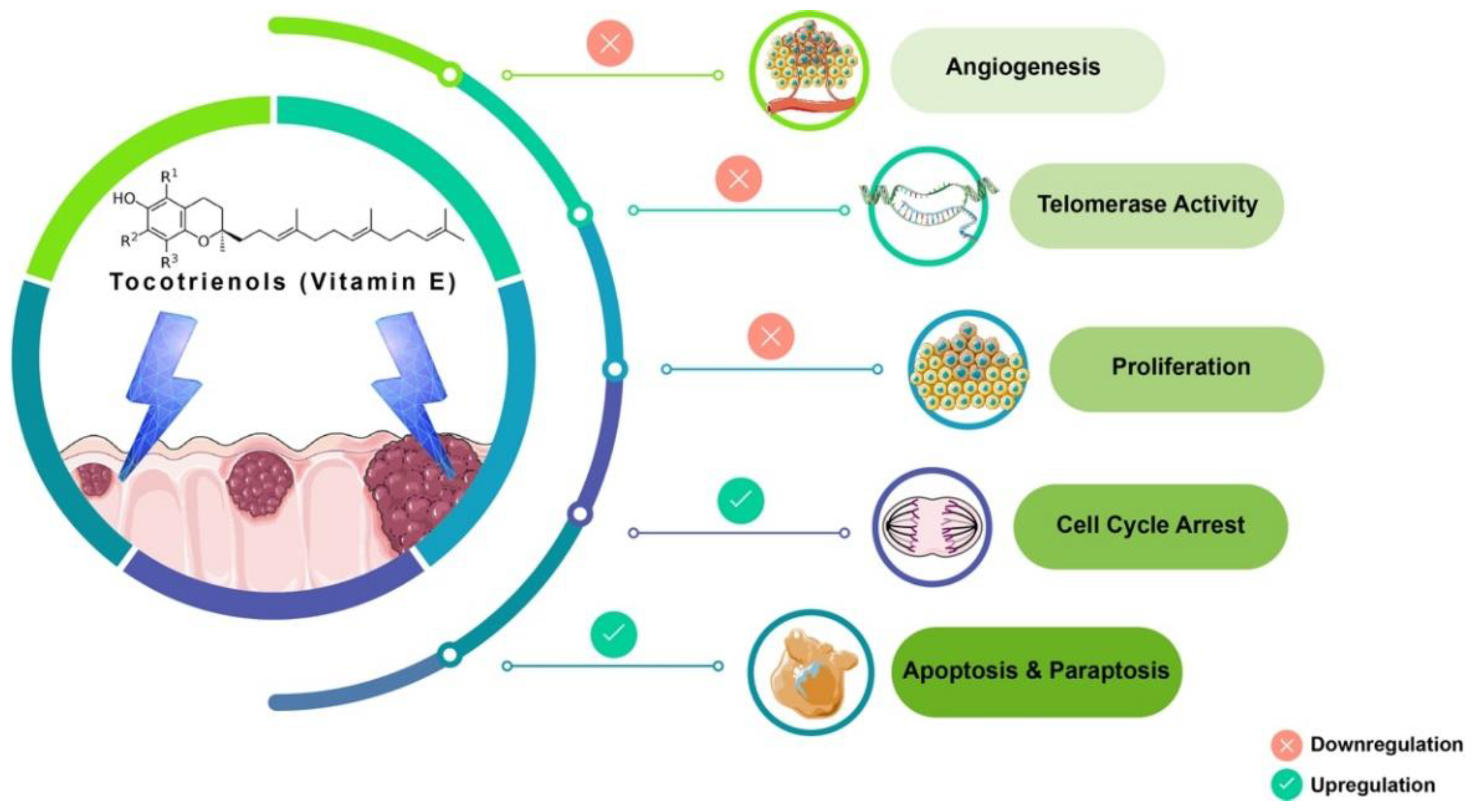
3.4. Target Biomarkers
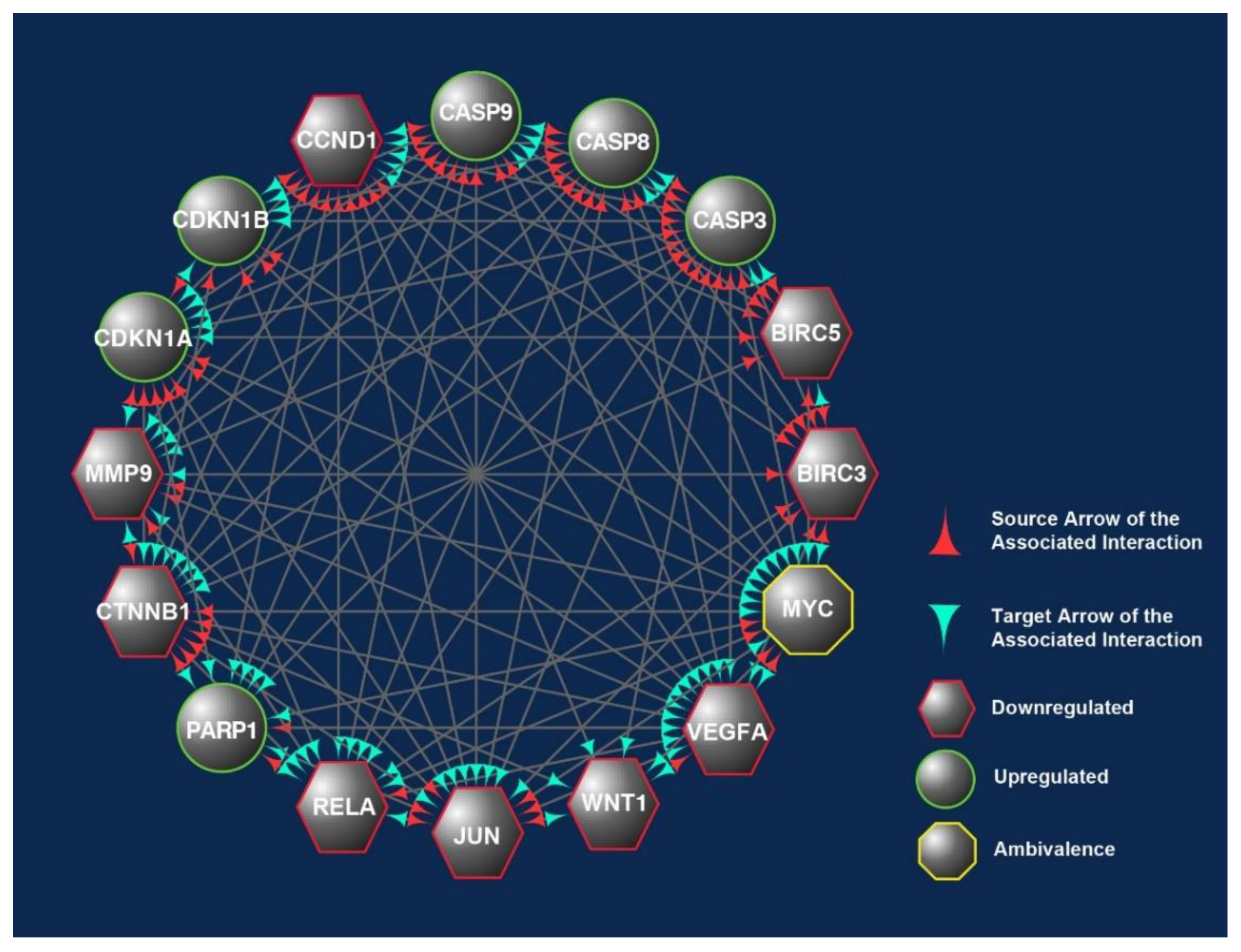
3.5. Differentially Expressed Candidate Proteins and Their Interactions
4. Discussion
5. Conclusions
Supplementary Materials
Author Contributions
Funding
Acknowledgments
Conflicts of Interest
Abbreviations
| Akt | protein kinase B |
| APAF1 | apoptotic protease activating factor 1 |
| APC | adenomatous polyposis coli |
| BAX | BCL2-Associated X Protein |
| Bcl-xL | B-cell lymphoma-extra large |
| BCL2 | B-cell Lymphoma 2 |
| BRAF | proto-oncogene, serine/threonine kinase |
| c-Jun | avian Sarma virus-17 oncogene (Jun comes from Japanese JU-NANA) |
| c-Myc | cellular homologue of avian myelocytomatosis virus |
| CASP3 | caspease-3 |
| CASP8 | caspease-8 |
| CASP9 | caspease-9 |
| CB | candidate biomarker |
| CC3 | Cleaved Caspase-3 |
| CDK-p21 | Cyclin-Dependent Kinase Inhibitor 1 |
| CDK-p72 | Cyclin-Dependent Kinase Inhibitor 1B |
| CDK4 | cyclin-dependent kinase 4 |
| CI | combinational index |
| cIAP1 | cellular inhibitor of apoptosis protein-1 |
| cIAP2 | Baculoviral IAP Repeat-Containing Protein 3 |
| CIN | Chromosomal Instability |
| COX-2 | Cyclooxygenase 2 |
| CRC | colorectal carcinoma |
| CRISPR | clusters of regularly interspaced short palindromic repeats |
| CXCR-4 | C-X-C chemokine receptor type 4 |
| Cyt c | cytochrome complex |
| DCC | deleted in colorectal carcinoma |
| DR | death receptor |
| EGFR | epidermal growth factor receptor |
| EMT | epithelial–mesenchymal transition |
| ERKs | extracellular signal-regulated kinases |
| FAP | Familial Adenomatous Polyposis |
| FCC | Familial Colorectal Cancer |
| FOLFIRI | folinic acid, fluorouracil and irinotecan |
| GADD45 | Growth Arrest and DNA Damage-inducible |
| GDT | Gamma-Delta Tocotrienol |
| HDI | Human Development Index |
| HIF-1α | hypoxia-inducible factor 1-alpha |
| HME | High MYC Expression |
| HMGCR | HMG-CoA reductase |
| HNPCC | Hereditary Non-Polyposis Colorectal Cancer |
| hTERT | human telomerase reverse transcriptase |
| IC50 | half maximum inhibitory concentration |
| ICAM-1 | intercellular adhesion molecule 1 |
| IL-8 | interleukin-8 |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| KO | Knockout |
| KRAS | Kirsten rat sarcoma viral oncogene homolog |
| LEF | lymphoid enhancer-binding factor |
| LME | low MYC Expression |
| LNM | lymph node metastasis |
| MCRC | metastatic colorectal cancer |
| miRNAs | micro-ribonucleic acids |
| MLH1 | MutL homolog 1 |
| MMP-7 | Matrix metalloproteinase-7 or Matrilysin |
| MMP-9 | Matrix Metallopeptidase 9 |
| MSH2 | MutS homolog 2 |
| MSI | Microsatellite Instability |
| MSI-H | Microsatellite Instability High-Frequency |
| MSI-L | Microsatellite Instability Low-Frequency |
| NF-κB | nuclear factor kappa-light-chain-enhancer of activated B cells |
| OS | overall survival |
| p-value | calculated probability |
| p38 MAPK | p38 mitogen-activated protein kinases |
| p53 | tumor suppressor protein (MW = 53 kD) |
| PARP-1 | poly [ADP-Ribose] polymerase 1 |
| PFS | progression-free survival |
| PKC | protein kinase C |
| PPI | protein-protein interaction |
| PRISMA | preferred reporting items for systematic reviews and meta-analyses |
| RhoA | transforming protein |
| Ser473 | serine kinase |
| SMAD4 | such as mothers against decapentaplegic homolog 4 |
| T3s | tocotrienols |
| TBE | TCF4/LEF Binding Elements |
| TCF | T-cell factor |
| TCF4 | Transcription Factor 4 |
| TGF-RII | transforming growth factor receptor II |
| Thr308 | threonine kinase |
| TRAIL | tumor Necrosis Factor-Related Apoptosis-Inducing Ligand |
| TRF | tocotrienol-rich fraction |
| TSG | tumor suppressor genes |
| TT | target therapy |
| VEGF | vascular endothelial growth factor |
| VEGFA | vascular endothelial growth factor A |
| Wnt | Wingless-related integration site |
| Wnt-1 | Wingless-related integration site-1 |
| αToc | alpha-tocopherol |
| γT3 | gamma-tocotrienol |
| δT3 | delta-tocotrienol |
References
- Ferlay, J.; Soerjomataram, I.; Dikshit, R.; Eser, S.; Mathers, C.; Rebelo, M.; Parkin, D.M.; Forman, D.; Bray, F. Cancer Incidence and Mortality Worldwide: Sources, Methods and Major Patterns in GLOBOCAN 2012. Int. J. Cancer 2015, 136, E359–E386. [Google Scholar] [CrossRef] [PubMed]
- Arnold, M.; Sierra, M.S.; Laversanne, M.; Soerjomataram, I.; Jemal, A.; Bray, F. Global patterns and trends in colorectal cancer incidence and mortality. Gut 2017, 66, 683–691. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Aggarwal, V.; Kashyap, D.; Sak, K.; Tuli, H.S.; Jain, A.; Chaudhary, A.; Garg, V.K.; Sethi, G.; Yerer, M.B. Molecular mechanisms of action of tocotrienols in cancer: Recent trends and advancements. Int. J. Mol. Sci. 2019, 20, 656. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Ahn, K.S.; Sethi, G.; Krishnan, K.; Aggarwal, B.B. γ-Tocotrienol inhibits nuclear factor-κb signaling pathway through inhibition of receptor-interacting protein and TAK1 leading to suppression of antiapoptotic gene products and potentiation of apoptosis. J. Biol. Chem. 2007, 282, 809–820. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Eitsuka, T.; Nakagawa, K.; Miyazawa, T. Down-regulation of telomerase activity in DLD-1 human colorectal adenocarcinoma cells by tocotrienol. Biochem. Biophys. Res. Commun. 2006, 348, 170–175. [Google Scholar] [CrossRef]
- Shibata, A.; Nakagawa, K.; Sookwong, P.; Tsuduki, T.; Tomita, S.; Shirakawa, H.; Komai, M.; Miyazawa, T. Tocotrienol inhibits secretion of angiogenic factors from human colorectal adenocarcinoma cells by suppressing hypoxia-inducible factor-1α. J. Nutr. 2008, 138, 2136–2142. [Google Scholar] [CrossRef] [Green Version]
- Shibata, A.; Nakagawa, K.; Sookwong, P.; Tsuzuki, T.; Oikawa, S.; Miyazawa, T. Tumor anti-angiogenic effect and mechanism of action of δ-tocotrienol. Biochem. Pharmacol. 2008, 76, 330–339. [Google Scholar] [CrossRef] [PubMed]
- Brew, R.; Erikson, J.S.; West, D.C.; Kinsella, A.R.; Slavin, J.; Christmas, S. Interleukin-8 as an autocrine growth factor for human colon carcinoma cells in vitro. Cytokine 2000, 12, 78–85. [Google Scholar] [CrossRef]
- Ananthula, S.; Parajuli, P.; Behery, F.; Alayoubi, A.Y.; El Sayed, K.; Nazzal, S.; Sylvester, P.W. Oxazine derivatives of γ- and δ-tocotrienol display enhanced anticancer activity in vivo. Anticancer. Res. 2014, 34, 2715–2726. [Google Scholar]
- Xu, W.-L.; Liu, J.-R.; Liu, H.-K.; Qi, G.-Y.; Sun, X.-R.; Sun, W.-G.; Chen, B.-Q. Inhibition of proliferation and induction of apoptosis by γ-tocotrienol in human colon carcinoma HT-29 cells. Nutrition 2009, 25, 555–566. [Google Scholar] [CrossRef]
- Tham, S.-Y.; Loh, H.-S.; Mai, C.-W.; Fu, J.-Y. Tocotrienols modulate a life or death decision in cancers. Int. J. Mol. Sci. 2019, 20, 372. [Google Scholar] [CrossRef] [Green Version]
- Rudner, J.; Jendrossek, V.; Lauber, K.; Daniel, P.T.; Wesselborg, S.; Belka, C. Type I and type II reactions in TRAIL-induced apoptosis–results from dose–response studies. Oncogene 2004, 24, 130–140. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Arksey, H.; O’Malley, L. Scoping studies: Towards a methodological framework. Int. J. Soc. Res. Methodol. 2005, 8, 19–32. [Google Scholar] [CrossRef] [Green Version]
- Page, M.J.; McKenzie, J.; Bossuyt, P.; Boutron, I.; Hoffmann, T.; Mulrow, C.; Shamseer, L.; Tetzlaff, J.; Akl, E.; Brennan, S.E.; et al. The PRISMA 2020 Statement: An Updated Guideline for Reporting Systematic Reviews. BMJ 2021, 372, n71. [Google Scholar] [CrossRef] [PubMed]
- Miller, S.A.; Forrest, J.L. Enhancing your practice through evidence-based decision making: PICO, learning how to ask good questions. J. Évid. Based Dent. Pr. 2001, 1, 136–141. [Google Scholar] [CrossRef]
- Shibata, A.; Nakagawa, K.; Sookwong, P.; Tsuduki, T.; Asai, A.; Miyazawa, T. α-Tocopherol attenuates the cytotoxic effect of δ-tocotrienol in human colorectal adenocarcinoma cells. Biochem. Biophys. Res. Commun. 2010, 397, 214–219. [Google Scholar] [CrossRef] [PubMed]
- Yang, Z.; Xiao, H.; Jin, H.; Koo, P.T.; Tsang, D.J.; Yang, C.S. Synergistic actions of atorvastatin with γ-tocotrienol and celecoxib against human colon cancer HT29 and HCT116 cells. Int. J. Cancer 2009, 126, 852–863. [Google Scholar] [CrossRef] [Green Version]
- Kannappan, R.; Ravindran, J.; Prasad, S.; Sung, B.; Yadav, V.R.; Reuter, S.; Chaturvedi, M.M.; Aggarwal, B.B. γ-Tocotrienol promotes TRAIL-induced apoptosis through reactive oxygen species/extracellular signal-regulated kinase/p53–Mediated upregulation of death receptors. Mol. Cancer Ther. 2010, 9, 2196–2207. [Google Scholar] [CrossRef] [Green Version]
- Zhang, J.-S.; Li, D.-M.; He, N.; Liu, Y.-H.; Wang, C.-H.; Jiang, S.-Q.; Chen, B.-Q.; Liu, J.-R. A paraptosis-like cell death induced by δ-tocotrienol in human colon carcinoma SW620 cells is associated with the suppression of the Wnt signaling pathway. Toxicology 2011, 285, 8–17. [Google Scholar] [CrossRef]
- Xu, W.; Du, M.; Zhao, Y.; Wang, Q.; Sun, W.; Chen, B. γ-Tocotrienol inhibits cell viability through suppression of β-catenin/Tcf signaling in human colon carcinoma HT-29 cells. J. Nutr. Biochem. 2012, 23, 800–807. [Google Scholar] [CrossRef]
- Zhang, J.-S.; Li, D.-M.; Ma, Y.; He, N.; Gu, Q.; Wang, F.-S.; Jiang, S.-Q.; Chen, B.-Q.; Liu, J.-R. γ-Tocotrienol induces paraptosis-like cell death in human colon carcinoma SW620 cells. PLoS ONE 2013, 8, e57779. [Google Scholar] [CrossRef]
- Shibata, A.; Nakagawa, K.; Tsuduki, T.; Miyazawa, T. δ-Tocotrienol treatment is more effective against hypoxic tumor cells than normoxic cells: Potential implications for cancer therapy. J. Nutr. Biochem. 2015, 26, 832–840. [Google Scholar] [CrossRef] [PubMed]
- Yusof, K.M.; Makpol, S.; Jamal, R.; Harun, R.; Mokhtar, N.; Ngah, W.Z.W. γ-Tocotrienol and 6-Gingerol in combination synergistically induce cytotoxicity and apoptosis in HT-29 and SW837 human colorectal cancer cells. Molecules 2015, 20, 10280–10297. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Abubakar, I.B.; Lim, K.-H.; Kam, T.-S.; Loh, H.-S. Synergistic cytotoxic effects of combined δ-tocotrienol and jerantinine B on human brain and colon cancers. J. Ethnopharmacol. 2016, 184, 107–118. [Google Scholar] [CrossRef] [PubMed]
- Eitsuka, T.; Tatewaki, N.; Nishida, H.; Nakagawa, K.; Miyazawa, T. A combination of δ-tocotrienol and ferulic acid synergistically inhibits telomerase activity in DLD-1 human colorectal adenocarcinoma cells. J. Nutr. Sci. Vitaminol. 2016, 62, 281–287. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Prasad, S.; Gupta, S.C.; Tyagi, A.K.; Aggarwal, B.B. γ-Tocotrienol suppresses growth and sensitises human colorectal tumours to capecitabine in a nude mouse xenograft model by down-regulating multiple molecules. Br. J. Cancer 2016, 115, 814–824. [Google Scholar] [CrossRef] [Green Version]
- Husain, K.; Zhang, A.; Shivers, S.C.; Davis-Yadley, A.H.; Coppola, D.; Yang, C.S.; Malafa, M.P. Chemoprevention of azoxymethane-induced colon carcinogenesis by Delta-tocotrienol. Cancer Prev. Res. 2019, 12, 357–366. [Google Scholar] [CrossRef] [Green Version]
- Draw Venn Diagram. Available online: http://bioinformatics.psb.ugent.be/webtools/Venn/ (accessed on 11 August 2021).
- Jensen, L.J.; Kuhn, M.; Stark, M.; Chaffron, S.; Creevey, C.; Muller, J.; Doerks, T.; Julien, P.; Roth, A.; Simonovic, M.; et al. STRING 8--a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2008, 37, D412–D416. [Google Scholar] [CrossRef]
- Otasek, D.; Morris, J.H.; Bouças, J.; Pico, A.R.; Demchak, B. Cytoscape automation: Empowering workflow-based network analysis. Genome Biol. 2019, 20, 1–15. [Google Scholar] [CrossRef] [Green Version]
- Loganathan, R.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotrienols promote apoptosis in human breast cancer cells by inducing poly(ADP -ribose) polymerase cleavage and inhibiting nuclear factor kappa-B activity. Cell Prolif. 2013, 46, 203–213. [Google Scholar] [CrossRef]
- Ramdas, P.; Rajihuzzaman, M.; Veerasenan, S.D.; Selvaduray, K.R.; Nesaretnam, K.; Radhakrishnan, A.K. Tocotri-enol-treated MCF-7 human breast cancer cells show down-regulation of API5 and up-regulation of MIG6 genes. Cancer Genom. Proteom. 2011, 8, 19–31. [Google Scholar]
- Meganathan, P.; Jabir, R.S.; Fuang, H.G.; Bhoo-Pathy, N.; Choudhury, R.B.; Taib, N.A.; Nesaretnam, K.; Chik, Z. A new formulation of Gamma Delta Tocotrienol has superior bioavailability compared to existing Tocotrienol-Rich Fraction in healthy human subjects. Sci. Rep. 2015, 5, 13550. [Google Scholar] [CrossRef] [PubMed]
- Barretina, J.; Caponigro, G.; Stransky, N.; Venkatesan, K.; Margolin, A.A.; Kim, S.; Wilson, C.J.; Lehár, J.; Kryukov, G.V.; Sonkin, D.; et al. The Cancer Cell Line Encyclopedia enables predictive modelling of anticancer drug sensitivity. Nature 2012, 483, 603–607. [Google Scholar] [CrossRef] [PubMed]
- Ronen, J.; Hayat, S.; Akalin, A. Evaluation of colorectal cancer subtypes and cell lines using deep learning. Life Sci. Alliance 2019, 2, e201900517. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Flis, S.; Spłwiński, J. Inhibitory effects of 5-fluorouracil and oxaliplatin on human colorectal cancer cell survival are synergistically enhanced by sulindac sulfide. Anticancer. Res. 2009, 29, 435–441. [Google Scholar]
- Hein, M.; Graver, S. Tumor cell response to bevacizumab single agent therapy in vitro. Cancer Cell Int. 2013, 13, 94. [Google Scholar] [CrossRef] [Green Version]
- Ahmed, D.; Eide, P.W.; Eilertsen, I.A.; Danielsen, S.A.; Eknaes, M.; Hektoen, M.; Lind, G.E.; Lothe, R.A. Epigenetic and genetic features of 24 colon cancer cell lines. Oncogenesis 2013, 2, e71. [Google Scholar] [CrossRef]
- Burt, R.W. Colon cancer screening. Gastroenterology 2000, 119, 837–853. [Google Scholar] [CrossRef]
- Grady, W.M. Genomic instability and colon cancer. Cancer Metastasis Rev. 2004, 23, 11–27. [Google Scholar] [CrossRef]
- Rajagopalan, H.; Nowak, M.A.; Vogelstein, B.; Lengauer, C. The significance of unstable chromosomes in colorectal cancer. Nat. Rev. Cancer 2003, 3, 695–701. [Google Scholar] [CrossRef]
- Nojadeh, J.N.; Sharif, S.B.; Sakhinia, E. Microsatellite instability in colorectal cancer. EXCLI J. 2018, 17, 159–168. [Google Scholar] [CrossRef]
- Lynch, H.T.; De La Chapelle, A. Hereditary colorectal cancer. N. Engl. J. Med. 2003, 348, 919–932. [Google Scholar] [CrossRef]
- Anwar, S.; Hall, C.; White, J.; Deakin, M.; Farrell, W.; Elder, J. Hereditary non-polyposis colorectal cancer: An updated review. Eur. J. Surg. Oncol. (EJSO) 2000, 26, 635–645. [Google Scholar] [CrossRef] [PubMed]
- Zhang, Z.; Wang, M.; Zhou, L.; Feng, X.; Cheng, J.; Yü, Y.; Gong, Y.; Zhu, Y.; Li, C.; Tian, L.; et al. Increased HMGB1 and cleaved caspase-3 stimulate the proliferation of tumor cells and are correlated with the poor prognosis in colorectal cancer. J. Exp. Clin. Cancer Res. 2015, 34, 1–9. [Google Scholar] [CrossRef] [Green Version]
- Kim, H.S.; Lee, J.W.; Soung, Y.H.; Park, W.S.; Kim, S.Y.; Park, J.Y.; Cho, Y.G.; Kim, C.J.; Jeong, S.W.; Nam, S.W.; et al. Inactivating mutations of caspase-8 gene in colorectal carcinomas. Gastroenterology 2003, 125, 708–715. [Google Scholar] [CrossRef]
- Duiker, E.W.; Meijer, A.; Van Der Bilt, A.R.M.; Meersma, G.J.; Kooi, N.; Van Der Zee, A.G.J.; De Vries, E.G.; De Jong, S. Drug-induced caspase 8 upregulation sensitises cisplatin-resistant ovarian carcinoma cells to rhTRAIL-induced apoptosis. Br. J. Cancer 2011, 104, 1278–1287. [Google Scholar] [CrossRef] [PubMed]
- Shen, X.-G.; Wang, C.; Li, Y.; Wang, L.; Zhou, B.; Xu, B.; Jiang, X.; Zhou, Z.-G.; Sun, X.-F. Downregulation of caspase-9 is a frequent event in patients with stage II colorectal cancer and correlates with poor clinical outcome. Color. Dis. 2010, 12, 1213–1218. [Google Scholar] [CrossRef] [PubMed]
- Ogino, S.; Kawasaki, T.; Kirkner, G.; Ogawa, A.; Dorfman, I.; Loda, M.; Fuchs, C. Down-regulation of p21 (CDKN1A/CIP1) is inversely associated with microsatellite instability and CpG island methylator phenotype (CIMP) in colorectal cancer. J. Pathol. 2006, 210, 147–154. [Google Scholar] [CrossRef] [PubMed]
- Wang, H.; Birkenbach, M.; Hart, J. Expression of jun family members in human colorectal adenocarcinoma. Carcinogenesis 2000, 21, 1313–1317. [Google Scholar] [CrossRef] [PubMed]
- Strippoli, A.; Cocomazzi, A.; Basso, M.; Cenci, T.; Ricci, R.; Pierconti, F.; Cassano, A.; Fiorentino, V.; Barone, C.; Bria, E.; et al. c-MYC expression is a possible keystone in the colorectal cancer resistance to EGFR inhibitors. Cancers 2020, 12, 638. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Wang, S.; Liu, Z.; Wang, L.; Zhang, X. NF-κB signaling pathway, inflammation and colorectal cancer. Cell. Mol. Immunol. 2009, 6, 327–334. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Slattery, M.L.; Mullany, L.E.; Sakoda, L.; Samowitz, W.S.; Wolff, R.K.; Stevens, J.R.; Herrick, J.S. The NF-κB signalling pathway in colorectal cancer: Associations between dysregulated gene and miRNA expression. J. Cancer Res. Clin. Oncol. 2018, 144, 269–283. [Google Scholar] [CrossRef] [Green Version]
- Balcerczak, E.; Pasz-Walczak, G.; Kumor, P.; Panczyk, M.; Kordek, R.; Wierzbicki, R.; Mirowski, M. Cyclin D1 protein and CCND1 gene expression in colorectal cancer. Eur. J. Surg. Oncol. (EJSO) 2005, 31, 721–726. [Google Scholar] [CrossRef]
- Mazeda, I.; Martins, S.F.; Garcia, E.A.; Rodrigues, M.; Longatto, A. VEGF expression in colorectal cancer metastatic lymph nodes: Clinicopathological correlation and prognostic significance. Gastrointest. Disord. 2020, 2, 267–280. [Google Scholar] [CrossRef]
- Mirabelli-Primdahl, L.; Gryfe, R.; Kim, H.; Millar, A.; Luceri, C.; Dale, D.; Holowaty, E.; Bapat, B.; Gallinger, S.; Redston, M. Beta-catenin mutations are specific for colorectal carcinomas with microsatellite instability but occur in endometrial carcinomas irrespective of mutator pathway. Cancer Res. 1999, 59, 3346–3351. [Google Scholar]
- Zhou, M.; Liu, X.; Li, Z.; Huang, Q.; Li, F.; Li, C.-Y. Caspase-3 regulates the migration, invasion and metastasis of colon cancer cells. Int. J. Cancer 2018, 143, 921–930. [Google Scholar] [CrossRef]
- Kanehisa, M.; Sato, Y.; Kawashima, M.; Furumichi, M.; Tanabe, M. KEGG as a reference resource for gene and protein annotation. Nucleic Acids Res. 2016, 44, D457–D462. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Montalto, F.I.; De Amicis, F. Cyclin D1 in Cancer: A molecular connection for cell cycle control, adhesion and invasion in tumor and stroma. Cells 2020, 9, 2648. [Google Scholar] [CrossRef]
- Lanza, G.; Ferracin, M.; Gafà, R.; Veronese, A.; Spizzo, R.; Pichiorri, F.; Liu, C.-G.; Calin, G.; Croce, C.M.; Negrini, M. mRNA/microRNA gene expression profile in microsatellite unstable colorectal cancer. Mol. Cancer 2007, 6, 1–11. [Google Scholar] [CrossRef] [Green Version]
- Kim, M.; Lee, H.E.; Lee, H.S.; Yang, H.-K.; Kim, W.H. Expression of apoptosis-related proteins and its clinical implication in surgically resected gastric carcinoma. Virchows Arch. 2011, 459, 503–510. [Google Scholar] [CrossRef]
- Taylor, R.; Cullen, S.P.; Martin, S. Apoptosis: Controlled demolition at the cellular level. Nat. Rev. Mol. Cell Biol. 2008, 9, 231–241. [Google Scholar] [CrossRef] [PubMed]
- Asadi, M.; Shanaehbandi, D.; Kermani, T.A.; Sanaat, Z.; Zafari, V.; Hashemzadeh, S. Expression level of caspase genes in colorectal cancer. Asian Pac. J. Cancer Prev. 2018, 19, 1277–1280. [Google Scholar] [CrossRef]
- Cacina, C.; YaylIm-Eraltan, I.; Arikan, S.; Saglam, E.K.; Zeybek, U.; Isbir, T. Association between CDKN1A Ser31Arg and C20T gene polymorphisms and colorectal cancer risk and prognosis. Vivo 2010, 24, 179–183. [Google Scholar]
- Lee, T.I.; Young, R.A. Transcriptional regulation and its misregulation in disease. Cell 2013, 152, 1237–1251. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Thomas, R.; Weihua, Z. Rethink of EGFR in cancer with its kinase independent function on board. Front. Oncol. 2019, 9, 800. [Google Scholar] [CrossRef]
- Arango, D.; Mariadason, J.M.; Wilson, A.J.; Yang, W.; Corner, G.; Nicholas, C.; Aranes, M.J.; Augenlicht, L.H. c-Myc overexpression sensitises colon cancer cells to camptothecin-induced apoptosis. Br. J. Cancer 2003, 89, 1757–1765. [Google Scholar] [CrossRef] [PubMed] [Green Version]
- Kalkat, M.; De Melo, J.; Hickman, K.A.; Lourenco, C.; Redel, C.; Resetca, D.; Tamachi, A.; Tu, W.B.; Penn, L.Z. MYC deregulation in primary human cancers. Genes 2017, 8, 151. [Google Scholar] [CrossRef] [Green Version]
- Rochlitz, C.F.; Herrmann, R.; De Kant, E. Overexpression and amplification of c-myc during progression of human colorectal cancer. Oncology 1996, 53, 448–454. [Google Scholar] [CrossRef]
- Khare, V.; Tabassum, S.; Chatterjee, U.; Chatterjee, S.; Ghosh, M.K. RNA helicase p68 deploys β-catenin in regulating RelA/p65 gene expression: Implications in colon cancer. J. Exp. Clin. Cancer Res. 2019, 38, 1–19. [Google Scholar] [CrossRef] [Green Version]
- Vasaikar, S.; Huang, C.; Wang, X.; Petyuk, V.A.; Savage, S.R.; Wen, B.; Dou, Y.; Zhang, Y.; Shi, Z.; Arshad, O.A.; et al. Proteogenomic analysis of human colon cancer reveals new therapeutic opportunities. Cell 2019, 177, 1035–1049.e19. [Google Scholar] [CrossRef] [Green Version]


| Study | Year | CRC Cell Lines | T3s | Protein(s) and Gene(s) Modulated | Main Outcomes | ||
|---|---|---|---|---|---|---|---|
| Shibata et al. [16] | 2010 | DLD-1 | δT3 |  | CDK-p21(Cdkn1A); CDK-p72 (Cdkn1b); GADD45 alpha (GADD45A); Caspase-3 (CASP3); Caspase-7 (CASP7); Caspase-9 (CASP9); APAF1; elegans CED-4 |  | Cell cycle arrest; Apoptosis signals |
| Yang et al. [17] | 2010 | HT-29 HCT-116 | δT3 or γT3 |  | PARP-1 |  | Cell cycle arrest; Apoptosis (Cleavage; PARP activation and DNA fragmentation) |
 | RhoA; HMGCR |  | G-protein geranylgeranylation. | ||||
| Kannappan et al. [18] | 2010 | HCT-116 HT-29 | γT3 |  | TRAIL; ERKs; Caspase-3; Caspase-8; Caspase-9; PARP-1 |  | Cell death; apoptosis, DR4 and DR5; ROS |
 | cIAP2; Bcl-xL; p38 MAPK |  | Cell survival proteins | ||||
| Zhang et al. [19] | 2011 | SW620 | δT3 |  | c-myc |  | Swelling of mitochondria/ER; paraptosis-like cell death |
 | β-catenin; Wnt-1; Cyclin D1; c-Jun; MMP-7 |  | Cell viability | ||||
| Xu et al. [20] | 2012 | HT-29 | γT3 |  | Wnt (No specification); β-catenin; TCF; Survivin; Cyclin D1; Myc |  | Shrunk/floated cells; apoptotic changes; apoptosis |
 | Adhesive ability; cell proliferation | ||||||
| Zhang et al. [21] | 2013 | SW620 HCT-8 | δT3 |  | Caspase-3 |  | Cell size; round cells; paraptosis-based cell death; cytoplasmic vacuolization |
 | β-catenin; Cyclin D1; c-Jun; Wnt-1 |  | Proliferation | ||||
| Shibata et al. [22] | 2015 | DLD-1 | δT3 |  | Caspase-3; Caspase-9; CDK-p21 (Cdkn1a); CDK-p72 (Cdkn1b) |  | Apoptosis; cell cycle arrest; hypoxia genes/proteins expression > Normoxia genes/proteins |
 | Akt–Thr308 and Ser473; CDK4; HIF-1α |  | Cell proliferation | ||||
| Yusof et al. [23] | 2015 | HT-29 SW837 | γT3 | No significant effect |  | Distortion and shrinkage of cells; pyknosis and apoptotic bodies, chemoprevention | |
 | Proliferation | ||||||
| Abubakar et al. [24] | 2016 | HT-29 | δT3 |  | Caspase-3; Caspase-8 |  | Apoptosis |
| ~ | Caspase-9 | ||||||
| Eitsuka et al. [25] | 2016 | DLD-1 | δT3 |  | hTERT |  | Cellular telomerase activity; Proliferation |
| Prasad et al. [26] | 2016 | HCT-116 HT-29 Caco-2 | γT3 |  | Survivin; cIAP1; cIAP2; Cyclin D1; c-Myc; MMP-9; CXCR-4; VEGF; ICAM-1; NF-κB/p65 |  | Apoptosis; cell cycle arrest |
 | Proliferation; colony formation; expression of tumorigenic and metastasis proteins | ||||||
| Husain et al. [27] | 2019 | HCT-116 HT-29 SW480 SW620 | δT3 |  | PARP1; Phosphatidylserine |  | Apoptosis; cell cycle arrest |
 | E-cadherin; Vimentin; MMP-9; VEGF; NF-κB/p65; β-catenin |  | Colony formation; EMT; angiogenesis; migration; invasion and metastasis | ||||
 Downregulat.ed;
Downregulat.ed;  Upregulated; ~ Uncertain; δT3: delta-tocotrienol; γT3: gamma-tocotrienol; IC50: half-maximum inhibitory concentration; CASP: caspase; DR: death receptors; EMT: epithelial-mesenchymal transition; ER: endoplasmic reticulum; hr: hour; PARP: poly (ADP-ribose) polymerase; ROS: reactive oxygen species; T3s: tocotrienols.
Upregulated; ~ Uncertain; δT3: delta-tocotrienol; γT3: gamma-tocotrienol; IC50: half-maximum inhibitory concentration; CASP: caspase; DR: death receptors; EMT: epithelial-mesenchymal transition; ER: endoplasmic reticulum; hr: hour; PARP: poly (ADP-ribose) polymerase; ROS: reactive oxygen species; T3s: tocotrienols.| Pathways Involved | CBs Modulation by T3s | Reported Effects in Colon Cancer Patients | Ref. |
|---|---|---|---|
| Apoptosis | Caspase 3 ( ) ) | ● Irradiated CRC cells from patients with lower levels of caspase-3 was associated with poor prognosis | [45] |
Caspase 8 ( ) ) | ● Higher prevalence of mutations in the caspase-8 genes in invasive carcinomas; reduce apoptotic activity | [46,47] | |
Caspase 9 ( ) ) | ● Expression of the caspase-9 gene downregulated in CRC tissue compared to surrounding normal mucosa | [48] | |
| Transcriptional dysregulation in cancer | CDKN1A (p21) ( ) ) | ● P21 was downregulated in 50% (371/737) of CRC samples | [49] |
Jun family ( ) ) | ● Higher c-Jun expression observed in human colorectal adenocarcinomas | [50] | |
c-MYC ( ) ) | ● CRC patients with higher MYC expression significantly shorter progression-free survival time and overall survival | [51] | |
RELA (NF-kB/p65) ( ) ) | ● Reduced RELA expression, resulting in deceased activation of the NF-κB signaling pathway, which inhibited carcinogenesis | [52,53] | |
| Cancer progression | CCND1 ( ) ) | ● CCND1 gene was detected in tumors from about 50% (54 out of 111) of CRC patients; absent in normal mucosa | [54] |
VEGFA ( ) ) | ● Elevated expression of the VEGF family, especially of VEGFA, was reported in CRC patients with LNM | [55] | |
CTNNB1 ( ) ) | ● CTNNB1 codes for β-catenin, which supports tumor growth ● A significant link between mutations in CTNNB1 gene and MSI | [56] |
 Downregulat.ed;
Downregulat.ed;  Upregulated; CB: candidate biomarkers; CRC: colorectal carcinoma; LNM: lymph node metastasis; MSI: microsatellite instability; T3s: tocotrienols. Italics refer to the gene name of the protein.
Upregulated; CB: candidate biomarkers; CRC: colorectal carcinoma; LNM: lymph node metastasis; MSI: microsatellite instability; T3s: tocotrienols. Italics refer to the gene name of the protein.Publisher’s Note: MDPI stays neutral with regard to jurisdictional claims in published maps and institutional affiliations. |
© 2021 by the authors. Licensee MDPI, Basel, Switzerland. This article is an open access article distributed under the terms and conditions of the Creative Commons Attribution (CC BY) license (https://creativecommons.org/licenses/by/4.0/).
Share and Cite
Khalid, A.Q.; Bhuvanendran, S.; Magalingam, K.B.; Ramdas, P.; Kumari, M.; Radhakrishnan, A.K. Clinically Relevant Genes and Proteins Modulated by Tocotrienols in Human Colon Cancer Cell Lines: Systematic Scoping Review. Nutrients 2021, 13, 4056. https://doi.org/10.3390/nu13114056
Khalid AQ, Bhuvanendran S, Magalingam KB, Ramdas P, Kumari M, Radhakrishnan AK. Clinically Relevant Genes and Proteins Modulated by Tocotrienols in Human Colon Cancer Cell Lines: Systematic Scoping Review. Nutrients. 2021; 13(11):4056. https://doi.org/10.3390/nu13114056
Chicago/Turabian StyleKhalid, Ali Qusay, Saatheeyavaane Bhuvanendran, Kasthuri Bai Magalingam, Premdass Ramdas, Mangala Kumari, and Ammu Kutty Radhakrishnan. 2021. "Clinically Relevant Genes and Proteins Modulated by Tocotrienols in Human Colon Cancer Cell Lines: Systematic Scoping Review" Nutrients 13, no. 11: 4056. https://doi.org/10.3390/nu13114056
APA StyleKhalid, A. Q., Bhuvanendran, S., Magalingam, K. B., Ramdas, P., Kumari, M., & Radhakrishnan, A. K. (2021). Clinically Relevant Genes and Proteins Modulated by Tocotrienols in Human Colon Cancer Cell Lines: Systematic Scoping Review. Nutrients, 13(11), 4056. https://doi.org/10.3390/nu13114056








